Transcriptome-Based Identification, Characterization, Evolutionary Analysis, and Expression Pattern Analysis of the WRKY Gene Family and Salt Stress Response in Panax ginseng
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources and Methods of Analysis
2.2. Identification of PgWRKY TFs
2.3. Phylogeny and Conserved Domain Protein Sequence Analysis of PgWRKY TFs
2.4. Prediction of Natural Selection in WRKY Superfamily in Ginseng
2.5. GO (Gene Ontology) Functional Categorization Analysis of PgWRKYs
2.6. The Expression Pattern Analysis of PgWRKYs
2.7. Response of PgWRKY Genes to Salt Stress
3. Results
3.1. Identification and Classification of PgWRKY TFs
3.2. Multiple Sequence Alignment Analysis of PgWRKY Proteins
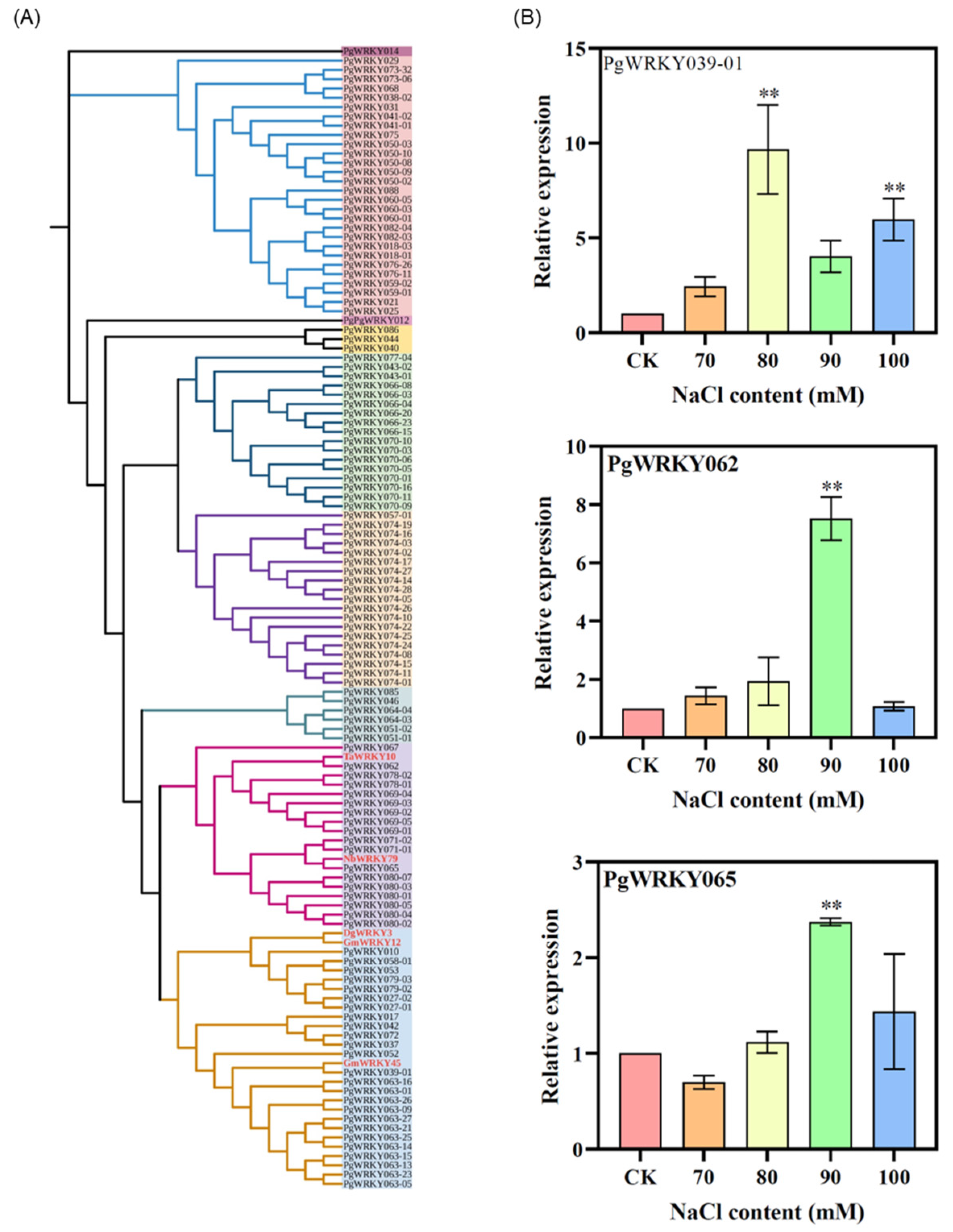
3.3. Phylogenetic Analysis of PgWRKYs
3.4. Ka/Ks Analysis of Natural Selection in PgWRKYs
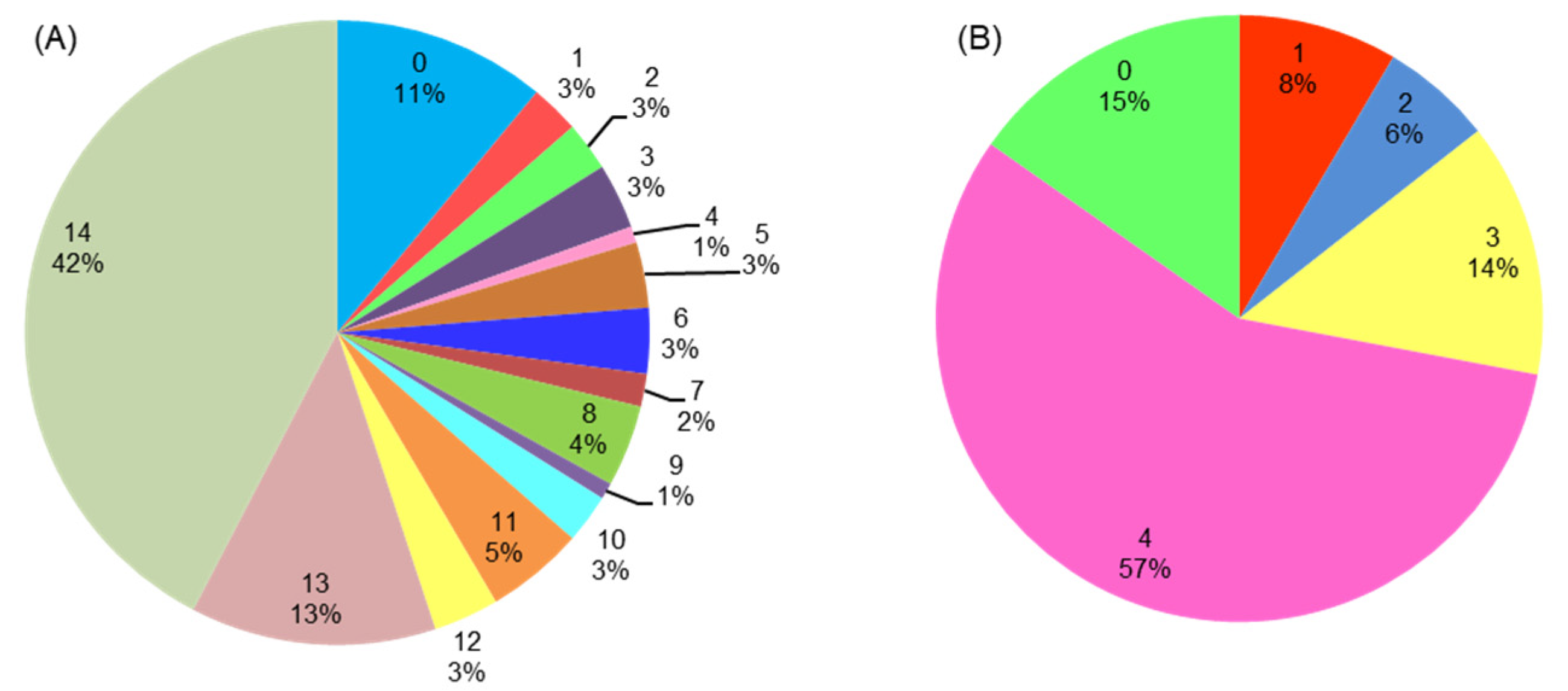
3.5. GO (Gene Ontology) Functional Categorization Analysis of PgWRKYs
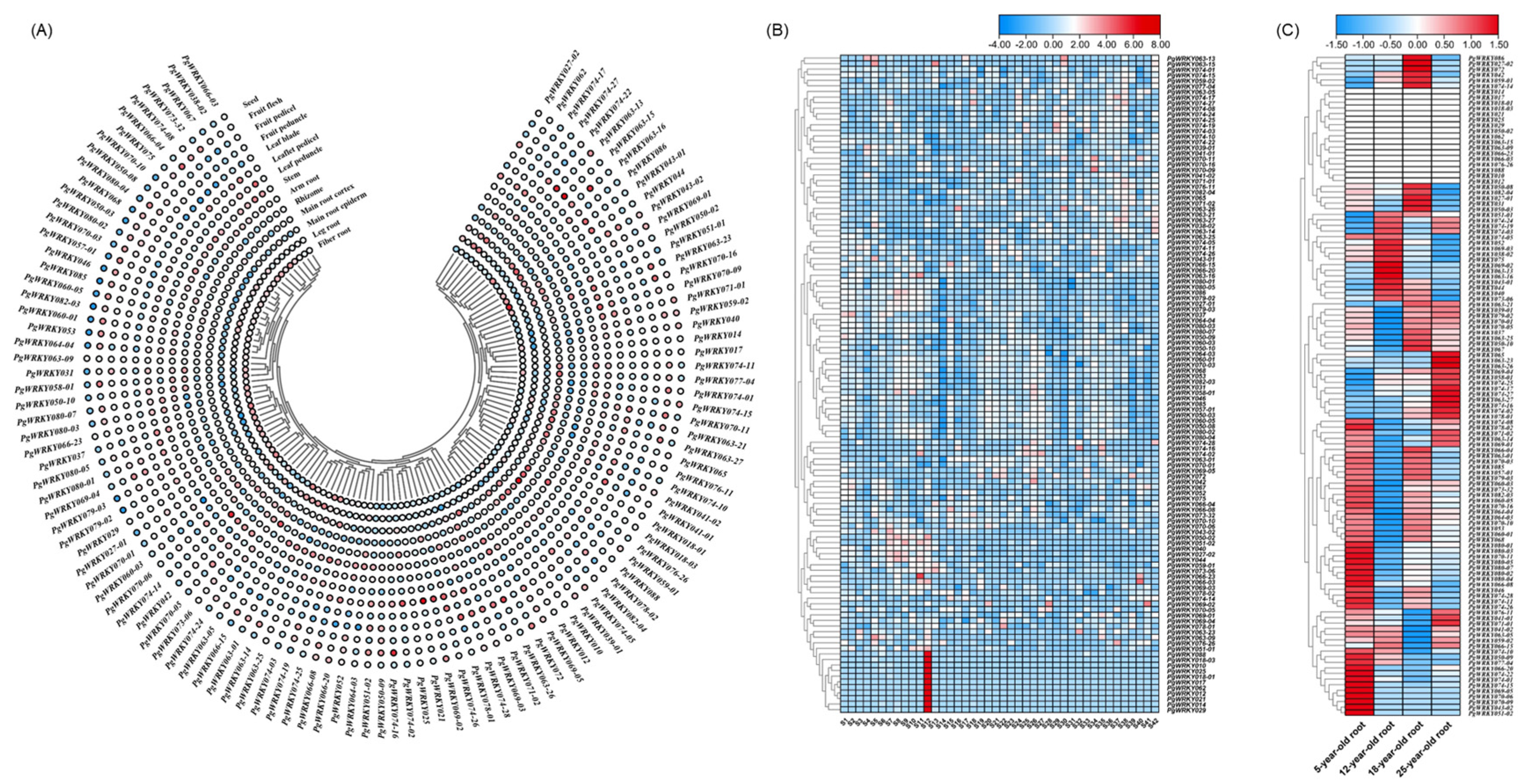
3.6. Expression Pattern and Characteristics Analysis of PgWRKYs
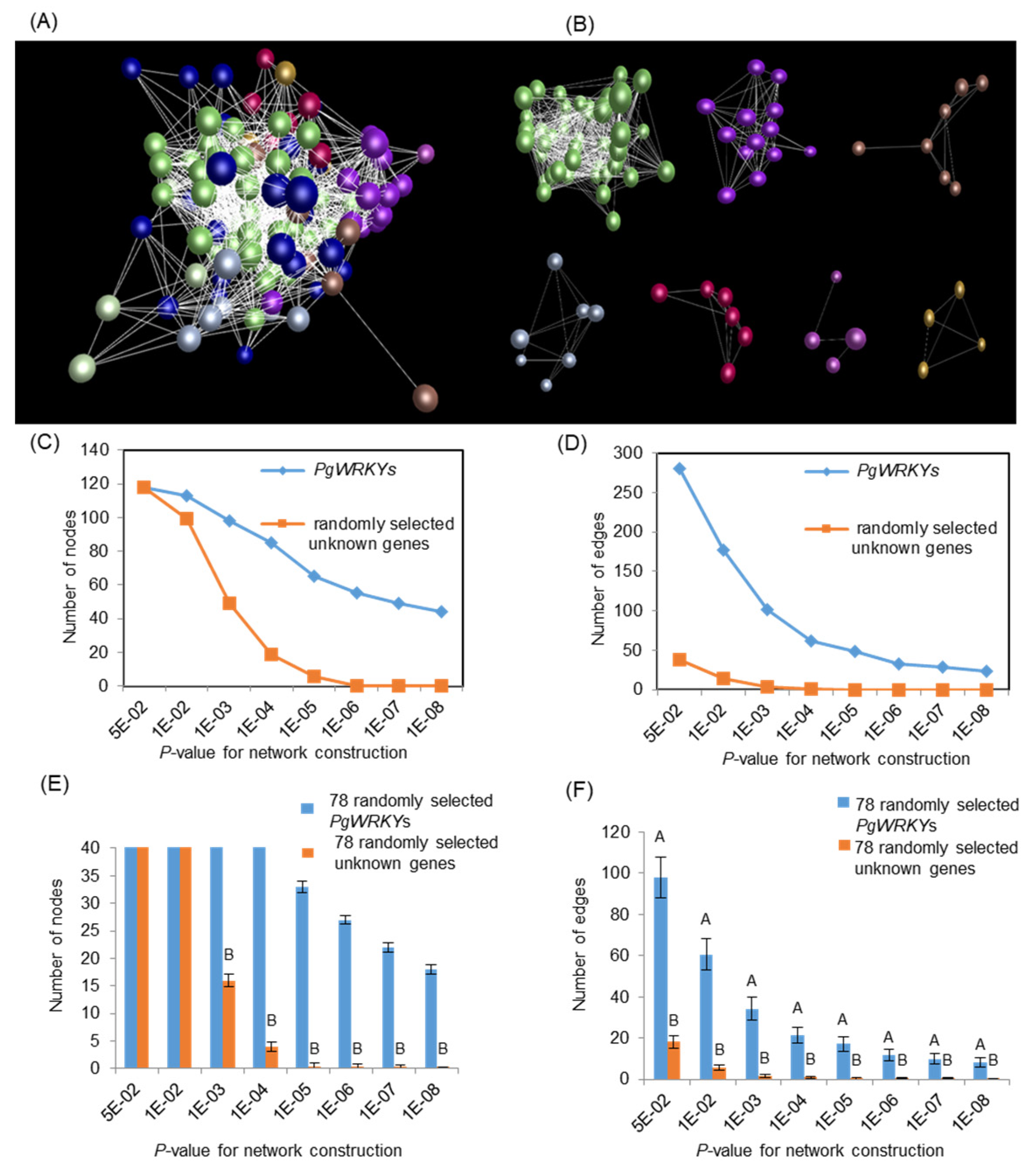
3.7. Co-Expression Network Analysis of PgWRKYs
3.8. Response of PgWRKY Genes to Salt Stress in Ginseng Adventitious Roots
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Jiang, Y.; Yu, D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 2011, 1, 303–313. [Google Scholar] [CrossRef] [Green Version]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Yu, D.Q.; Li, G.X.; Zhang, S.Q.; Zheng, S.J. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 2014, 79, 13–27. [Google Scholar] [CrossRef]
- Karim, A.; Jiang, Y.; Guo, L.; Ling, Z.; Ye, S.; Duan, Y.; Li, C.; Luo, K. Isolation and characterization of a subgroup IIa WRKY transcription factor PtrWRKY40 from Populus trichocarpa. Tree Physiol. 2015, 35, 1129–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, R.; Dong, Y.; Liu, X.; Feng, S.; Wang, C.; Ma, X.; Liu, J.; Liang, Q.; Bao, Y.; Xu, S.; et al. JrWRKY21 interacts with JrPTI5L to activate the expression of JrPR5L for resistance to Colletotrichum gloeosporioides in walnut. Plant J. 2022, 111, 1152–1166. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-speciic transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef]
- Liu, J.J.; Ekramoddoullah, A.K. Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome 2009, 52, 77–88. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Torres, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 50 upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, R.; Jiang, S.Y.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Peng, Y.L.; Guo, Z.J. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008, 18, 508–521. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef]
- Hui, D.Q.; Iqbal, J.; Lehmann, K.; Gase, K.; Saluz, H.P.; Baldwin, I.T. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuate: V. microarray analysis and further characterization of large-scale changes in herbivore-induced mRNAs. Plant Physiol. 2003, 131, 1877–1893. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.X.; Palmqvist, S.; Olsson, H.; Borén, M.; Ahlandsberg, S.; Jansson, C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003, 15, 2076–2092. [Google Scholar] [CrossRef] [Green Version]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Hao, Y.J.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, Z.B.; Zhang, J.S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef]
- Pnueli, L.; Hallak, H.E.; Rozenberg, M.; Cohen, M.; Goloubinoff, P.; Kaplan, A.; Mittler, R. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J. 2002, 31, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21 and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY Transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Mao, S.; Gao, Y.; Zhu, L.; Wu, D.; Cui, Y.; Li, J.; Qian, W. Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in Brassica napus. PLoS ONE 2016, 11, e0157558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Wang, D.; Yang, C.; Kong, N.; Shi, Z.; Zhao, P.; Nan, Y.; Nie, T.; Wang, R.; Ma, H.; et al. Genome-wide identification of the potato WRKY transcription factor family. PLoS ONE 2017, 12, e0181573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.; Nan, Z. Genome-wide identification and analysis of WRKY transcription factors in Medicago truncatula. Hereditas 2014, 36, 152–168. [Google Scholar]
- Hichri, I.; Muhovski, Y.; Žižková, E.; Dobrev, P.I.; Gharbi, E.; Franco-Zorrilla, J.M.; Lopez-Vidriero, I.; Solano, R.; Clippe, A.; Errachid, A.; et al. The Solanum lycopersicum WRKY3 Transcription Factor SlWRKY3 Is Involved in Salt Stress Tolerance in Tomato. Front. Plant Sci. 2017, 8, 1343. [Google Scholar] [CrossRef]
- Xu, Y.H.; Wang, J.W.; Wang, S.; Wang, J.Y.; Chen, X.Y. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Jiang, S.; Sun, C.; Lin, Y.; Yin, R.; Wang, Y.; Zhang, M. The spatial and temporal transcriptomic landscapes of Ginseng, Panax ginseng CA Meyer. Sci. Rep. 2015, 5, 18283. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Cao, H.; Xiu, H.; Luo, T.; Li, J.; Chen, X.; Luo, Z. Transcriptomics-based identification of WRKY genes and characterization of a salt and hormone-responsive PgWRKY1 gene in Panax ginseng. Acta Biochim. Biophys. Sin. 2015, 48, 117–131. [Google Scholar] [CrossRef] [Green Version]
- Xiu, H.; Nuruzzaman, M.; Guo, X.; Cao, H.; Huang, J.; Chen, X.; Luo, Z. Molecular cloning and expression analysis of eight PgWRKY genes in Panax ginseng responsive to salt and hormones. Int. J. Mol. Sci. 2016, 17, 319. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Lanczycki, C.J. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Procter, J.; Martin, D.; Clamp, M.; Barton, G. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, 609–612. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Harris, M.A. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Theocharidis, A.; Van Dongen, S.; Enright, A.J.; Freeman, T.C. Network visualization and analysis of gene expression data using BioLayout Express 3D. Nat. Protoc. 2009, 4, 1535. [Google Scholar] [CrossRef]
- Wang, C.; Deng, P.; Chen, L.; Wang, X.; Ma, H.; Hu, W.; Yao, N.; Feng, Y.; Chai, R.; Yang, G.; et al. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 2013, 8, e65120. [Google Scholar] [CrossRef]
- Liu, Q.L.; Zhong, M.; Li, S.; Pan, Y.Z.; Jiang, B.B.; Jia, Y.; Zhang, H.Q. Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, in tobacco enhances tolerance to salt stress. Plant Physiol. Biochem. 2013, 69, 27–33. [Google Scholar] [CrossRef]
- Nam, T.N.; Thia, L.H.; Mai, D.S.; Tuan, N.V. Overexpression of NbWRKY79 enhances salt stress tolerance in Nicotiana benthamiana. Acta Physiol. Plant. 2017, 39, 121. [Google Scholar] [CrossRef]
- Shi, W.Y.; Du, Y.T.; Ma, J.; Min, D.H.; Jin, L.G.; Chen, J.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; et al. The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in Soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Liu, X.; Ruan, H.; Zhang, J.; Xie, F.; Gai, J.; Yang, S. GmWRKY45 Enhances Tolerance to Phosphate Starvation and Salt Stress, and Changes Fertility in Transgenic Arabidopsis. Front. Plant Sci. 2020, 10, 1714. [Google Scholar] [CrossRef]
- Xu, J.; Chu, Y.; Liao, B.; Xiao, S.; Yin, Q.; Bai, R.; Su, H.; Dong, L.; Li, X.; Qian, J.; et al. Panax ginseng genome examination for ginsenoside biosynthesis. Gigascience 2017, 6, gix093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Century, K.; Reuber, T.L.; Ratcliffe, O.J. Regulating the regulators: The future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 2008, 147, 20–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Wang, M.; Zhang, X.; Hao, B.; Kaushik, S.K.; Pan, Y. WRKY gene family evolution in Arabidopsis thaliana. Genetica 2011, 139, 973. [Google Scholar] [CrossRef]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, P.; Rabara, R.C.; Langum, T.J.; Boken, A.K.; Rushton, D.L.; Boomsma, D.D.; Rohila, J.S. The WRKY transcription factor family in Brachypodium distachyon. BMC Genom. 2012, 13, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Niu, Y.; Xu, J.; Li, Y.; Luo, H.; Zhu, Y.; Chen, S. Discovery of WRKY transcription factors through transcriptome analysis and characterization of a novel methyl jasmonate-inducible PqWRKY1 gene from Panax quinquefolius. Plant Cell Tissue Organ Cult. 2013, 114, 269–277. [Google Scholar] [CrossRef]
- Fan, X.; Guo, Q.; Xu, P.; Gong, Y.; Shu, H.; Yang, Y.; Shen, X. Transcriptome-wide identification of salt-responsive members of the WRKY gene family in Gossypium aridum. PLoS ONE 2015, 10, e0126148. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Nan, Z.; Wang, X. The WRKY transcription factor genes in Lotus japonicus. Int. J. Genom. 2014, 2014, 420128. [Google Scholar]
- Baranwal, V.K.; Negi, N.; Khurana, P. Genome-wide identification and structural, functional and evolutionary analysis of WRKY components of mulberry. Sci. Rep. 2016, 6, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.Y.; Xu, Z.S.; Tian, C.; Huang, Y.; Wang, F.; Xiong, A.S. Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci. Rep. 2016, 6, 3101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musavizadeh, Z.; Najafi-Zarrini, H.; Kazemitabar, S.K.; Hashemi, S.H.; Faraji, S.; Barcaccia, G.; Heidari, P. Genome-Wide Analysis of Potassium Channel Genes in Rice: Expression of the OsAKT and OsKAT Genes under Salt Stress. Genes 2021, 12, 784. [Google Scholar] [CrossRef]
- Faraji, S.A.; Mehmood, F.; Malik, H.M.T.; Ahmed, I.; Heidari, P.; Poczai, P. The GASA Gene Family in Cacao (Theobroma cacao, Malvaceae): Genome Wide Identification and Expression Analysis. Agronomy 2021, 11, 1425. [Google Scholar]
- Blake, J.A. Ten quick tips for using the gene ontology. PLoS Comput. Biol. 2013, 9, e1003343. [Google Scholar] [CrossRef]
- Aken, O.V.; Zhang, B.; Law, S.; Narsai, R.; Whelan, J. AtWRKY40 and AtWRKY63 Modulate the Expression of Stress-Responsive Nuclear Genes Encoding Mitochondrial and Chloroplast Proteins. Plant Physiol. 2013, 162, 254–271. [Google Scholar] [CrossRef] [Green Version]

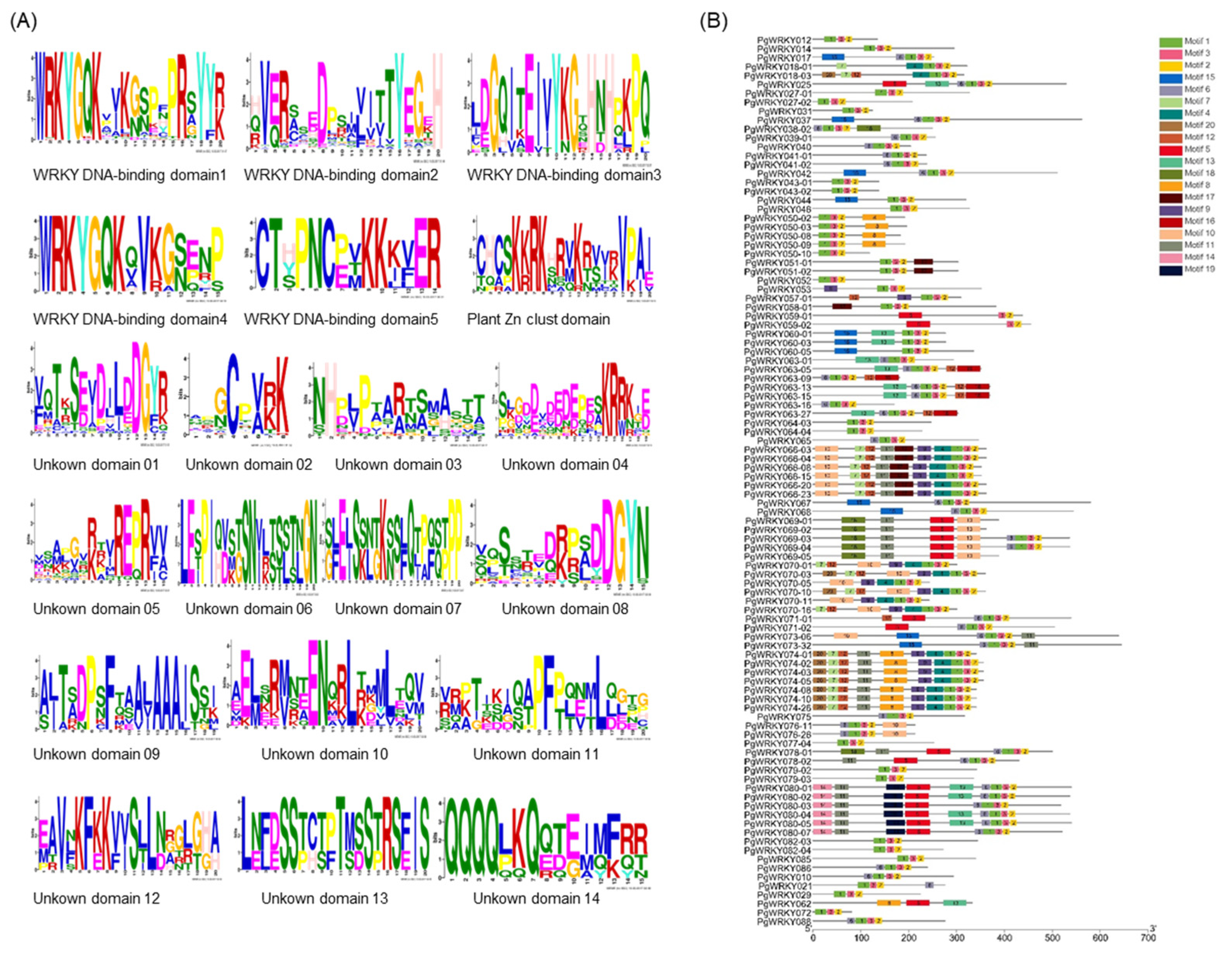
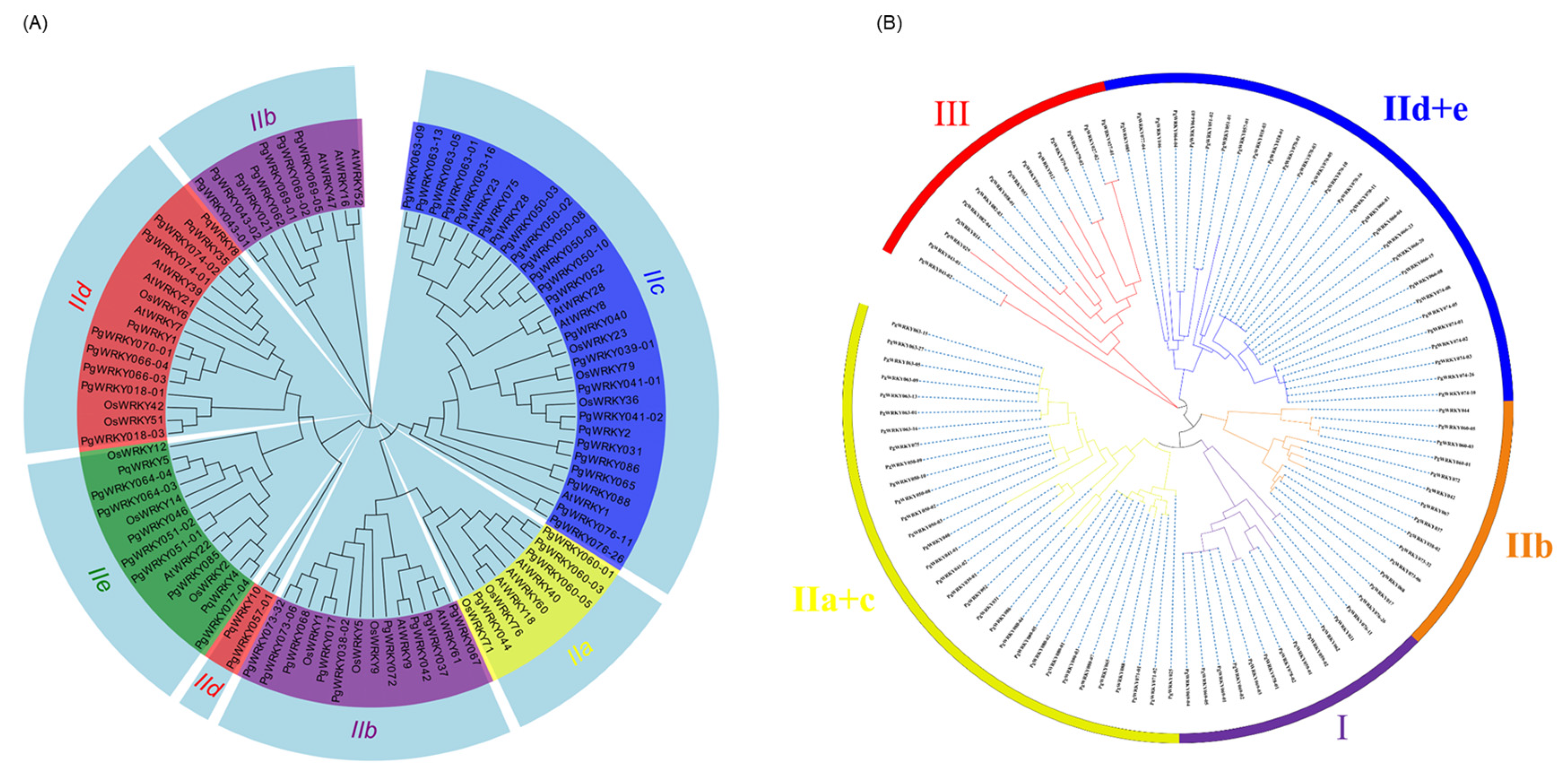
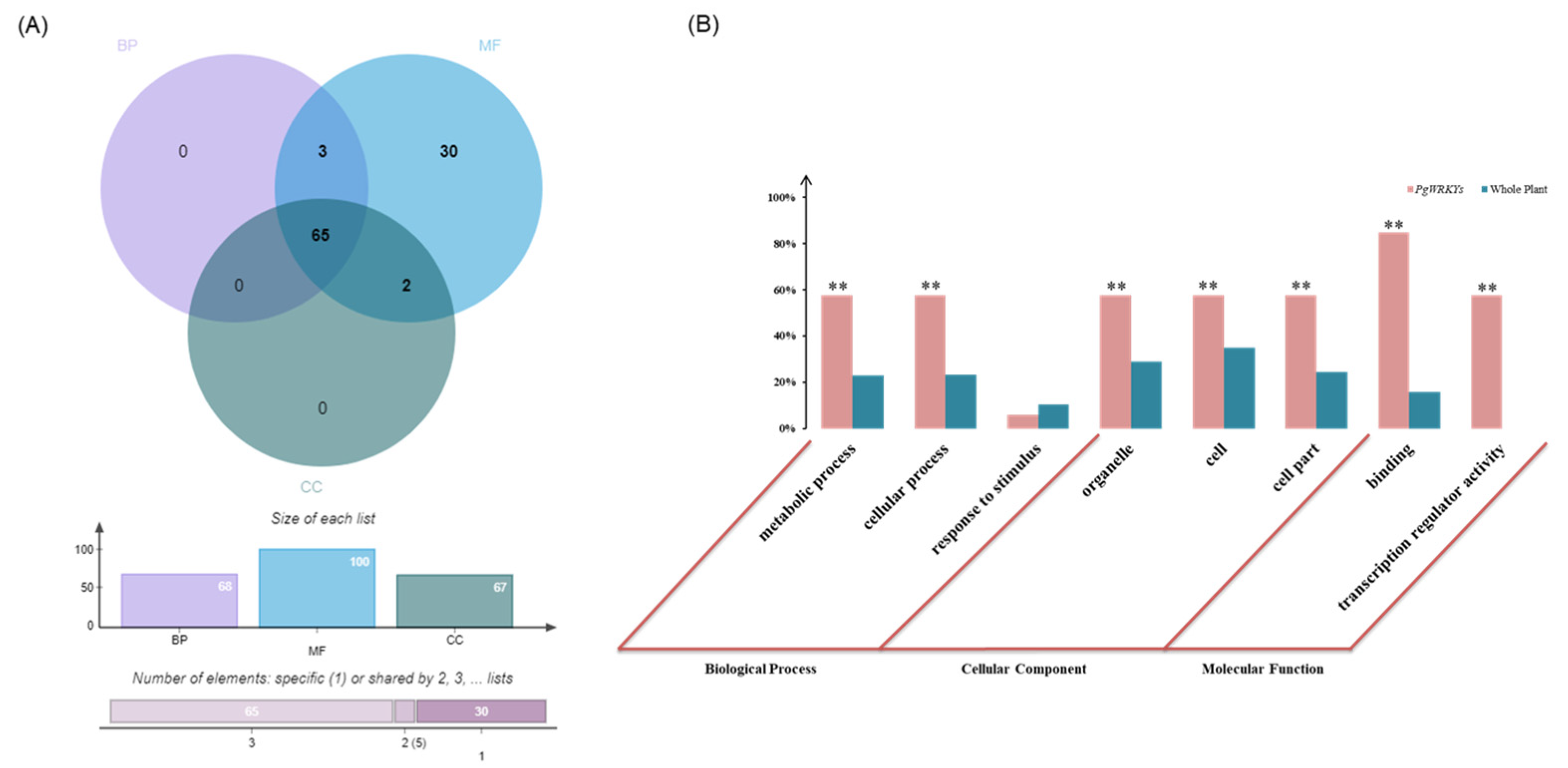
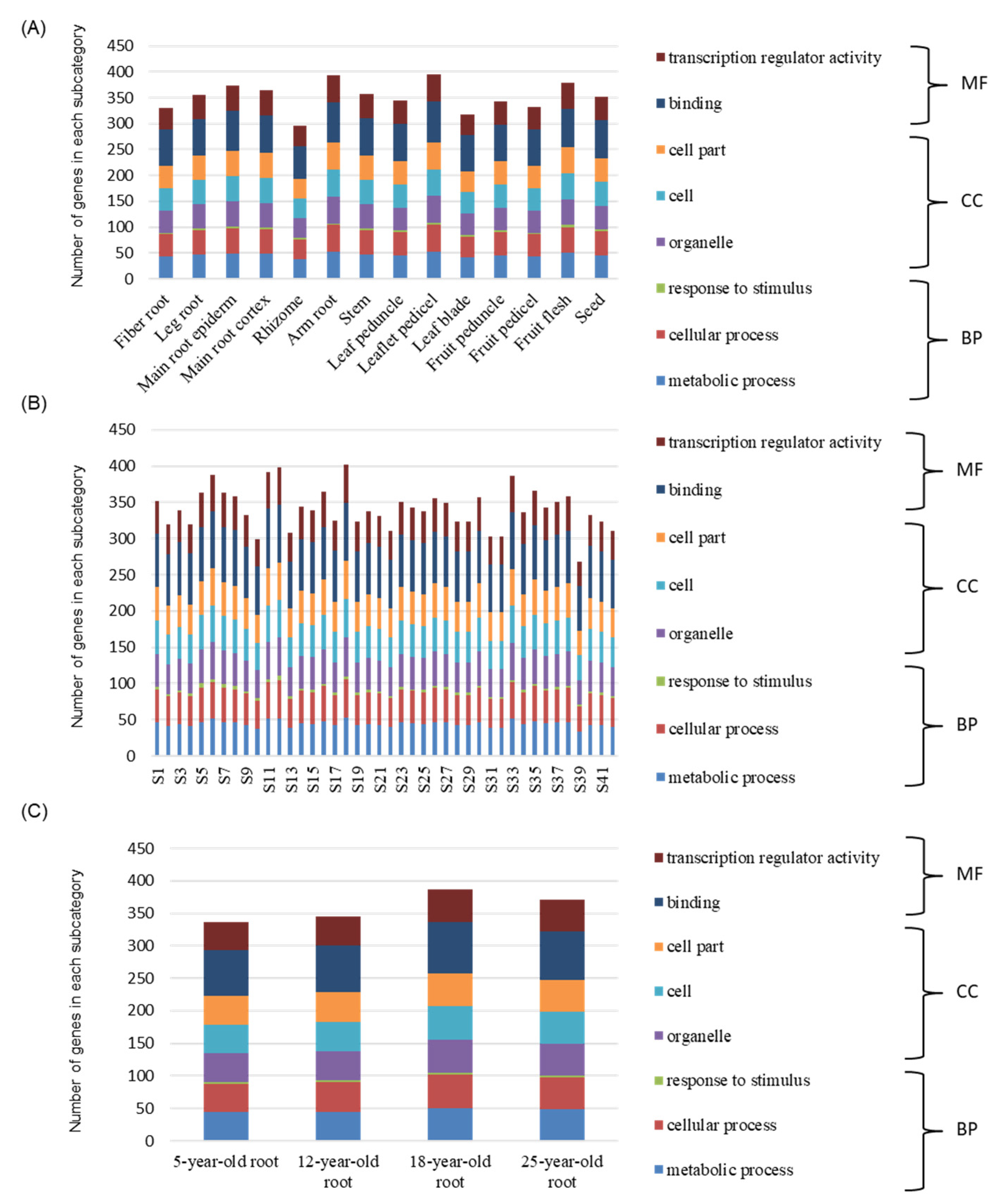
| Group Type | Group I | Group IIa + IIb | Group IIc | Group IId + IIe | Group III | Total |
|---|---|---|---|---|---|---|
| PgWRKY | 15 | 20 | 29 | 42 | 12 | 118 |
| DcWRKY | 17 | 20 | 18 | 29 | 11 | 95 |
| AtWRKY | 13 | 11 | 18 | 16 | 14 | 72 |
| OsWRKY | 15 | 12 | 15 | 18 | 36 | 96 |
| CsWRKY | 10 | 8 | 16 | 15 | 6 | 55 |
| BdWRKY | 17 | 9 | 21 | 16 | 23 | 86 |
| PtWRKY | 50 | 14 | 13 | 17 | 10 | 104 |
| PqWRKY | 23 | 4 | 8 | 5 | 5 | 55 |
| BnWRKY | 121 | 45 | 55 | 58 | 51 | 330 |
| GaWRKY | 17 | 22 | 30 | 28 | 12 | 109 |
| LjWRKY | 12 | 13 | 13 | 14 | 7 | 61 |
| MaWRKY | 10 | 11 | 9 | 13 | 8 | 51 |
| Gene 1 | Gene 2 | Group | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|
| PgWRKY059-01 | PgWRKY071-01 | I | 0.3429 | 2.9142 | 0.118 |
| PgWRKY057-01 | PgWRKY066-03 | IId | 1.5078 | 3.0856 | 0.489 |
| PgWRKY031 | PgWRKY075 | IIc | 2.5019 | 3.0983 | 0.807 |
| PgWRKY079-02 | PgWRKY053 | III | 0.6561 | 2.9124 | 0.225 |
| PgWRKY064-04 | PgWRKY051-01 | IIe | 0.8981 | 3.0321 | 0.296 |
| PgWRKY060-01 | PgWRKY044 | IIa | 0.1556 | 1.7321 | 0.0898 |
| PgWRKY069-02 | PgWRKY043-01 | IIb | 2.6637 | 3.8128 | 0.698 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Yu, E.; Hou, L.; Hua, P.; Zhao, M.; Wang, Y.; Hu, J.; Zhang, M.; Wang, K.; Wang, Y. Transcriptome-Based Identification, Characterization, Evolutionary Analysis, and Expression Pattern Analysis of the WRKY Gene Family and Salt Stress Response in Panax ginseng. Horticulturae 2022, 8, 756. https://doi.org/10.3390/horticulturae8090756
Liu T, Yu E, Hou L, Hua P, Zhao M, Wang Y, Hu J, Zhang M, Wang K, Wang Y. Transcriptome-Based Identification, Characterization, Evolutionary Analysis, and Expression Pattern Analysis of the WRKY Gene Family and Salt Stress Response in Panax ginseng. Horticulturae. 2022; 8(9):756. https://doi.org/10.3390/horticulturae8090756
Chicago/Turabian StyleLiu, Tao, En Yu, Lihe Hou, Panpan Hua, Mingzhu Zhao, Yanfang Wang, Jian Hu, Meiping Zhang, Kangyu Wang, and Yi Wang. 2022. "Transcriptome-Based Identification, Characterization, Evolutionary Analysis, and Expression Pattern Analysis of the WRKY Gene Family and Salt Stress Response in Panax ginseng" Horticulturae 8, no. 9: 756. https://doi.org/10.3390/horticulturae8090756
APA StyleLiu, T., Yu, E., Hou, L., Hua, P., Zhao, M., Wang, Y., Hu, J., Zhang, M., Wang, K., & Wang, Y. (2022). Transcriptome-Based Identification, Characterization, Evolutionary Analysis, and Expression Pattern Analysis of the WRKY Gene Family and Salt Stress Response in Panax ginseng. Horticulturae, 8(9), 756. https://doi.org/10.3390/horticulturae8090756







