Advances in Functional Genomics for Watermelon and Melon Breeding: Current Progress and Future Perspectives
Abstract
1. Introduction
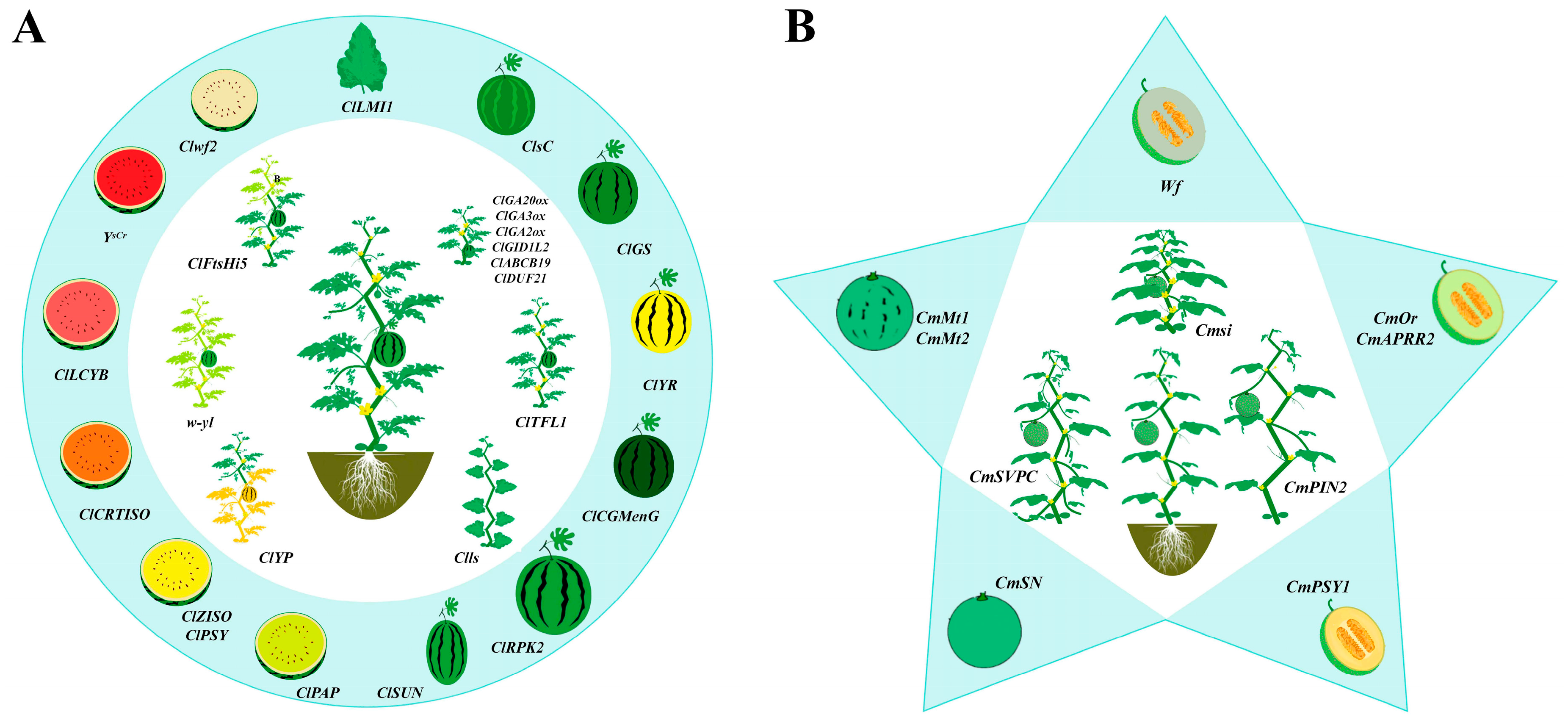
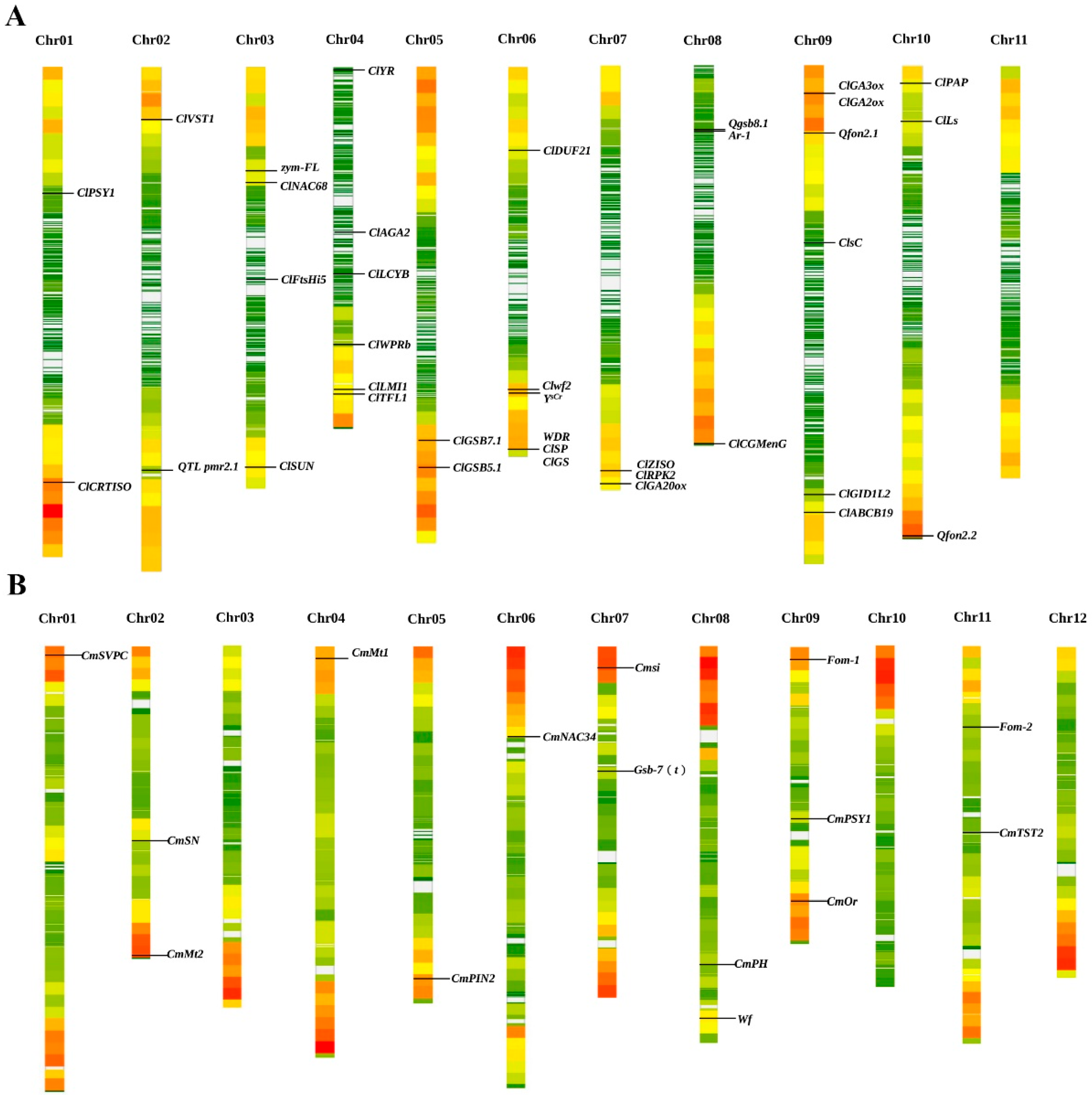
2. Research Progress on Gene Mapping and Functional Characterization of Plant Architecture-Related Traits
2.1. Short-Internode or Dwarfing Genes
2.2. Lateral Branch
2.3. Leaf Shape
2.4. Leaf Color
3. Advances in Functional Genes of Fruit Quality Traits
3.1. Rind Color and Stripe Patterns
3.2. Flesh Color
3.3. Sugar and Acid Metabolism
4. Progress in Disease Resistance-Related Genes
4.1. Fusarium Wilt (FW)
4.2. Powdery Mildew (PM)
4.3. Gummy Stem Blight (GSB)
4.4. Other Diseases
5. Future Perspectives
5.1. Systematic Discovery and Characterization of Superior Trait Genes
5.2. Deepening Functional Analysis and Regulatory Mechanism Elucidation
5.3. Accelerating Molecular Breeding System Development and Smart Breeding Integration
5.4. Epigenetics: A Frontier in Molecular Breeding of Watermelon and Melon
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, D.; Zhang, Z.; Unver, T.; Zhang, B. CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 2020, 29, 207–221. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.; Batchelor, W.; Xiong, L.; Yan, J. Crop phenomics and high-throughput phenotyping: Past decades, current challenges, and future perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yang, M.; Zhao, L.; Zhu, Z.; Liu, F.; Sun, H.; Sun, C.; Tan, L. HIGH-TILLERING AND DWARF 12 modulates photosynthesis and plant architecture by affecting carotenoid biosynthesis in rice. J. Exp. Bot. 2021, 72, 1212–1224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, R.; Xing, Y.; Xu, Y.; Xiong, D.; Wang, Y.; Yao, S. Separable regulation of POW1 in grain size and leaf angle development in rice. Plant Biotechnol. J. 2021, 19, 2517–2531. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Wang, Y.; Yang, H.; Niu, H.; Liu, D.; Yang, S.; Zhu, H.; Sun, S.; Yang, L. Development of branchless watermelon near isogenic lines by marker assisted selection. Hortic. Plant J. 2022, 8, 627–636. [Google Scholar] [CrossRef]
- Dou, J.; Yang, H.; Sun, D.; Yang, S.; Sun, S.; Zhao, S.; Lu, X.; Zhu, H.; Liu, D.; Ma, C.; et al. The branchless gene Clbl in watermelon encoding a TERMINAL FLOWER 1 protein regulates the number of lateral branches. Theor. Appl. Genet. 2022, 135, 65–79. [Google Scholar] [CrossRef]
- Mohr, H.C. Mode of inheritance of the bushy growth characteristics in watermelon. In Proceedings of the Association of Southern Agricultural Workers; University of Florida: Gainesville, FL, USA, 1956; Volume 53, p. 174. [Google Scholar]
- Liu, P.B.W.; Loy, J. Inheritance and morphology of two dwarf mutants in watermelon. J. Am. Soc. Hortic. Sci. 1972, 97, 745–748. [Google Scholar] [CrossRef]
- Dyutin, K.E.; Afanas’eva, E.A. Inheritance of the short vine trait in watermelon. Cytol. Genet. (Tsitologiya Genet.) 1987, 21, 71–73. [Google Scholar]
- Huang, H.; Zhang, X.; Wei, Z.; Li, Q.; Li, X. Inheritance of male-sterility and dwarfism in watermelon [Citrullus lanatus, (Thunb.) Matsum. and Nakai]. Sci. Hortic. 1998, 74, 175–181. [Google Scholar] [CrossRef]
- Guner, N.; Wehner, T.C. The genes of watermelon. HortScience 2004, 39, 1175–1182. [Google Scholar] [CrossRef]
- Dong, W.; Wu, D.; Li, G.; Wu, D.; Wang, Z. Next-generation sequencing from bulked segregant analysis identifies a dwarfism gene in watermelon. Sci. Rep. 2018, 8, 2908. [Google Scholar] [CrossRef]
- Wei, C.; Zhu, C.; Yang, L.; Zhao, W.; Ma, R.; Li, H.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. A point mutation resulting in a 13 bp deletion in the coding sequence of Cldf leads to a GA-deficient dwarf phenotype in watermelon. Hortic. Res. 2019, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Gebremeskel, H.; Dou, J.; Li, B.; Zhao, S.; Muhammad, U.; Lu, X.; He, N.; Liu, W. Molecular mapping and candidate gene analysis for GA3 responsive short internode in watermelon (Citrullus lanatus). Int. J. Mol. Sci. 2019, 21, 290. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, H.; Fan, M.; He, Y.; Guo, P. A mutation in the intron splice acceptor site of a GA3ox gene confers dwarf architecture in watermelon (Citrullus lanatus L.). Sci. Rep. 2020, 10, 14915. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.; Yun, H.; Rhee, S.; Seo, M.; Lee, G. Exploring molecular markers and candidate genes responsible for watermelon dwarfism. Hortic. Environ. Biotechnol. 2020, 61, 173–182. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, J.; Amanullah, S.; Ding, Z.; Cui, H.; Luan, F.; Gao, P. Fine mapping of Cla015407 controlling plant height in watermelon. J. Am. Soc. Hortic. Sci. 2021, 146, 196–205. [Google Scholar] [CrossRef]
- Liu, J.; Gao, P.; Wang, X.; Liu, H.; Ma, S.; Wang, J.; Luan, F. Genetic analysis and mapping of a short-internode gene (cladw) in watermelon (Citrullus lanatus L.). Euphytica 2022, 218, 119. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, M.; Sun, S.; Yang, S.; Li, J.; Li, H.; Yang, H.; Zhang, K.; Hu, J.; Liu, D.; et al. A single nucleotide deletion in an ABC transporter gene leads to a dwarf phenotype in watermelon. Front. Plant Sci. 2019, 10, 1399. [Google Scholar] [CrossRef]
- Sun, P.; Zhao, H.; Cao, L.; Zhang, T.; Zhang, H.; Yang, T.; Zhao, B.; Jiang, Y.; Dong, J.; Chen, T.; et al. A DUF21 domain-containing protein regulates plant dwarfing in watermelon. Plant Physiol. 2024, 196, 3091–3104. [Google Scholar] [CrossRef]
- Paris, H.S.; Nerson, H.; Karchi, Z. Genelics of internode length in melons. J. Hered. 1984, 75, 403–406. [Google Scholar] [CrossRef]
- Knavel, D.E. Inheritance of a Short-internode Mutant of ‘Mainstream’ Muskmelon. HortScience 1990, 25, 1274–1275. [Google Scholar] [CrossRef]
- Hwang, J.; Oh, J.; Kim, Z.; Staub, J.E.; Chung, S.M.; Park, Y. Fine genetic mapping of a locus controlling short internode length in melon (Cucumis melo L.). Mol. Breed. 2014, 34, 949–961. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, J.; Liu, S.; Ding, Z.; Luan, F.; Gao, P. Bulked-segregant analysis identified a putative region related to short internode length in melon. HortScience 2019, 54, 1293–1298. [Google Scholar] [CrossRef]
- Zink, F.W. UC SR-91 Bush, UC Top Mark Bush, and UC Perlita Bush muskmelon breeding lines. HortScience 1978, 13, 486. [Google Scholar] [CrossRef]
- Halsey, L.H. UF G508, G509, G510, G511, G515 muskmelon breeding lines. HortScience 1980, 15, 538. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, K.; Zhu, H.; Zhang, X.; Yan, W.; Xu, N.; Liu, D.; Hu, J.; Wu, Y.; Weng, Y.; et al. Melon short internode (CmSi) encodes an ERECTA-like receptor kinase regulating stem elongation through auxin signaling. Hortic. Res. 2020, 7, 202. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Grandío, E.G.; Serra, F.; Marcel, F.; Rodríguez-Buey, M.L.; Schmitz, G.; Theres, K.; Bendahmane, A.; Dopazo, H.; Cubas, P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 2011, 67, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, T.; Kyozuka, J.; Oikawa, T.; Kyozuka, J. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in Rice. Plant Cell 2009, 21, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizu-Sato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H.; et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 2011, 23, 3276–3287. [Google Scholar] [CrossRef]
- Duan, E.; Wang, Y.; Li, X.; Lin, Q.; Zhang, T.; Wang, Y.; Zhou, C.; Zhang, H.; Jiang, L.; Wang, J.; et al. OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell 2019, 31, 1026–1042. [Google Scholar] [CrossRef] [PubMed]
- Shao, G.; Lu, Z.; Xiong, J.; Wang, B.; Jing, Y.; Meng, X.; Liu, G.; Ma, H.; Liang, Y.; Chen, F.; et al. Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol. Plant 2019, 12, 1090–1102. [Google Scholar] [CrossRef]
- Zhao, B.; Wu, T.; Ma, S.; Jiang, D.; Bie, X.; Sui, N.; Zhang, X.; Wang, F. TaD27-B gene controls the tiller number in hexaploid wheat. Plant Biotechnol. J. 2020, 18, 513–525. [Google Scholar] [CrossRef]
- Schumacher, K.; Schmitt, T.; Rossberg, M.; Schmitz, G.; Theres, K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 1999, 96, 290–295. [Google Scholar] [CrossRef]
- Schmitz, G.; Tillmann, E.; Carriero, F.; Fiore, C.; Cellini, F.; Theres, K. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 2002, 99, 1064–1069. [Google Scholar] [CrossRef]
- Weng, L.; Bai, X.; Zhao, F.; Li, R.; Xiao, H. Manipulation of flowering time and branching by overexpression of the tomato transcription factor SlZFP2. Plant Biotechnol. J. 2016, 14, 2310–2321. [Google Scholar] [CrossRef]
- Silva Ferreira, D.; Kevei, Z.; Kurowski, T.; de Noronha Fonseca, M.E.; Mohareb, F.; Boiteux, L.S.; Thompson, A.J. BIFURCATE FLOWER TRUSS: A novel locus controlling inflorescence branching in tomato contains a defective MAP kinase gene. J. Exp. Bot. 2018, 69, 2581–2593. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, H.; Zhao, J.; Pan, Y.; Cheng, S.; Lietzow, C.D.; Wen, C.; Zhang, X.; Weng, Y. LITTLELEAF(LL) encodes a WD40 repeat domain-containing protein associated with organ size variation in cucumber. Plant J. 2018, 95, 834–847. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, Y.; Ge, D.; Wang, Z.; Song, W.; Gu, R.; Che, G.; Cheng, Z.; Liu, R.; Zhang, X. CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proc. Natl. Acad. Sci. USA 2019, 116, 17105–17114. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.M.; Wagler, T.N.; Quijada, P.; Doebley, J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 2006, 38, 594–597. [Google Scholar] [CrossRef]
- Tanaka, M.; Takei, K.; Kojima, M.; Sakakibara, H.; Mori, H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006, 45, 1028–1036. [Google Scholar] [CrossRef]
- Chen, X.; Xia, X.; Guo, X.; Zhou, Y.; Shi, K.; Zhou, J.; Yu, J. Apoplastic H2O2 plays a critical role in axillary bud outgrowth by altering auxin and cytokinin homeostasis in tomato plants. New Phytol. 2016, 211, 1266–1278. [Google Scholar] [CrossRef]
- Shinohara, N.; Taylor, C.; Leyser, O.; Scheres, B. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 2013, 11, e1001474. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Q.; Yan, J.; Sun, K.; Liang, Y.; Jia, M.; Meng, X.; Fang, S.; Wang, Y.; Jing, Y.; et al. ζ-Carotene isomerase suppresses tillering in rice through the coordinated biosynthesis of strigolactone and abscisic acid. Mol. Plant 2020, 13, 1784–1801. [Google Scholar] [CrossRef]
- Yi, L.; Zhou, W.; Zhou, Q.; Chen, Z.; Zhang, Y.; Dai, Z.; Wang, Y. Fine mapping identifies ClTFL1 encodes a TERMINAL FLOWER 1 protein as putative candidate gene for inflorescence architecture and tendril development and in watermelon. J. Plant Growth Regul. 2022, 42, 4150–4160. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, A.; He, W.; Li, Q.; Zhao, B.; Zhao, H.; Ke, X.; Guo, Y.; Sun, P.; Yang, T.; et al. GRAS family member LATERAL SUPPRESSOR regulates the initiation and morphogenesis of watermelon lateral organs. Plant Physiol. 2023, 193, 2592–2604. [Google Scholar] [CrossRef]
- Dou, J.; Kang, Q.; Li, T.; Umer, M.J.; Alharthi, B.; Liu, D.; Yang, S.; Niu, H.; Ma, C.; Zhu, H.; et al. Construction and application of a new watermelon germplasm with the phenotype of dwarf and branchless. Funct. Integr. Genom. 2023, 23, 310. [Google Scholar] [CrossRef] [PubMed]
- Zalapa, J.E.; Staub, J.E.; Mccreight, J.D. Variance component analysis of plant architectural traits and fruit yield in melon. Euphytica 2008, 162, 129–143. [Google Scholar] [CrossRef]
- Ohara, T.; Wako, T.; Kojima, A.; Yoshida, T.; Ishiuchi, D. Breeding of sup-pressed- branching melon fine ‘Melon chukanbohon nou 4’ (‘Melon- Parental line 4’) and its characteristics. Acta Hortic. 2002, 588, 227–231. [Google Scholar] [CrossRef]
- Fukino, N.; Ohara, T.; Sugiyama, M.; Kubo, N.; Hirai, M.; Sakata, Y.; Matsumoto, S. Mapping of a gene that confers short lateral branching (slb) in melon (Cucumis melo L.). Euphytica 2012, 187, 133–143. [Google Scholar] [CrossRef]
- Fang, S.; Zhao, J.; Guo, K.; Duan, Y.; Wang, F.; Nie, L.; Zhao, W. Identification of SHORT VEGETATIVE PHASE (SVP)-like genes and necessary responsibility of CmSVPc for the development of lateral branches in melon (Cucumis melo L.). Sci. Hortic. 2023, 312, 111845. [Google Scholar] [CrossRef]
- Wei, C.; Chen, X.; Wang, Z.; Liu, Q.; Li, H.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Genetic mapping of the LOBED LEAF 1 (ClLL1) gene to a 127.6-kb region in watermelon (Citrullus lanatus L.). PLoS ONE 2017, 12, e0180741. [Google Scholar] [CrossRef]
- Duan, S.; Guo, Y.; Wang, Y.; Umer, M.J.; Liu, D.; Yang, S.; Niu, H.; Sun, S.; Yang, L.; Dou, J.; et al. HD-Zip transcription factor is responsible for no-lobed leaf in watermelon (citrullus lanatus L.). Phyton-Int. J. Exp. Bot. 2023, 92, 18. [Google Scholar] [CrossRef]
- Ni, X.; Huang, J.; Ali, B.; Zhou, W.; Zhao, J. Genetic analysis and fine mapping of the LOBEDLEAF 1 (BnLL1) gene in rapeseed (Brassica napus L.). Euphytica 2015, 204, 29–38. [Google Scholar] [CrossRef]
- Andres, R.J.; Coneva, V.; Frank, M.H.; Tuttle, J.R.; Samayoa, L.F.; Han, S.W.; Kaur, B.; Zhu, L.; Fang, H.; Bowman, D.T.; et al. Modifications to a LATE MERISTEM IDENTITY1 gene are responsible for the major leaf shapes of Upland cotton (Gossypium hirsutum L.). Proc. Natl. Acad. Sci. USA 2017, 114, E57–E66. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, J.; Sambandam, C.N. Inheritance of leaf shape in muskmelon (Cucumis melo L.) I. A qualitative approach. Annamalai Univ. Agric. Res. Annu. 1985, 12, 53–58. [Google Scholar]
- Gao, X.; Ning, X.; Wang, Y.; Wang, X.; Yan, W.; Zhang, Z.; Li, G. Fine mapping of a gene that confers palmately lobed leaf (pll) in melon (Cucumis melo L.). Euphytica 2014, 200, 337–347. [Google Scholar] [CrossRef]
- Li, C.; Xu, Y.; Ma, J.; Jin, J.; Huang, D.; Yao, M.; Ma, C.; Chen, L. Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘Anji Baicha’ (Camellia sinensis). BMC Plant Biol. 2016, 16, 195. [Google Scholar] [CrossRef]
- Guo, P.; Huang, Z.; Zhao, W.; Lin, N.; Wang, Y.; Shang, F. Mechanisms for leaf color changes in Osmanthus fragrans ‘Ziyan Gongzhu’ using physiology, transcriptomics and metabolomics. BMC Plant Biol. 2023, 23, 453. [Google Scholar] [CrossRef]
- Cheng, L.; Lai, K.J.D. Golden2-like (GLK2) Transcription Factor: Developmental Control of Tomato Fruit Photosynthesis and Its Contribution to Ripe Fruit Characteristics. Master’s Thesis, University of California Davis, Davis, CA, USA, 2013. [Google Scholar]
- Zhu, J.; Yin, Y.; Lu, J.; Warner, T.A.; Xu, X.; Lyu, M.; Wang, X.; Guo, C.; Cheng, T.; Zhu, Y.; et al. The relationship between wheat yield and sun-induced chlorophyll fluorescence from continuous measurements over the growing season. Remote Sens. Environ. 2023, 298, 18. [Google Scholar] [CrossRef]
- Zhu, K.; Chen, H.; Mei, X.; Lu, S.; Xie, H.; Liu, J.; Chai, L.; Xu, Q.; Wurtzel, E.T.; Ye, J.; et al. Transcription factor CsMADS3 coordinately regulates chlorophyll and carotenoid pools in Citrus hesperidium. Plant Physiol. 2023, 193, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, X.; Zhang, X.; Zhang, H.; Zhao, X. Mutation mechanism of leaf color in plants: A Review. Forests 2020, 11, 851. [Google Scholar] [CrossRef]
- Barham, W.S. A study of the Royal Golden watermelon with emphasis on the inheritance of the chlorotic condition characteristic of this variety. Proc. Am. Soc. Hort. Sci. 1956, 67, 487. [Google Scholar]
- Abdelhafez, A.A. Inheritance of marker genes for leaf colour and shape in watermelon, Citrullus lanatus, Thumb. Acta Agron. Acad. Sci. Hung. 1983, 343–348. [Google Scholar]
- Xu, B.; Zhang, C.; Gu, Y.; Cheng, R.; Huang, D.; Liu, X.; Sun, Y. Physiological and transcriptomic analysis of a yellow leaf mutant in watermelon. Sci. Rep. 2023, 13, 9647. [Google Scholar] [CrossRef]
- Zhu, Y.; Yuan, G.; Wang, Y.; An, G.; Li, W.; Liu, J.; Sun, D. Mapping and functional verification of leaf yellowing genes in watermelon during whole growth period. Front. Plant Sci. 2022, 13, 1049114. [Google Scholar] [CrossRef] [PubMed]
- Kidanemariam, H.G. Genetic and Molecular Mechanisms of Delayed Green Leaf Color and Short Internode Length in Watermelon (Citrullus lanatus). Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2020. (In Chinese). [Google Scholar]
- Han, H.; Zhou, Y.; Liu, H.; Chen, X.; Wang, Q.; Zhuang, H.; Sun, X.; Ling, Q.; Zhang, H.; Wang, B.; et al. Transcriptomics and metabolomics analysis provides insight into leaf color and photosynthesis variation of the yellow-green leaf mutant of Hami melon (Cucumis melo L.). Plants 2023, 12, 1623. [Google Scholar] [CrossRef]
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018, 16, 1363–1374. [Google Scholar] [CrossRef]
- Yu, X.; Xiao, J.; Chen, S.; Yu, Y.; Ma, J.; Lin, Y.; Li, R.; Lin, J.; Fu, Z.; Zhou, Q.; et al. Metabolite signatures of diverse Camellia sinensis tea populations. Nat. Commun. 2020, 11, 5586. [Google Scholar] [CrossRef]
- Weetman, M. Inheritance and Correlation of Shape, Size, Color and Time of Maturity in the Watermelon (Citrullus vulgaris Schrad.). Ph.D. Thesis, Iowa State University, Ames, IA, USA, 1935. [Google Scholar]
- Poole, C. Genetics of cultivated cucurbits. J. Hered. 1944, 35, 122–128. [Google Scholar] [CrossRef]
- Lou, L.; Wehner, T. Qualitative inheritance of external fruit traits in watermelon. HortScience 2016, 51, 487–496. [Google Scholar] [CrossRef]
- Yang, H.; Park, S.; Park, Y.; Lee, G.; Kang, S.; Kim, Y. Linkage analysis of the three loci determining rind color and stripe pattern in watermelon. Hortic. Sci. Technol. 2015, 33, 559–565. [Google Scholar] [CrossRef]
- Park, S.; Kim, K.; Kang, S.; Yang, H. Rapid and practical molecular marker development for rind traits in watermelon. Hortic. Environ. Biotechnol. 2016, 57, 385–391. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhao, S.; Sun, H.; Wang, X.; Wu, S.; Lin, T.; Ren, Y.; Gao, L.; Deng, Y.; Zhang, J.; et al. Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat. Genet. 2019, 51, 1616–1623. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, M.; Xu, N.; Yang, S.; Dou, J.; Liu, D.; Zhu, L.; Zhu, H.; Hu, J.; Ma, C.; et al. Fine mapping a ClGS gene controlling dark-green stripe rind in watermelon. Sci. Hortic. 2022, 291, 110583. [Google Scholar] [CrossRef]
- Liu, D.; Liang, J.; Liu, Q.; Chen, Y.; Duan, S.; Sun, D.; Zhu, H.; Dou, J.; Niu, H.; Yang, S.; et al. The pseudo-type response regulator gene Clsc regulates rind stripe coloration in watermelon. J. Integr. Agric. 2025, 24, 147–160. [Google Scholar] [CrossRef]
- Zhen, Y.; Fu, Y.; Dai, X.; Chen, Y.; Guo, C.; Zhang, R.; Huang, X.; Feng, M.; Yan, X.; Wang, Z.; et al. The KNOX transcription factor ClSP activates ClAPRR2 to regulate dark green stripe formation in watermelon. Plant Biotechnol. J. 2025, 23, 3012–3023. [Google Scholar] [CrossRef]
- Zhen, Y.; Ma, R.; Cheng, D.; Yan, X.; He, Y.; Wang, C.; Pan, X.; Yin, L.; Zhang, X.; Wei, C. Candidate gene analysis of watermelon stripe pattern locus ClSP ongoing recombination suppression. Theor. Appl. Genet. 2021, 134, 3263–3277. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, S.; Dou, J.; Ali, A.; Gebremeskel, H.; Gao, L.; He, N.; Lu, X.; Liu, W. Genetic mapping and development of molecular markers for a candidate gene locus controlling rind color in watermelon. Theor. Appl. Genet. 2019, 132, 2741–2753. [Google Scholar] [CrossRef]
- Liu, D.; Yang, H.; Yuan, Y.; Zhu, H.; Zhang, M.; Wei, X.; Sun, D.; Wang, X.; Yang, S.; Yang, L. Comparative transcriptome analysis provides insights into yellow rind formation and preliminary mapping of the Clyr (yellow rind) gene in watermelon. Front. Plant Sci. 2020, 11, 192. [Google Scholar] [CrossRef]
- Ma, J.; Yuan, G.; Xu, X.; Zhang, H.; Qiu, Y.; Zhang, H. Identification and molecular marker development for peel color gene in melon (Cucumis melo L.). J. Integr. Agric. 2025, 24, 2589–2600. [Google Scholar] [CrossRef]
- Liu, L.; Sun, T.; Liu, X.; Guo, Y.; Huang, X.; Gao, P.; Wang, X. Genetic analysis and mapping of a striped rind gene (st3) in melon (Cucumis melo L.). Euphytica 2019, 215, 20. [Google Scholar] [CrossRef]
- Gao, M.; Liang, X.; Liu, X.; Guo, Y.; Liu, X.; Liu, J.; Gao, Y. Research progress on fruit stripe genes in Cucurbitaceae crops. Mol. Plant Breed. 2021, 19, 2922–2932. [Google Scholar]
- Périn, C.; Hagen, L.; De Conto, V.; Katzir, N.; Danin-Poleg, Y.; Portnoy, V.; Baudracco-Arnas, S.; Chadoeuf, J.; Dogimont, C.; Pitrat, M. A reference map of Cucumis melo based on two recombinant inbred line populations. Theor. Appl. Genet. 2002, 104, 1017–1034. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Ruggieri, V.; Pérez, S.; Alexiou, K.; Fernández, M.; Jahrmann, T.; Pujol, M.; Garcia-Mas, J. QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map. BMC Plant Biol. 2018, 18, 324. [Google Scholar] [CrossRef]
- Liang, X.; Li, Q.; Cao, L.; Du, X.; Qiang, J.; Hou, J.; Li, X.; Zhu, H.; Yang, S.; Liu, D.; et al. Natural allelic variation in the EamA-like transporter, CmSN, is associated with fruit skin netting in melon. Theor. Appl. Genet. 2023, 136, 192. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xing, S.; Sun, G.; Shang, J.; Yao, J.L.; Li, N.; Zhou, D.; Wang, Y.; Lu, Y.; Bi, J.; et al. Multi-omics analyses unveil dual genetic loci governing four distinct watermelon flesh color phenotypes. Mol. Hortic. 2025, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Bang, H.; Kim, S.; Leskovar, D.; King, S. Development of a codominant CAPS marker for allelic selection between canary yellow and red watermelon based on SNP in lycopene β-cyclase (LCYB) gene. Mol. Breed. 2007, 20, 63–72. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, H.; Guo, S.; Ren, Y.; Li, M.; Wang, J.; Zhang, H.; Gong, G.; Xu, Y. Decreased protein abundance of lycopene beta-cyclase contributes to red flesh in domesticated watermelon. Plant Physiol. 2020, 183, 1171–1183. [Google Scholar] [CrossRef]
- Liu, S.; Gao, Z.; Wang, X.; Luan, F.; Dai, Z.; Yang, Z.; Zhang, Q. Nucleotide variation in the phytoene synthase (ClPsy1) gene contributes to golden flesh in watermelon (Citrullus lanatus L.). Theor. Appl. Genet. 2022, 135, 185–200. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, H.; Guo, S.; Ren, Y.; Li, M.; Wang, J.; Yu, Y.; Zhang, H.; Gong, G.; He, H.; et al. ClZISO mutation leads to photosensitive flesh in watermelon. Theor. Appl. Genet. 2022, 135, 1565–1578. [Google Scholar] [CrossRef]
- Jin, B.; Lee, J.; Kweon, S.; Cho, Y.; Choi, Y.; Lee, S.; Park, Y. Analysis of flesh color-related carotenoids and development of a CRTISO gene-based DNA marker for prolycopene accumulation in watermelon. Hortic. Environ. Biotechnol. 2019, 60, 399–410. [Google Scholar] [CrossRef]
- Li, N.; Shang, J.; Wang, J.; Zhou, D.; Ma, S. Discovery of the genomic region and candidate genes of the Scarlet Red Flesh Color (Yscr) locus in watermelon (Citrullus lanatus L.). Front. Plant Sci. 2020, 11, 116. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Zhou, W.; Zhang, Y.; Chen, Z.; Wu, N.; Wang, Y.; Dai, Z. Genetic mapping of a single nuclear locus determines the white flesh color in watermelon (Citrullus lanatus L.). Front. Plant Sci. 2023, 14, 1090009. [Google Scholar] [CrossRef]
- Pei, S.; Liu, Z.; Wang, X.; Luan, F.; Dai, Z.; Yang, Z.; Zhang, Q.; Liu, S. Quantitative trait loci and candidate genes responsible for pale green flesh colour in watermelon (Citrullus lanatus). Plant Breed. 2021, 140, 349–359. [Google Scholar] [CrossRef]
- Tzuri, G.; Zhou, X.; Chayut, N.; Yuan, H.; Portnoy, V.; Meir, A.; Sa’ar, U.; Baumkoler, F.; Mazourek, M.; Lewinsohn, E.; et al. A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis melo). Plant J. 2015, 82, 267–279. [Google Scholar] [CrossRef]
- Galpaz, N.; Gonda, I.; Shem-Tov, D.; Barad, O.; Tzuri, G.; Lev, S.; Fei, Z.; Xu, Y.; Mao, L.; Jiao, C.; et al. Deciphering genetic factors that determine melon fruit-quality traits using RNA-Seq-based high-resolution QTL and eQTL mapping. Plant J. 2018, 94, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Monforte, A.; Oliver, M.; Gonzalo, M.; Alvarez, J.; Dolcet-Sanjuan, R.; Arús, P. Identification of quantitative trait loci involved in fruit quality traits in melon (Cucumis melo L.). Theor. Appl. Genet. 2004, 108, 750–758. [Google Scholar] [CrossRef]
- Zhao, G.; Lian, Q.; Zhang, Z.; Fu, Q.; He, Y.; Ma, S.; Ruggieri, V.; Monforte, A.J.; Wang, P.; Julca, I.; et al. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 2019, 51, 1607–1615. [Google Scholar] [CrossRef]
- Duan, X.; Jiang, C.; Zhao, Y.; Gao, G.; Li, M.; Qi, H. Transcriptome and metabolomics analysis revealed that CmWRKY49 regulating CmPSY1 promotes β-carotene accumulation in orange fleshed oriental melon. Hortic. Plant J. 2022, 8, 650–666. [Google Scholar] [CrossRef]
- Ren, Y.; Sun, H.; Zong, M.; Guo, S.; Ren, Z.; Zhao, J.; Li, M.; Zhang, J.; Tian, S.; Wang, J.; et al. Localization shift of a sugar transporter contributes to phloem unloading in sweet watermelons. New Phytol. 2020, 227, 1858–1871. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, M.; Guo, S.; Sun, H.; Zhao, J.; Zhang, J.; Liu, G.; He, H.; Tian, S.; Yu, Y.; et al. Evolutionary gain of oligosaccharide hydrolysis and sugar transport enhanced carbohydrate partitioning in sweet watermelon fruits. Plant Cell 2021, 33, 1554–1573. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Zhang, J.; Ren, Y.; Li, M.; Tian, S.; Yu, Y.; Zuo, Y.; Gong, G.; Zhang, H.; et al. The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Hortic. Res. 2021, 8, 214. [Google Scholar] [CrossRef]
- Cheng, J.; Wen, S.; Xiao, S.; Lu, B.; Ma, M.; Bie, Z. Overexpression of the tonoplast sugar transporter CmTST2 in melon fruit increases sugar accumulation. J. Exp. Bot. 2018, 69, 511–523. [Google Scholar] [CrossRef]
- Dou, J.; Zhao, S.; Lu, X.; He, N.; Zhang, L.; Ali, A.; Kuang, H.; Liu, W. Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatus L.). Theor. Appl. Genet. 2018, 131, 947–958. [Google Scholar] [CrossRef]
- Qiu, B.; Zhang, T.; Zhang, S.; Qu, Q.; Zhu, Z.; Liu, S.; Song, Z.; Xia, L.; Yang, Z.; Zhang, Q.; et al. BSA-seq and quantitative trait locus mapping reveals a major effective QTL for carpel number in watermelon (Citrullus lanatus). Plant Breed. 2022, 141, 460–470. [Google Scholar] [CrossRef]
- Li, Q.; Hao, X.; Guo, Z.; Qu, K.; Gao, M.; Song, G.; Yin, Z.; Yuan, Y.; Dong, C.; Niu, J.; et al. Screening and resistance locus identification of the mutant fcrZ22 resistant to crown rot caused by Fusarium pseudograminearum. Plant Dis. 2024, 108, 426–433. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, Z.; Miao, J.; Liu, Q.; Ma, C.; Tian, X.; He, J.; Bi, H.; Yao, W.; Li, T.; et al. Pm57 from Aegilops searsii encodes a tandem kinase protein and confers wheat powdery mildew resistance. Nat. Commun. 2024, 15, 4796. [Google Scholar] [CrossRef]
- Bruton, B.D. Soilborne diseases in Cucurbitaceae: Pathogen virulence and host resistance. In Cucurbitaceae’ 98: Evaluation and Enhancement of Cucurbit Germplasm; Mc Creight, J.D., Ed.; ASHS Press: Alexandria, VA, USA, 1998; pp. 143–166. [Google Scholar]
- Egel, D.S.; Martyn, R.D. Fusarium Wilt of Watermelon and Other Cucurbits. Plant Health Instructor. 2007. Available online: https://www.apsnet.org/edcenter/disandpath/fungalasco/pdlessons/Pages/FusariumWatermelon.aspx (accessed on 29 July 2025).
- Wehner, T.C. Watermelon. In Vegetables I: Asteraceae, Brassicaceae, Chenopodicaceae, and Cucurbitaceae; Prohens, J., Nuez, F., Eds.; Springer: New York, NY, USA, 2008; pp. 381–418. [Google Scholar]
- Zhou, X.G.; Everts, K.L.; Bruton, B.D. Race 3, a new and highly virulent race of Fusarium oxysporum f. sp. niveum causing Fusarium wilt in watermelon. Plant Dis. 2010, 94, 92–98. [Google Scholar] [CrossRef]
- Lambel, S.; Lanini, B.; Vivoda, E.; Fauve, J.; Patrick Wechter, W.; Harris-Shultz, K.R.; Massey, L.; Levi, A. A major QTL associated with Fusarium oxysporum race 1 resistance identified in genetic populations derived from closely related watermelon lines using selective genotyping and genotyping-by-sequencing for SNP discovery. Theor. Appl. Genet. 2014, 127, 2105–2115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, M.; Tan, J.; Huang, M.; Chu, X.; Li, Y.; Han, X.; Fang, T.; Tian, Y.; Jarret, R.; et al. Telomere-to-telomere Citrullus super-pangenome provides direction for watermelon breeding. Nat. Genet. 2024, 56, 1750–1761. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Jiao, D.; Gong, G.; Zhang, H.; Guo, S.; Zhang, J.; Xu, Y. Genetic analysis and chromosome mapping of resistance to Fusarium oxysporum f. sp. Niveum (FON) race 1 and race 2 in watermelon (Citrullus lanatus L.). Mol. Breed. 2015, 35, 183. [Google Scholar] [CrossRef] [PubMed]
- Branham, S.E.; Patrick, W.W.; Ling, K.S.; Chanda, B.; Massey, L.; Zhao, G.; Guner, N.; Bello, M.; Kabelka, E.; Fei, Z.; et al. QTL mapping of resistance to Fusarium oxysporum f. sp. niveum race 2 and Papaya ringspot virus in Citrullus amarus. Theor. Appl. Genet. 2020, 133, 677–687. [Google Scholar] [CrossRef]
- Meru, G.; McGregor, C.E. A genetic locus associated with resistance to Fusarium oxysporum f. sp. niveum race 2 in Citrullus lanatus-type watermelon. J. Am. Soc. Hortic. Sci. 2016, 141, 617–622. [Google Scholar] [CrossRef]
- Branham, S.E.; Levi, A.; Farnham, M.W.; Patrick, W.W. A GBS-SNP-based linkage map and quantitative trait loci (QTL) associated with resistance to Fusarium oxysporum f. sp. niveum race 2 identified in Citrullus lanatus var. citroides. Theor. Appl. Genet. 2017, 130, 319–330. [Google Scholar] [CrossRef]
- Branham, S.E.; Levi, A.; Wechter, W.P. QTL mapping identifies novel source of resistance to fusarium wilt race 1 in Citrullus amarus. Plant Dis. 2019, 103, 984–989. [Google Scholar] [CrossRef]
- Ganaparthi, V.R.; Wechter, P.; Levi, A.; Branham, S.E. Mapping and validation of Fusarium wilt race 2 resistance QTL from Citrullus amarus line USVL246-FR2. Theor. Appl. Genet. 2024, 137, 91. [Google Scholar] [CrossRef]
- Risser, G. A proposed nomenclature of Fusarium oxysporum f. sp. melonis races and resistance genes in Cucumis melo [Muskmelon, fungal diseases]. Phytopathology 1976, 66, 1105–1106. [Google Scholar] [CrossRef]
- Zink, F.W.; Gubler, W.D. 1985. Inheritance of resistance in muskmelon to fusarium wilt. J. Am. Soc. Hortic. Sci. 1985, 110, 600–604. [Google Scholar] [CrossRef]
- Tezuka, T.; Waki, K.; Kuzuya, M.; Ishikawa, T.; Takatsu, Y.; Miyagi, M. Development of new DNA markers linked to the Fusarium wilt resistance locus Fom-1 in melon. Plant Breed. 2011, 130, 261–267. [Google Scholar] [CrossRef]
- Oumouloud, A.; Arnedo-Andres, M.S.; Gonzalez-Torres, R.; Alvarez, J.M. Inheritance of resistance to Fusarium oxysporum f. sp. melonis races 0 and 2 in melon accession Tortuga. Euphytica 2010, 176, 183–189. [Google Scholar] [CrossRef]
- Brotman, Y.; Normantovich, M.; Goldenberg, Z.; Zvirin, Z.; Kovalski, I.; Stovbun, N.; Doniger, T.; Bolger, A.M.; Troadec, C.; Bendahmane, A.; et al. Dual resistance of melon to Fusarium oxysporum races 0 and 2 and to Papaya ring-spot virus is controlled by a pair of head-to-head-oriented NB-LRR genes of unusual architecture. Mol. Plant 2013, 6, 235–238. [Google Scholar] [CrossRef] [PubMed]
- El Otmani, M.; Oumouloud, A.; Álvarez, J.M. Molecular characterization of Fom-1 gene and development of functional markers for molecular breeding of resistance to Fusarium race 2 in melon. Euphytica 2015, 205, 491–501. [Google Scholar] [CrossRef]
- Kim, K.H.; Ahn, S.G.; Hwang, J.H.; Choi, Y.M.; Moon, H.S.; Park, Y.H. Inheritance of resistance to powdery mildew in the watermelon and development of a molecular marker for selecting resistant plants. Hortic. Environ. Biotechnol. 2013, 54, 134–140. [Google Scholar] [CrossRef]
- Han, B.K.; Rhee, S.J.; Jang, Y.J.; Sim, T.Y.; Kim, Y.J.; Park, T.S.; Lee, G.P. Identification of a causal pathogen of watermelon powdery mildew in Korea and development of a genetic linkage marker for resistance in watermelon (Citrullus lanatus). Hortic. Sci. Technol. 2016, 34, 912–923. [Google Scholar] [CrossRef]
- de Souza Gama, R.N.C.; Santos, C.A.F.; de Cassia Souza Dias, R.; de Souza, R.R.C.; de Queiroz, M.A. Microsatellite markers linked to powdery mildew resistance locus in watermelon. Aust. J. Crop Sci. 2015, 9, 92–97. [Google Scholar]
- Ben-Naim, Y.; Cohen, Y. Inheritance of resistance to powdery mildew race 1W in watermelon. Phytopathology 2015, 105, 1446–1457. [Google Scholar] [CrossRef]
- Kim, K.H.; Hwang, J.H.; Han, D.Y.; Park, M.; Kim, S.; Choi, D.; Kim, Y.; Lee, G.P.; Kim, S.T.; Park, Y.H. Major quantitative trait loci and putative candidate genes for powdery mildew resistance and fruit-related traits revealed by an intraspecific genetic map for watermelon (Citrullus lanatus var. lanatus). PLoS ONE 2015, 10, e0145665. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, X.; Liu, S.; Li, X.; Xue, L.; Bai, T.; Xu, B.; Li, G.; Sun, Y.; Zhang, X. Fine mapping of ClLOX, a QTL for powdery mildew resistance in watermelon (Citrullus lanatus L.). Theor. Appl. Genet. 2024, 137, 51. [Google Scholar] [CrossRef]
- Cui, L.; Siskos, L.; Wang, C.; Schouten, H.J.; Visser, R.G.F.; Bai, Y. Breeding melon (Cucumis melo) with resistance to powdery mildew and downy mildew. Hortic. Plant J. 2022, 8, 545–561. [Google Scholar] [CrossRef]
- Cao, Y.; Diao, Q.; Chen, Y.; Jin, H.; Zhang, Y.; Zhang, H. Development of KASP markers and identification of a QTL underlying powdery mildew resistance in melon (Cucumis melo L.) by bulked segregant analysis and RNA-Seq. Front. Plant Sci. 2021, 11, 593207. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Yuan, Y.; Real, N.; Tang, M.; Ren, J.; Wei, J.; Liu, B.; Zhang, X. Fine mapping and identification of candidate genes associated with powdery mildew resistance in melon (Cucumis melo L.). Hortic. Res. 2024, 11, uhae222. [Google Scholar] [CrossRef] [PubMed]
- Rennberger, G.; Gerard, P.; Keinath, A.P. Occurrence of foliar pathogens of watermelon on commercial farms in South Carolina estimated with stratified cluster sampling. Plant Dis. 2018, 102, 2285–2295. [Google Scholar] [CrossRef]
- Huang, C.J.; Lai, Y.R. First report of Stagonosporopsis citrulli causing gummy stem blight of watermelon in Taiwan. J. Plant Pathol. 2019, 101, 417. [Google Scholar] [CrossRef]
- Mao, X.; Wu, Z.; Zhao, F.; Yang, X.; Zhou, M.; Hou, Y. Bioactivity and resistance risk of fluxapyroxad, a novel SDHI fungicide, in Didymella bryoniae. Plant Dis. 2024, 108, 658–665. [Google Scholar] [CrossRef]
- Norton, J. Inheritance of resistance to gummy stem blight [caused by Didymella bryoniae] in watermelon. HortScience 1979, 14, 630–632. [Google Scholar] [CrossRef]
- Gusmini, G.; Rivera-Burgos, L.A.; Wehner, T.C. Inheritance of resistance to gummy stem blight in watermelon. HortScience 2017, 52, 1477–1482. [Google Scholar] [CrossRef]
- Hassan, M.Z.; Rahim, M.A.; Jung, H.J.; Park, J.I.; Kim, H.T.; Nou, I.S. Genome-wide characterization of NBS-encoding genes in watermelon and their potential association with gummy stem blight resistance. Int. J. Mol. Sci. 2019, 20, 902. [Google Scholar] [CrossRef]
- Ren, R.; Xu, J.; Zhang, M.; Liu, G.; Yao, X.; Zhu, L.; Hou, Q. Identification and molecular mapping of a gummy stem blight resistance gene in wild watermelon (Citrullus amarus) Germplasm PI 189225. Plant Dis. 2020, 104, 16–24. [Google Scholar] [CrossRef]
- Gimode, W.; Bao, K.; Fei, Z.; McGregor, C. QTL associated with gummy stem blight resistance in watermelon. Theor. Appl. Genet. 2021, 134, 573–584. [Google Scholar] [CrossRef]
- Zuniga, T.; Jantz, J.; Zitter, T.; Jahn, M. Monogenic dominant resistance to gummy stem blight in two melon (Cucumis melo) accessions. Plant Dis. 1999, 83, 1105–1107. [Google Scholar] [CrossRef]
- Wako, T.; Sakata, Y.; Sugiyama, M.; Ohara, T.; Ishiuchi, D.; Kojima, A. Identification of melon accessions resistant to gummy stem blight and genetic analysis of the resistance using an efficient technique for seedling test. Acta Hortic. 2002, 588, 161–164. [Google Scholar] [CrossRef]
- Frantz, J.; Jahn, M. Five independent loci each control monogenic resistance to gummy stem blight in melon (Cucumis melo L.). Theor. Appl. Genet. 2004, 108, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, C.; Tian, J.; Qiu, Y.; Geng, L.; Wang, J. Identification and Fine Mapping of Gummy Stem Blight Resistance Gene Gsb-7(t) in Melon. Phytopathology 2023, 113, 858–865. [Google Scholar] [CrossRef]
- Yang, J.; Deng, G.; Lian, J.; Garraway, J.; Niu, Y.; Hu, Z.; Yu, J.; Zhang, M. The chromosome-scale genome of melon dissects genetic architecture of important agronomic traits. iScience 2020, 23, 101422. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Deng, G.; Mou, H.; Xu, Y.; Chen, L.; Yang, J.; Zhang, M. A re-sequencing-based ultra-dense genetic map reveals a gummy stem blight resistance-associated gene in Cucumis melo. DNA Res. 2018, 25, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Suvanprakorn, K.; Norton, J.D. Inheritance of resistance to race 2 anthracnose in watermelon. J. Am. Soc. Hortic. Sci. 1980, 105, 197–199. [Google Scholar] [CrossRef]
- Jang, Y.J.; Seo, M.; Hersh, C.P.; Rhee, S.J.; Kim, Y.; Lee, G.P. An evolutionarily conserved non-synonymous SNP in a leucine-rich repeat domain determines anthracnose resistance in watermelon. Theor. Appl. Genet. 2019, 132, 473–488. [Google Scholar] [CrossRef]
- Ling, K.S.; Harris, K.; Meyer, J.D.F.; Levi, A.; Guner, N.; Wehner, T.C.; Bendahmane, A.; Havey, M.J. Non-synonymous single nucleotide polymorphisms in the watermelon eIF4E gene are closely associated with resistance to Zucchini yellow mosaic virus. Theor. Appl. Genet. 2009, 120, 191–200. [Google Scholar] [CrossRef]
- Cai, L.; Liu, J.; Wang, S.; Gong, Z.; Yang, S.; Xu, F.; Hu, Z.; Zhang, M.; Yang, J. The coiled-coil protein gene WPRb confers recessive resistance to Cucumber green mottle mosaic virus. Plant Physiol. 2023, 191, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, X.; Reddy, U.; Sun, H.; Bao, K.; Gao, L.; Mao, L.; Patel, T.; Ortiz, C.; Abburi, V.L.; et al. Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1,365 accessions in the U.S. National Plant Germplasm System watermelon collection. Plant Biotechnol. J. 2019, 17, 2246–2258. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.S.; Wu, S.; Pérez-Escobar, O.A.; Silber, M.V.; Fei, Z.; Chomicki, G. A chromosome-level genome of a Kordofan melon illuminates the origin of domesticated watermelons. Proc. Natl. Acad. Sci. USA 2021, 118, e2101486118. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, S.C.; Zhang, Y.L.; Tan, J.S.; Li, X.P.; Chu, X.; Xu, B.H.; Tian, Y.; Sun, Y.D.; Li, B.S.; et al. A telomere-to-telomere gap-free reference genome of watermelon and its mutation library provide important resources for gene discovery and breeding. Mol. Plant 2022, 15, 1268–1284. [Google Scholar] [CrossRef]
- Li, G.; Tang, L.; He, Y.; Xu, Y.; Bendahmane, A.; Garcia-Mas, J.; Lin, T.; Zhao, G. The haplotype-resolved T2T reference genome highlights structural variation underlying agronomic traits of melon. Hortic. Res. 2023, 10, uhad182. [Google Scholar] [CrossRef]
- Mo, C.; Wang, H.; Wei, M.; Zeng, Q.; Zhang, X.; Feim, Z.; Zhang, Y.; Kong, Q. Complete genome assembly provides a high-quality skeleton for pan-NLRome construction in melon. Plant J. 2024, 118, 2249–2268. [Google Scholar] [CrossRef]
- Chen, X.; Li, H.; Dong, Y.; Xu, Y.; Xu, K.; Zhang, Q.; Yao, Z.; Yu, Q.; Zhang, H.; Zhang, Z. A wild melon reference genome provides novel insights into the domestication of a key gene responsible for melon fruit acidity. Theor. Appl. Genet. 2024, 137, 144. [Google Scholar] [CrossRef]
- Duan, S.; Wang, D.; Kang, Q.; Yan, H.; Cui, J.; Zhang, M.; Liu, D.; Yang, S.; Zhu, Y.; Niu, H.; et al. The development of liquid-phase chip by target sequencing and their application in watermelon molecular breeding. Hortic. Plant J. 2025. [Google Scholar] [CrossRef]
- Yu, Q.; Li, S.; Su, X.; Chen, X.; Dong, Y.; Yao, Z.; Jiang, N.; Chai, S.; Zhang, Z. Melon 2 k array: A versatile 2 k liquid SNP chip for melon genetics and breeding. Hortic. Plant J. 2025, 11, 314–322. [Google Scholar] [CrossRef]
- Pan, W.; Cheng, Z.; Han, Z.; Yang, H.; Zhang, W.; Zhang, H. Efficient genetic transformation and CRISPR/Cas9-mediated genome editing of watermelon assisted by genes encoding developmental regulators. J. Zhejiang Univ. Sci. B 2022, 23, 339–344. [Google Scholar] [CrossRef]
- Feng, Q.; Xiao, L.; He, Y.; Liu, M.; Wang, J.; Tian, S.; Zhang, X.; Yuan, L. Highly efficient, genotype-independent transformation and gene editing in watermelon (Citrullus lanatus) using a chimeric ClGRF4-GIF1 gene. J. Integr. Plant Biol. 2021, 63, 2038–2042. [Google Scholar] [CrossRef]
- Cao, L.; Wei, W.; Shen, J.; Xu, Z.; Li, Z. Study on the optimization of transformation systems in watermelon. Veg. Res. 2022, 2, 12. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, H.; Lu, X.; Anees, M.; He, N.; Yang, D.; Chen, Z.; Hong, Z.; Zhang, J.; Liu, W. Streamlined Agrobacterium rhizogenes-mediated hairy root transformation for efficient CRISPR/Cas9-based gene editing evaluation in diverse Citrullus cultivars. Hortic. Plant J. 2025, 11, 816–826. [Google Scholar] [CrossRef]
- Gu, Y.; Qin, Y.; Hua, S.; Shi, J.; Yang, C.; Peng, Y.; Zhu, L.; Dong, W. Novel methods for genetic transformation of watermelon (Citrullus lanatus) without tissue culture via Agrobacterium rhizogenes. Mol. Breed. 2025, 45, 22. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, N.; Feng, J.; Liu, Y.; Wang, H.; Deng, S.; Dong, W.; Liu, X.; Lv, B.; Sun, J.; et al. Enhancing genetic transformation efficiency in cucurbit crops through AtGRF5 overexpression: Mechanistic insights and applications. J. Integr. Plant Biol. 2025, 67, 1843–1860. [Google Scholar] [CrossRef]
- Li, X.; Cao, C.; Liu, Y.; Bolaños-Villegas, P.; Wang, J.; Zhou, R.; Hou, J.; Li, Q.; Mao, W.; Wang, P.; et al. Enhancing genetic transformation efficiency of melon (Cucumis melo L.) through an extended sucrose-removal co-culture. Plant Cell Rep. 2025, 44, 123. [Google Scholar] [CrossRef]
- Gupta, C.; Salgotra, R.K. Epigenetics and its role in effecting agronomical traits. Front. Plant Sci. 2022, 13, 925688. [Google Scholar] [CrossRef]
- Abdulraheem, M.I.; Xiong, Y.; Moshood, A.Y.; Cadenas-Pliego, G.; Zhang, H.; Hu, J. Mechanisms of plant epigenetic regulation in response to plant stress: Recent discoveries and implications. Plants 2024, 13, 163. [Google Scholar] [CrossRef]
- Chachar, S.; Chachar, M.; Riaz, A.; Shaikh, A.A.; Li, X.; Li, X.; Guan, C.; Zhang, P. Epigenetic modification for horticultural plant improvement comes of age. Sci. Hortic. 2022, 292, 110633. [Google Scholar] [CrossRef]
- Xue, Y.; Cao, X.; Chen, X.; Deng, X.; Deng, X.W.; Ding, Y.; Dong, A.; Duan, C.-G.; Fang, X.; Gong, L.; et al. Epigenetics in the modern era of crop improvements. Sci. China Life Sci. 2025, 68, 1570–1609. [Google Scholar] [CrossRef]
- Zhao, G.; Zou, C.; Li, K.; Wang, K.; Li, T.; Gao, L.; Zhang, X.; Wang, H.; Yang, Z.; Liu, X.; et al. The Aegilops tauschii genome reveals multiple impacts of transposons. Nat. Plants 2017, 3, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, P.; Guo, W.; Liu, H.; Li, X.; Zhang, Q.; Du, Z.; Hu, G.; Han, X.; Pu, L.; et al. A deep learning approach to automate whole-genome prediction of diverse epigenomic modifications in plants. New Phytol. 2021, 232, 880–897. [Google Scholar] [CrossRef]
- Cho, Y.; Kadam, U.; Park, B.; Amariillis, S.; Nguyen, T.K.; Can, T.M.; Lee, K.O.; Park, S.J.; Chung, W.S.; Hong, C. Recent progress in single-cell transcriptomic studies in plants. Plant Biotechnol. Rep. 2025, 19, 91–103. [Google Scholar] [CrossRef]
- Liu, J.; Zhong, X. Population epigenetics: DNA methylation in the plant omics era. Plant Physiol. 2024, 194, 2039–2048. [Google Scholar] [CrossRef]
- Liu, P.; Liu, R.; Xu, Y.; Zhang, C.; Niu, Q.; Lang, Z. DNA cytosine methylation dynamics and functional roles in horticultural crops. Hortic. Res. 2023, 10, uhad170. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, Q.; Zhao, K.; Cao, D.; Cao, Z.; Zhao, K.; Ma, Q.; Zhai, G.; Hu, S.; Li, Z.; et al. Dynamic DNA methylation modification in peanut seed development. iScience 2023, 26, 107062. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Troadec, C.; Boualem, A.; Rajab, M.; Fernandez, R.; Morin, H.; Pitrat, M.; Dogimont, C.; Bendahmane, A. A transposon-induced epigenetic change leads to sex determination in melon. Nature 2009, 461, 1135–1138. [Google Scholar] [CrossRef]
- Wu, T.; Liu, B.; Xiong, T.; Yan, M.; Zhang, J.; Yang, Y.; Hu, G. Mechanisms governing melon fruit skin pigmentation: Insights from transcriptome sequencing and whole-genome bisulfite sequencing analyses. Sci. Hortic. 2024, 333, 113283. [Google Scholar] [CrossRef]
- Zhu, F.; Li, M.; Yan, M.; Qiao, F.; Jiang, X. Integrated transcriptome analysis and single-base resolution methylomes of watermelon (Citrullus lanatus) reveal epigenome modifications in response to osmotic stress. Front. Plant Sci. 2021, 12, 769712. [Google Scholar] [CrossRef]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, E4511–E4519. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 156. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawy, I.; Liang, D.; Xu, K. Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J. Exp. Bot. 2015, 66, 7359–7376. [Google Scholar] [CrossRef] [PubMed]
| Diseases | Races | Resistance Sources | QTL a | Notes |
|---|---|---|---|---|
| FW | Fon 1 | HMw017 | Fo1.1 **, Fo1.2 *, Fo1.3 *, Fo1.4 *, Fo1.5 **, Fo1.6 *, Fo1.7 * | |
| Fon 1 | PI 296341-FR | Fo1.1 ** | derived from PI 296341 | |
| Fon 1 | USVL246-FR2 | Fon1-9 ** | derived from PI 482246 | |
| Fon 2 | PI 296341-FR | fon2.1 *, fon2.2 * | derived from PI 296341 | |
| Fon 2 | USVL252-FR2 | Fon2-1 **, Fon2-2, Fon2-5, Fon2-6, Fon2-8.1, Fon2-8.2, Fon2-11 | derived from PI 482252 | |
| Fon 2 | UGA147 | Fon11 ** | derived from PI 169233 | |
| Fon 2 | USVL246-FR2 | Fon2-2 *, Fon2-5, Fon2-8, Fon2-9 **, Fon2-10 | derived from PI 482246 | |
| Fon 2 | USVL246-FR2 | Fon2-1, Fon2-6, Fon2-8, Fon2-9.1, Fon2-9.2 ** | derived from PI 482246 | |
| PM | 1 W | Arka Manik | pmr2.1 ** | |
| 2 WF | R23 | pm-lox | ||
| GSB | Isolate JS002 | PI 189225 | gsb8.1 ** | |
| Isolate 12178A | PI 482276 | gsb3.1 *, 5.1 *, 7.1 ** | ||
| AR | Race 1 | DrHs7250 | n/a | |
| ZYMV | FL | PI 595203 | zym-FL | |
| CGMMV | n/a | PI 595203 | WPRb |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, H.; Tan, J.; Yan, W.; Liu, D.; Yang, L. Advances in Functional Genomics for Watermelon and Melon Breeding: Current Progress and Future Perspectives. Horticulturae 2025, 11, 1100. https://doi.org/10.3390/horticulturae11091100
Niu H, Tan J, Yan W, Liu D, Yang L. Advances in Functional Genomics for Watermelon and Melon Breeding: Current Progress and Future Perspectives. Horticulturae. 2025; 11(9):1100. https://doi.org/10.3390/horticulturae11091100
Chicago/Turabian StyleNiu, Huanhuan, Junyi Tan, Wenkai Yan, Dongming Liu, and Luming Yang. 2025. "Advances in Functional Genomics for Watermelon and Melon Breeding: Current Progress and Future Perspectives" Horticulturae 11, no. 9: 1100. https://doi.org/10.3390/horticulturae11091100
APA StyleNiu, H., Tan, J., Yan, W., Liu, D., & Yang, L. (2025). Advances in Functional Genomics for Watermelon and Melon Breeding: Current Progress and Future Perspectives. Horticulturae, 11(9), 1100. https://doi.org/10.3390/horticulturae11091100





