WUSCHEL Transcription Factor Regulates Floral Development in ‘Jizaomi’ Grapevine
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Cloning of VvWUS
2.3. Phylogenetic Tree, Multiple Sequence Alignment, and Promoter Cis-Acting Element Analysis
2.4. Expression Profiling of VvWUS
2.5. Subcellular Localization Analysis of VvWUS
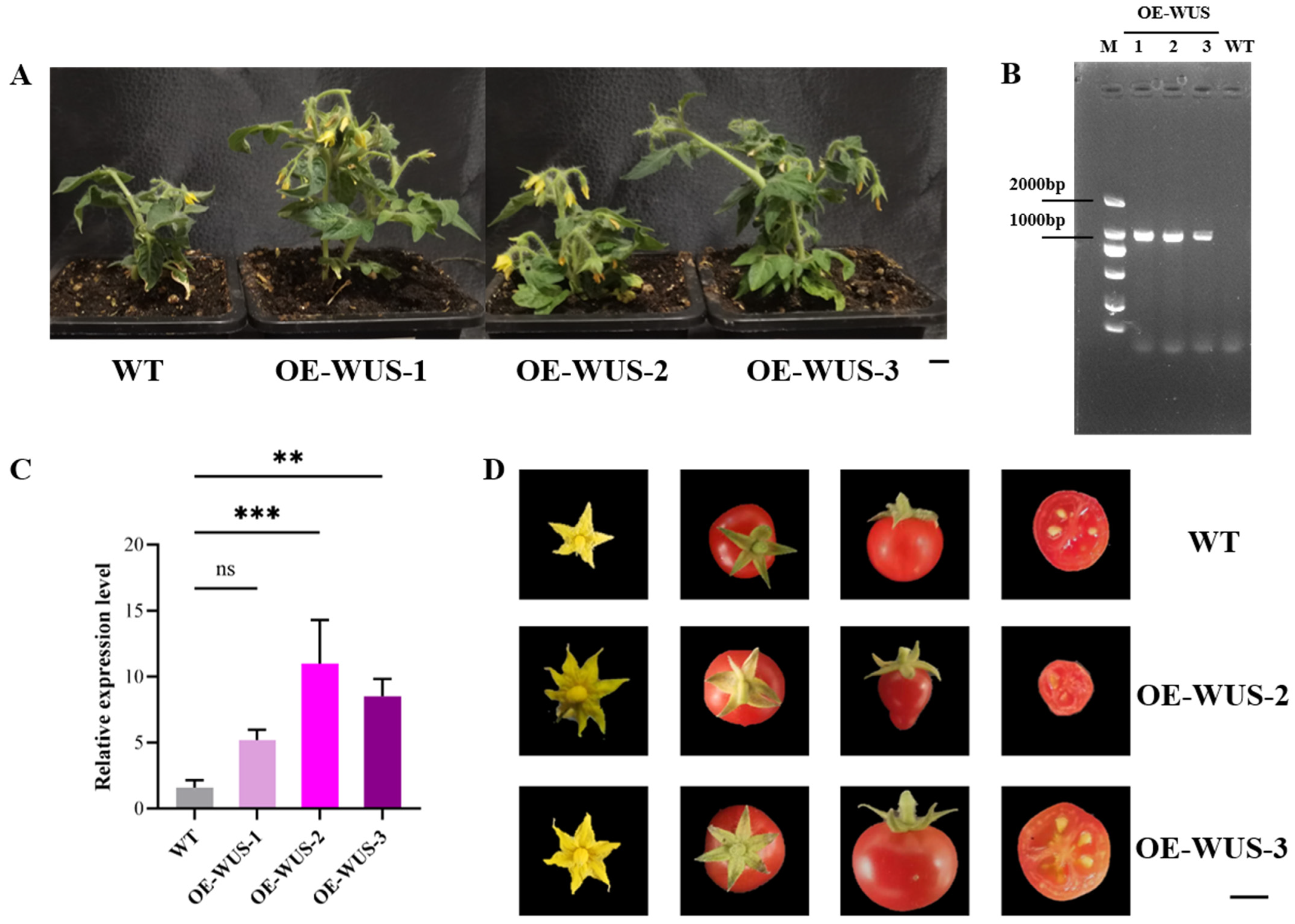
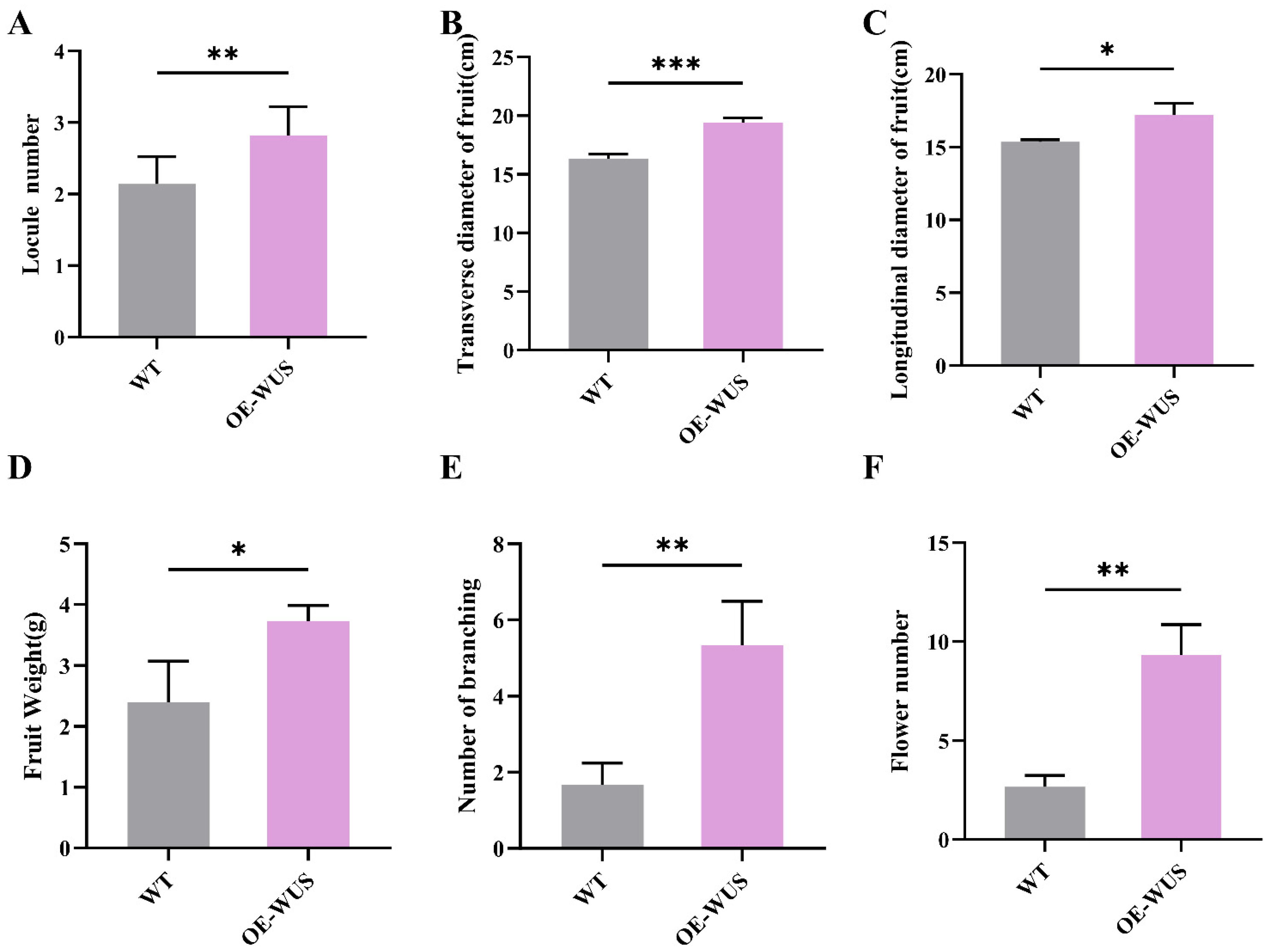
2.6. Agrobacterium-Mediated Tomato Transformation and Phenotypic Analysis of Transgenic Plants
2.7. Yeast One-Hybrid (Y1H) Assay
2.8. Dual-Luciferase (LUC) Assay
2.9. Statistical Analysis
2.10. Primers
3. Results
3.1. VvWUS Expression Profiling Across Vitis vinifera Tissues and Floral Development Stages
3.2. Phylogenetic, Sequence, and cis-Regulatory Element Analyses
3.3. Subcellular Localization of the VvWUS Transcription Factor
3.4. Functional Validation of VvWUS Through Heterologous Overexpression in Solanum lycopersicum
3.5. Molecular Regulation of VvAG by the VvWUS Transcription Factor in Grapevine
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Primer Name | Primer Sequence (5′-3′) | |
|---|---|---|
| VvWUS | F | ATGGAACCTCAACAACAGCTCCA |
| R | TCAGGGGGAATCCGGGGACCT | |
| qVvWUS | F | ATCAGCAGGTGGTGGTCGTG |
| R | AGTGGGAGCGTTTCAATCT | |
| Hyg | F | CGACAGATCCGGTCGGCATCTACTCTATTTCTT |
| R | TCTCGTGCTTTCAGCTTCGATGTAGGAGGG | |
| qVvUBQ | F | GCTCGCTGTTTTGCAGTTCTAC |
| R | AACATAGGTGAGGCCGCACTT | |
| 1300-VvWUS | F | GGGGTACCATGGAACCTCAACAAC |
| R | GCTCTAGAGGGGGAATCCGGG | |
| qSlAG | F | GGTTCAGTATCCGAGGCCAATGC |
| R | AATTGCTGAGGTGGAGGCACAAG | |
| Slactin | F | GCACCCTGTTCTGCTTACTG |
| R | CAAAGCATAACCCTCGTAAATAG | |
| AD-VvWUS | F | CGGAATTCATGGAACCTCAACAACAGCTCCA |
| R | CGGGATCCTCAGGGGGAATCCGG | |
| AbAi-VvAG | F | CCCAAGCTTACCCCATTAAAAATCGCTTTCAG |
| R | GCGTCGACTATTCTCTCTCTAATTAGCTCTGTATTTATGT | |
| 0800-VvAG | F | ACTCACTATAGGGCGAATTGGGTACCGGTTGAATCAGACCAAAGGGAT |
| R | GCGGCCGCTCTAGAACTAGTGGATCCTTTTCTCTGGGGGGAGACCT | |
References
- Fang, J.G.; Xu, W.D. Chinese-Bred Grape Varieties; China Forestry Publishing House: Beijing, China, 2019; pp. 2–5. [Google Scholar]
- Aichinger, E.; Kornet, N.; Friedrich, T.; Laux, T. Plant stem cell niches. Annu. Rev. Plant Biol. 2012, 63, 615–636. [Google Scholar] [CrossRef]
- Ferrandiz, C.; Fourquin, C.; Prunet, N.; Scutt, C.P.; Sundberg, E.; Trehin, C.; Vialette-Guiraud, A.C. Carpel development. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2010; Volume 55, pp. 1–73. [Google Scholar]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.-H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Lee, Z.H.; Hirakawa, T.; Yamaguchi, N.; Ito, T. The roles of plant hormones and their interactions with regulatory genes in determining meristem activity. Int. J. Mol. Sci. 2019, 20, 4065. [Google Scholar] [CrossRef] [PubMed]
- Gallois, J.-L.; Nora, F.R.; Mizukami, Y.; Sablowski, R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 2004, 18, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Ochatt, S.J.; Kumar, V. WUSCHEL: A master regulator in plant growth signaling. Plant Cell Rep. 2020, 39, 431–444. [Google Scholar] [CrossRef]
- Somssich, M.; Je, B.I.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef]
- Thomson, B.; Wellmer, F. Molecular regulation of flower development. Curr. Top. Dev. Biol. 2019, 131, 185–210. [Google Scholar]
- Laux, T.; Mayer, K.F.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Li, H.; Qi, M.; Sun, M.; Liu, Y.; Liu, Y.; Xu, T.; Li, Y.; Li, T. Tomato transcription factor SlWUS plays an important role in tomato flower and locule development. Front. Plant Sci. 2017, 8, 457. [Google Scholar] [CrossRef]
- Chu, Y.H.; Jang, J.C.; Huang, Z.; van der Knaap, E. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct 2019, 3, e00142. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Guan, P.; Liu, Z.; Wang, Y.; Xing, J.; Hu, J. The VvSUPERMAN-like gene is differentially expressed between bicarpellate and tricarpellate florets of Vitis vinifera L. Cv. ‘Xiangfei’ and its heterologous expression reduces carpel number in tomato. Plant Cell Physiol. 2020, 61, 1760–1774. [Google Scholar] [CrossRef]
- Zan, S.W.; Xie, H.H.; Zhang, Y.Q.; Wang, W.J.; Zhang, P.F.; Liang, J.J.; Wen, P.F. VvAGAMOUS Regulates Grape Carpel Development through VvCRABS CLAW. Biotechnol. Bull. 2025, 41, 208–217. [Google Scholar]
- Liang, C.; Yang, B.; Wei, Y.; Zhang, P.; Wen, P. SA incubation induced accumulation of flavan-3-ols through activated VvANR expression in grape leaves. Sci. Hortic. 2021, 287, 110269. [Google Scholar] [CrossRef]
- Atem, J.E.C.; Gan, L.; Yu, W.; Huang, F.; Wang, Y.; Baloch, A.; Nwafor, C.C.; Barrie, A.U.; Chen, P.; Zhang, C. Bioinformatics and functional analysis of EDS1 genes in Brassica napus in response to Plasmodiophora brassicae infection. Plant Sci. 2024, 347, 112175. [Google Scholar] [CrossRef]
- Heng-jiu, L.; Shu-chai, S.; Lü-yi, M.; Zhong, M. Cloning and functional analysis of ChaCBF1, a CBF/DREB1-like transcriptional factor from Corylus heterophylla× C. avellana. J. Beijing For. Univ. 2016, 38, 69–79. [Google Scholar]
- Xing, J.; Lu, L.; Wen, T.; Hu, J. The correlation between VvYABBY5 expression and the ontogeny of tricarpellate fruit in ‘Xiangfei’ grapevine (Vitis vinifera). J. Hortic. Sci. Biotechnol. 2016, 91, 308–315. [Google Scholar] [CrossRef]
- Zhao, W.; Wu, W.; Liu, Y.; Fan, L.; Meng, H.; Wang, L.; Wu, G. The TT2-type MYB transcription factor JrMYB12 positively regulates proanthocyanidin biosynthesis in red walnut. Sci. Hortic. 2023, 307, 111515. [Google Scholar] [CrossRef]
- Wang, M.; Wu, Y.; Zhan, W.; Wang, H.; Chen, M.; Li, T.; Bai, T.; Jiao, J.; Song, C.; Song, S. The apple transcription factor MdZF-HD11 regulates fruit softening by promoting Mdβ-GAL18 expression. J. Exp. Bot. 2024, 75, 819–836. [Google Scholar] [CrossRef]
- Wang, M.m.; Zhu, Q.g.; Deng, C.l.; Luo, Z.r.; Sun, N.j.; Grierson, D.; Yin, X.r.; Chen, K.s. Hypoxia-responsive ERF s involved in postdeastringency softening of persimmon fruit. Plant Biotechnol. J. 2017, 15, 1409–1419. [Google Scholar] [CrossRef]
- Liu, H.; Li, J.; Gong, P.; He, C. The origin and evolution of carpels and fruits from an evo-devo perspective. J. Integr. Plant Biol. 2023, 65, 283–298. [Google Scholar] [CrossRef]
- Song, S.; Huang, B.; Pan, Z.; Zhong, Q.; Yang, Y.; Chen, D.; Zhu, L.; Hu, G.; He, M.; Wu, C. The SlTPL3–SlWUS module regulates multi-locule formation in tomato by modulating auxin and gibberellin levels in the shoot apical meristem. J. Integr. Plant Biol. 2022, 64, 2150–2167. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, X.; Engstrom, E.M.; Nimchuk, Z.L.; Pruneda-Paz, J.L.; Tarr, P.T.; Yan, A.; Kay, S.A.; Meyerowitz, E.M. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 2015, 517, 377–380. [Google Scholar] [CrossRef]
- Cao, X.; He, Z.; Guo, L.; Liu, X. Epigenetic mechanisms are critical for the regulation of WUSCHEL expression in floral meristems. Plant Physiol. 2015, 168, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, L.; Hendelman, A.; Hutton, S.F.; McCandlish, D.M.; Lippman, Z.B. Idiosyncratic and dose-dependent epistasis drives variation in tomato fruit size. Science 2023, 382, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Chen, Z.; Liu, X.; Che, G.; Gu, R.; Zhao, J.; Wang, Z.; Hou, Y.; Zhang, X. CsLFY is required for shoot meristem maintenance via interaction with WUSCHEL in cucumber (Cucumis sativus). New Phytol. 2018, 218, 344–356. [Google Scholar] [CrossRef]
- Wu, H.; Liu, B.; Cao, Y.; Ma, G.; Zheng, X.; Zhu, H.; Sui, S. Genome-Wide Identification of WOX Gene Family in Chimonanthus praecox and a Functional Analysis of CpWUS. Plants 2025, 14, 1144. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Liu, C.Y.; Zhao, X.Q.; Wang, Y.Y.; Yan, M.; Yuan, Z.H. Cloning and Spatiotemporal Expression Analysis of Flower Organ Development-Related Genes PgWUS and PgBEL1 in Pomegranate. Acta Hortic. Sin. 2021, 48, 355–366. [Google Scholar]
- Zhao, R.N.; Zou, R.; Yamamoto, N.; Chen, Z.Y.; Wu, Y.C.; Jiang, J.; Yang, Z.J. Genome-Wide Identification of Wheat WUS Gene Subfamily and Expression Analysis in Stamens and Pistils. J. Agric. Biotechnol. 2024, 32, 2462–2473. [Google Scholar]
- Zhang, Y.D.; Wang, X.; Wang, L.P.; Tong, J.; Wu, Z.H.; Gao, Y.K. Identification, Evolution and Expression Analysis of WOX Transcription Factor Family in Pepper. J. Henan Agric. Sci. 2023, 52, 130–141. [Google Scholar]
- Lenhard, M.; Bohnert, A.; Jürgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef]
- Sun, B.; Looi, L.-S.; Guo, S.; He, Z.; Gan, E.-S.; Huang, J.; Xu, Y.; Wee, W.-Y.; Ito, T. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 2014, 343, 1248559. [Google Scholar] [CrossRef]
- Wu, J.; Liu, Z.; Hu, J.; Guan, P. VvWUS increases carpel number by upregulating VvAG2 expression in grapevine. Plant Cell Rep. 2025, 44, 136. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Xu, H.; Shi, W.; Fu, J.; Wen, P.; Liang, J.; Zhang, P. WUSCHEL Transcription Factor Regulates Floral Development in ‘Jizaomi’ Grapevine. Horticulturae 2025, 11, 1099. https://doi.org/10.3390/horticulturae11091099
Sun Z, Xu H, Shi W, Fu J, Wen P, Liang J, Zhang P. WUSCHEL Transcription Factor Regulates Floral Development in ‘Jizaomi’ Grapevine. Horticulturae. 2025; 11(9):1099. https://doi.org/10.3390/horticulturae11091099
Chicago/Turabian StyleSun, Zedong, Huan Xu, Wenxuan Shi, Jialin Fu, Pengfei Wen, Jinjun Liang, and Pengfei Zhang. 2025. "WUSCHEL Transcription Factor Regulates Floral Development in ‘Jizaomi’ Grapevine" Horticulturae 11, no. 9: 1099. https://doi.org/10.3390/horticulturae11091099
APA StyleSun, Z., Xu, H., Shi, W., Fu, J., Wen, P., Liang, J., & Zhang, P. (2025). WUSCHEL Transcription Factor Regulates Floral Development in ‘Jizaomi’ Grapevine. Horticulturae, 11(9), 1099. https://doi.org/10.3390/horticulturae11091099






