Developing Chinese Sugar Beet Core Collection: Comprehensive Analysis Based on Morphology and Molecular Markers
Abstract
1. Introduction
2. Materials and Methods
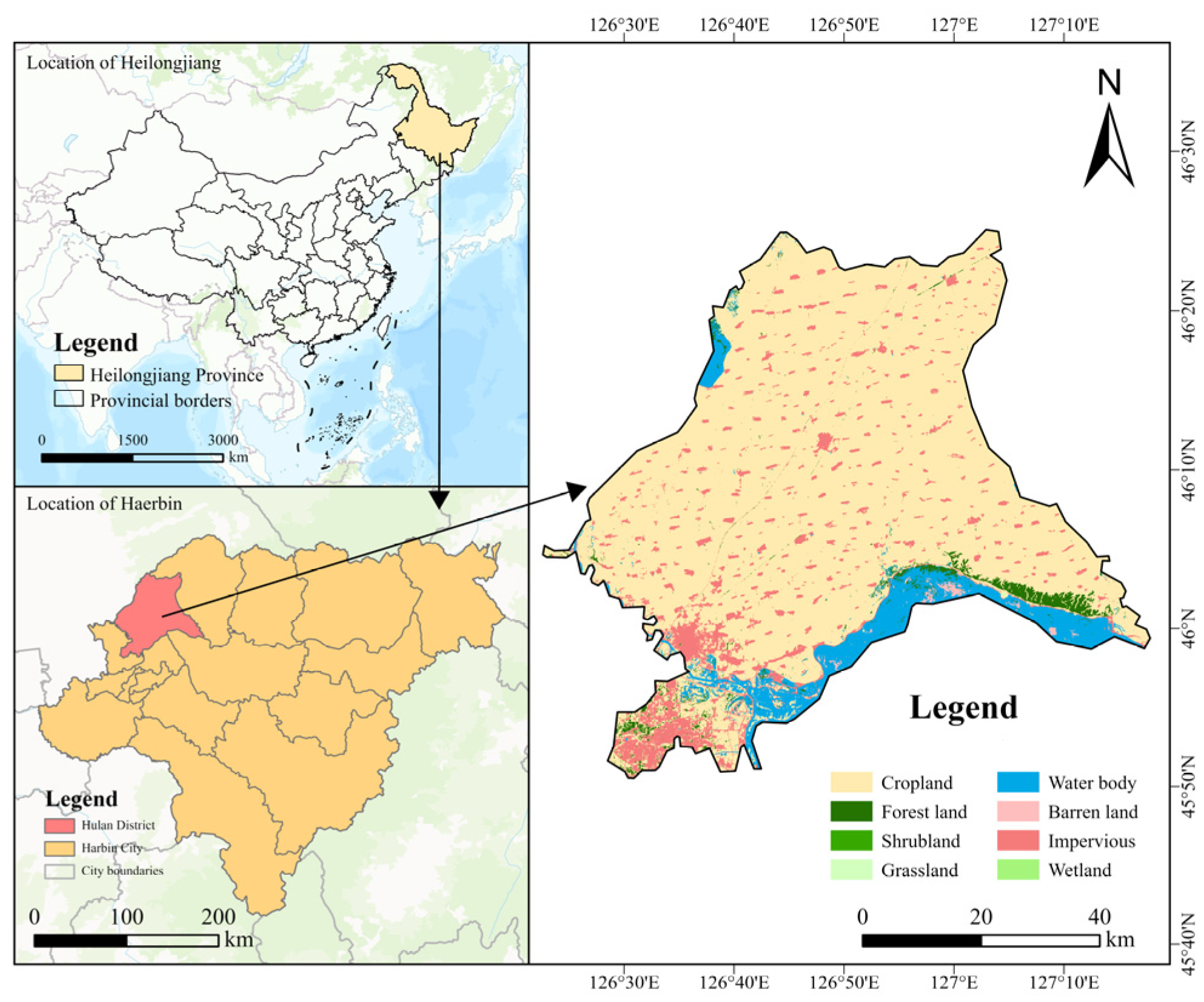
2.1. Plant Materials
2.2. Morphology Data Acquisition
2.3. Molecular Markers Data Acquisition
2.4. Data Handling
3. Results
3.1. Selection of Genetic Distances
3.2. Selection of Sampling Methods
3.3. Selection of Clustering Methods
3.4. Selection of Sampling Proportion
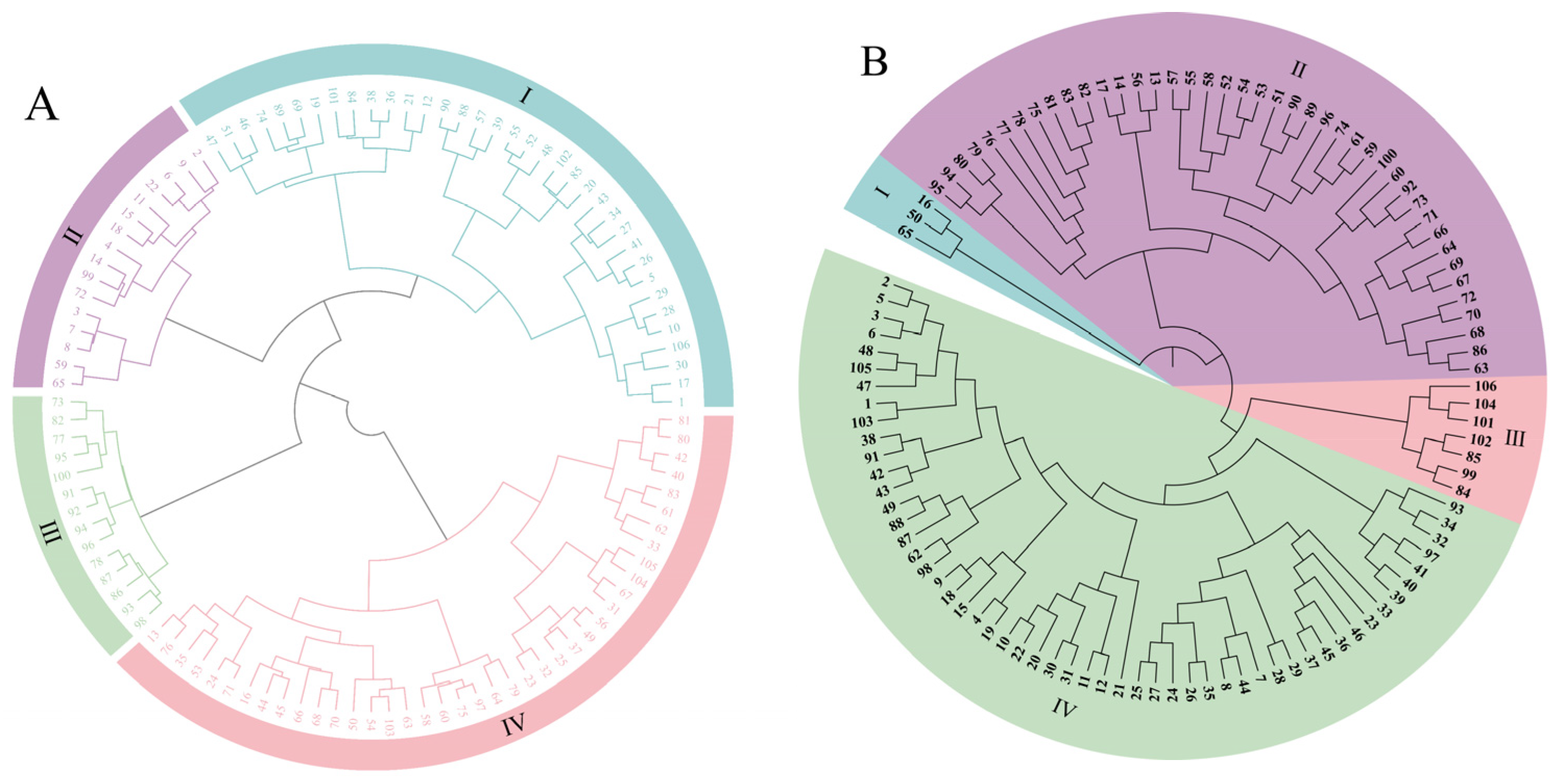
3.5. Clustering Analysis Based on Morphology and Molecular Markers
3.6. Construction of Core Collection
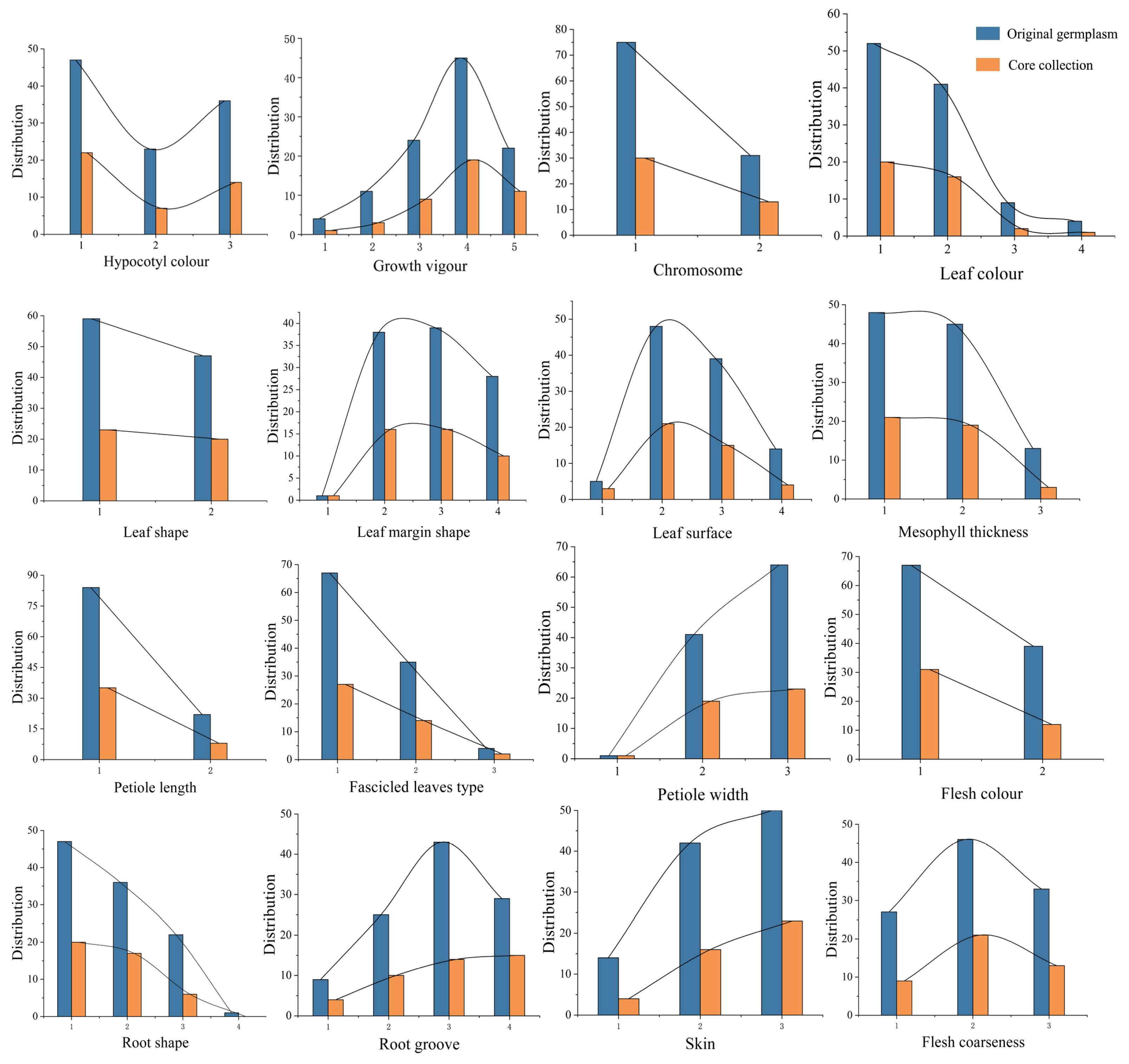
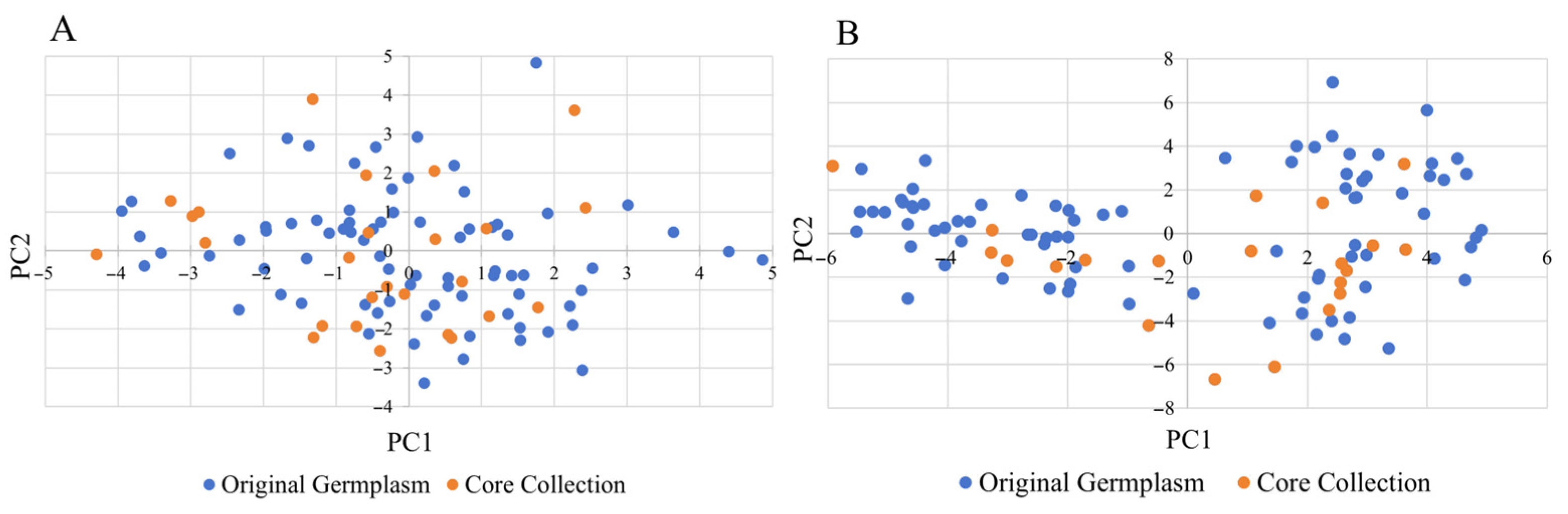
3.7. Representative Evaluation of Core Collection Based on Morphology
3.8. Representative Evaluation of Core Collection Based on SRAP Molecular Markers
3.9. Finalization of the Core Collection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keller, I.; Neuhaus, H. Innovations and threats facing the storage of sugar in sugar beet. Curr. Opin. Plant Biol. 2025, 85, 102721. [Google Scholar] [CrossRef]
- Jain, S.; Kumar, S. A comprehensive review of bioethanol production from diverse feedstocks: Current advancements and economic perspectives. Energy 2024, 296, 131130. [Google Scholar] [CrossRef]
- Pathak, A.; Srivastava, S.; Misra, V.; Mall, A.; Srivastava, S. Evolution and history of sugar beet in the world: An overview. In Sugar Beet Cultivation, Management and Processing; Springer: Singapore, 2022; pp. 3–10. [Google Scholar] [CrossRef]
- Geng, G.; Yang, J. Sugar Beet Production and Industry in China. Sugar Tech 2015, 17, 13–21. [Google Scholar] [CrossRef]
- Gu, R.; Fan, S.; Wei, S.; Li, J.; Zheng, S.; Liu, G. Developments on Core Collections of Plant Genetic Resources: Do We Know Enough? Forests 2023, 14, 926. [Google Scholar] [CrossRef]
- Odong, T.; Jansen, J.; Eeuwijk, F.; Hintum, L. Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation. Theor. Appl. Genet. 2013, 126, 289–305. [Google Scholar] [CrossRef]
- Lyu, X.; Liu, G.; Li, Y.; Ji, Y.; Li, Y.; Li, Y.; Cheng, Y.; Wu, Z.; Zhang, X. Current Status and Prospects of Plant Core Germplasm Research. J. Plant Genet. Resour. 2025, 26, 1693–1707. [Google Scholar]
- Fregene, M.; Suarez, M.; Mkumbira, J.; Kulembeka, H.; Ndedya, E.; Kulaya, A.; Mitchel, S.; Gullberg, U.; Rosling, H.; Dixon, A. Simple sequence repeat marker diversity in cassava landraces: Genetic diversity and differentiation in an asexually propagated crop. Theor. Appl. Genet. 2003, 107, 1083–1093. [Google Scholar] [CrossRef]
- Amirul, I.F.; Beebe, S.; Muñoz, M.; Tohme, J.; Redden, R.; Basford, K. Using molecular markers to assess the effect of introgression on quantitative attributes of common bean in the Andean gene pool. Theor. Appl. Genet. 2004, 108, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Frankel, O.H. Genetic perspectives of germplasm conservation. In Genetic Manipulation: Impact on Man and Society; Arber, W., Limensee, K., Peacock, W.J., Stralinger, P., Eds.; Cambridge University Press: Cambridge, UK, 1984; pp. 161–170. [Google Scholar]
- Brown, A. Core collections: A practical approach to genetic resources management. Genome 1999, 31, 818–824. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Zheng, S.; Cheng, X.; Guo, M.; LI, S.; Wang, M. Construction of a core collection of tomato (Solanum lycopersicum) germplasm based on phenotypic traits and SNP markers. Sci. Hortic. 2025, 339, 113855. [Google Scholar] [CrossRef]
- Guo, L.; Liao, T.; Wang, Y.; Cao, J.; Liu, G. Construction of a DNA fingerprint map and a core collection of Platycladus orientalis. J. Am. Soc. Hortic. Sci. 2024, 149, 142–151. [Google Scholar] [CrossRef]
- Xiong, M.; Wang, Y.; Chen, D.; Wang, X.; Zhou, D.; Wei, Z. Assessment of genetic diversity and identification of core germplasm in single-flowered amaryllis (Hippeastrum hybridum) using SRAP markers. Biotechnol. Biotechnol. Equip. 2020, 34, 966–974. [Google Scholar] [CrossRef]
- Certel, B.; İkten, H.; Yilmaz, Y.; Kantar, F.; Çiftçi, V.; Gözen, V.; Tepe, A. Molecular Characterization of Cold Tolerant Germplasm of Phaseolus Beans with Sequence Related Amplified Polymorphism (Srap) and Retrotransposon-Based Interprimer Binding Sites (Ipbs) Markers. J. Anim. Plant Sci. 2023, 33, 620–632. [Google Scholar] [CrossRef]
- Wang, X.; Cao, L.; Gao, J.; Li, K. Strategy for the construction of a core collection for Pinus yunnanensis Franch. to optimize timber based on combined phenotype and molecular marker data. Genet. Resour. Crop Evol. 2021, 68, 3219–3240. [Google Scholar] [CrossRef]
- Zhai, N.; Tang, J.; Zhou, J.; Zhou, C.; Wang, J.; Yun, Y.; Han, S.; Wang, Y.; Yan, W.; Xing, N. Genetic Diversity Analysis and Core Germplasm Construction of Oryza rufipogon Griff. in Hainan. J. Plant Genet. Resour. 2024, 25, 1624–1636. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Hu, G.; Liu, Y.; Li, H.; Zhang, F.; Li, Y.; Wang, Y. Construction and Utilization of Applied Core Collection in Maize. J. Plant Genet. Resour. 2023, 24, 911–916. [Google Scholar] [CrossRef]
- Yan, T.; Zhang, W.; Zheng, J.; Guo, J.; Li, X.; Qiao, Y.; Chen, F.; Chang, F.; Zhang, J. Evaluation of Adult Stage Resistance of 57 Chinese Wheat Mini-Core Collections to Wheat Stripe Rust and Leaf Rust. J. Northeast. Agric. Sci. 2023, 48, 30–34. [Google Scholar] [CrossRef]
- Barański, R.; Grzebelus, D.; Frese, L. Estimation of genetic diversity in a collection of the Garden Beet Group. Euphytica 2001, 122, 19–29. [Google Scholar] [CrossRef]
- Abramoff, M.; Magalhães, P.; Ram, S. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Reeves, P.; Panella, L.; Richards, C. Retention of agronomically important variation in germplasm core collections: Implications for allele mining. Theor. Appl. Genet. 2012, 124, 1155–1171. [Google Scholar] [CrossRef]
- Cui, P. Descriptors and Date Standard for Beet (Beta vulgaris L.); China Agriculture Press: Beijing, China, 2006; pp. 8–21. [Google Scholar]
- Li, J.; Wang, S.; Yu, J.; Wang, L.; Zhou, S. A Modified CTAB Protocol for Plant DNA Extraction. Chin. Bull. Bot. 2013, 48, 72–78. [Google Scholar] [CrossRef]
- Chen, G.; Zhu, X.; Zhang, F.; Zhu, J. Quantitative genetic analysis station for the genetic analysis of complex traits. Chin. Sci. Bull. 2012, 57, 2721–2726. [Google Scholar] [CrossRef]
- Karen, B.; Nancy, L.; Gene, G.; George, M. IBM SPSS for Introductory Statistics. Psychol. Methods Stat. 2019, 15, 266. [Google Scholar] [CrossRef]
- Yeh, C.; Yang, C.; Boyle, T. POPGENE, version 1.3.1; Microsoft Window-Bases Freeware for Population Genetic Analysis; University of Alberta and the Centre for International Forestry Research: Edmonton, AB, Canada, 1999. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 30223027. [Google Scholar] [CrossRef]
- Kathy, M. Origin Update. Science 2000, 288, 1982. [Google Scholar] [CrossRef]
- Mondal, R.; Kumar, A.; Gnanesh, B.N. Crop germplasm: Current challenges, physiological-molecular perspective, and advance strategies towards development of climate-resilient crops. Heliyon 2023, 9, e12973. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, H.; Gowda, L.; Sastry, D. Plant genetic resources management: Collection, characterization, conservation and utilization. J. SAT Agric. Res. 2008, 6, 16. [Google Scholar]
- Zhu, H.; Song, P.; Gu, D.; Guo, Q.; Li, Y.; Sun, S.; Weng, Y.; Yang, M. Genome wide characterization of simple sequence repeats in watermelon genome and their application in comparative mapping and genetic diversity analysis. BMC Genom. 2016, 17, 557. [Google Scholar] [CrossRef]
- Cobb, N.; Clerck, G.; Greenberg, A.; Randy, C.; Susan, C. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, S.; Singh, K.B.; Prasad, M.; Thakur, J.K. Designing a Mini-Core Collection Effectively Representing 3004 Diverse Rice Accessions. Plant Commun. 2020, 1, 100049. [Google Scholar] [CrossRef]
- Han, P.; Tian, X.; Wang, Y.; Huang, C.; Ma, Y.; Zhou, X.; Yu, Y.; Zhang, D.; Xu, H.; Cao, Y.; et al. Construction of a core germplasm bank of upland cotton (Gossypium hirsutum L.) based on phenotype, genotype and favorable alleles. Genet. Resour. Crop Evol. 2022, 69, 2309–2411. [Google Scholar] [CrossRef]
- Katinas, L.; Crisci, J. Agriculture Biogeography: An emerging discipline in search of a conceptual framework. Prog. Phys. Geogr. Earth Environ. 2018, 42, 513–529. [Google Scholar] [CrossRef]
- Mahmoodi, R.; Dadpour, M.R.; Hassani, D.; Zeinalabedini, M.; Vendramin, E.; Micali, S.; Nahandi, F.Z. Development of a core collection in Iranian walnut (Juglans regia L.) germplasm using the phenotypic diversity. Sci. Hortic. 2019, 249, 439–448. [Google Scholar] [CrossRef]
- Wang, C.; Hu, J.; Xu, M.; Zhang, S. A strategy on constructing core collections by least distance stepwise sampling. Theor. Appl. Genet. 2007, 115, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Mank, J. The role of sex chromosomes in sexual dimorphism: Discordance between molecular and phenotypic data. J. Evol. Biol. 2015, 7, 1443–1453. [Google Scholar] [CrossRef]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2015, 36, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Aneja, B.; Yadav, N.; Chawla, V.; Yadav, C. Sequence-related amplified polymorphism (SRAP) molecular marker system and its applications in crop improvement. Mol. Breed. 2012, 30, 1635–1648. [Google Scholar] [CrossRef]
| Traits | Quantified Value | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Hypocotyl color | Red | Green | Mix | - | - |
| Growth vigor | Very weak | Weak | Medium | Vigorous | Very vigorous |
| Chromosome | Diploid | Tetraploid | - | - | - |
| Leaf color | Light green | Green | Dark green | Yellow green | - |
| Leaf shape | Share | Tongue | - | - | - |
| Leaf margin shape | Full margin | Small wave | Medium wave | Big wave | - |
| Leaf surface | Smooth | Wavy | Slight crease | More creases | - |
| Mesophyll thickness | Thin | Medium | Thick | - | - |
| Petiole width | Narrow | Medium | Wide | - | - |
| Petiole length | Short | Medium | - | - | - |
| Fascicled leaves type | Erect | Semicrawl | Crawl | - | - |
| Root shape | Cuneiform | Conical | Spindle | Regular | - |
| Root groove | None | Not obvious | Shallow | Deep | - |
| Skin | Very smooth | Smoother | Very rough | - | - |
| Flesh color | White | Light yellow | - | - | - |
| Flesh coarseness | Fine | Medium | Crude | - | - |
| Genetic Distance | Clustering Methods | Random Sampling Method | Deviation Sampling Method | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MD% | VD% | CR% | VR% | MD% | VD% | CR% | VR% | ||
| Euclidean distance | SL | 0 | 10.34 | 94.19 | 56.14 | 3.45 | 20.69 | 95.33 | 57.80 |
| CL | 0 | 6.90 | 91.10 | 57.40 | 0 | 13.79 | 93.80 | 58.03 | |
| MM | 0 | 3.45 | 91.62 | 56.47 | 0 | 13.45 | 91.62 | 56.47 | |
| CM | 3.45 | 6.90 | 94.22 | 55.58 | 0 | 10.34 | 95.27 | 57.28 | |
| UPA | 0 | 6.70 | 88.90 | 57.13 | 0 | 17.24 | 95.28 | 59.03 | |
| WPA | 0 | 3.45 | 89.64 | 56.38 | 0 | 17.24 | 93.80 | 58.16 | |
| FM | 0 | 6.90 | 93.40 | 55.64 | 0 | 13.79 | 93.63 | 58.43 | |
| WM | 0 | 6.90 | 91.91 | 55.39 | 0 | 17.24 | 93.66 | 58.24 | |
| Mahalanobis distance | SL | 3.45 | 13.79 | 93.75 | 56.11 | 3.45 | 13.79 | 96.39 | 58.53 |
| CL | 0 | 6.90 | 90.90 | 56.50 | 3.45 | 13.79 | 95.32 | 57.23 | |
| MM | 0 | 6.90 | 90.30 | 55.53 | 0 | 10.34 | 96.23 | 58.79 | |
| CM | 3.45 | 3.45 | 95.12 | 57.90 | 3.45 | 17.24 | 96.96 | 58.40 | |
| UPA | 0 | 3.45 | 89.53 | 56.09 | 6.90 | 17.24 | 92.72 | 55.81 | |
| WPA | 0 | 3.45 | 92.28 | 56.79 | 0 | 20.69 | 95.05 | 57.94 | |
| FM | 0 | 3.44 | 90.32 | 56.51 | 0 | 20.69 | 94.76 | 57.59 | |
| WM | 0 | 3.45 | 84.04 | 52.58 | 3.45 | 20.69 | 95.33 | 57.80 | |
| Genetic Distance | Clustering Methods | Random Sampling Method | Deviation Sampling Method | ||||||
|---|---|---|---|---|---|---|---|---|---|
| VP | He | H | A | VP | He | H | A | ||
| Nei and Li | SL | 0.93 | 0.55 | 0.37 | 3.02 | 0.93 | 0.55 | 0.37 | 3.02 |
| CL | 0.93 | 0.57 | 0.39 | 2.71 | 0.93 | 0.57 | 0.39 | 2.72 | |
| MM | 0.92 | 0.55 | 0.37 | 3.10 | 0.93 | 0.55 | 0.37 | 3.10 | |
| CM | 0.91 | 0.55 | 0.37 | 2.98 | 0.92 | 0.55 | 0.37 | 3.00 | |
| UPA | 0.93 | 0.57 | 0.38 | 2.83 | 0.92 | 0.58 | 0.40 | 2.72 | |
| WPA | 0.91 | 0.56 | 0.38 | 2.89 | 0.94 | 0.56 | 0.38 | 2.90 | |
| FM | 0.91 | 0.57 | 0.39 | 2.86 | 0.93 | 0.58 | 0.39 | 2.89 | |
| WM | 0.92 | 0.61 | 0.42 | 2.56 | 0.93 | 0.58 | 0.39 | 2.87 | |
| Jaccard | SL | 0.94 | 0.55 | 0.37 | 3.02 | 0.93 | 0.55 | 0.37 | 3.02 |
| CL | 0.94 | 0.57 | 0.39 | 2.71 | 0.92 | 0.56 | 0.38 | 2.71 | |
| MM | 0.94 | 0.55 | 0.36 | 3.06 | 0.92 | 0.54 | 0.36 | 3.06 | |
| CM | 0.93 | 0.54 | 0.36 | 3.06 | 0.92 | 0.53 | 0.35 | 3.30 | |
| UPA | 0.94 | 0.58 | 0.40 | 2.87 | 0.93 | 0.58 | 0.39 | 2.87 | |
| WPA | 0.94 | 0.56 | 0.38 | 2.89 | 0.92 | 0.56 | 0.38 | 2.89 | |
| FM | 0.93 | 0.58 | 0.39 | 2.87 | 0.92 | 0.58 | 0.39 | 2.87 | |
| WM | 0.93 | 0.57 | 0.37 | 2.86 | 0.92 | 0.56 | 0.36 | 2.86 | |
| Simple matching | SL | 0.93 | 0.54 | 0.43 | 2.40 | 0.94 | 0.54 | 0.42 | 2.40 |
| CL | 0.93 | 0.53 | 0.42 | 2.52 | 0.94 | 0.53 | 0.42 | 2.52 | |
| MM | 0.92 | 0.54 | 0.43 | 2.40 | 0.93 | 0.54 | 0.42 | 2.37 | |
| CM | 0.92 | 0.55 | 0.44 | 2.34 | 0.94 | 0.55 | 0.44 | 2.35 | |
| UPA | 0.93 | 0.55 | 0.43 | 2.38 | 0.92 | 0.54 | 0.43 | 2.36 | |
| WPA | 0.92 | 0.55 | 0.42 | 2.11 | 0.92 | 0.55 | 0.42 | 2.37 | |
| FM | 0.92 | 0.53 | 0.42 | 2.47 | 0.92 | 0.53 | 0.42 | 2.43 | |
| WM | 0.91 | 0.54 | 0.42 | 2.56 | 0.92 | 0.55 | 0.42 | 2.53 | |
| Sampling Proportion% | MD% | VD% | CR% | VR% |
|---|---|---|---|---|
| 5.00 | 0 | 20.69 | 77.99 | 56.93 |
| 10.00 | 0 | 24.13 | 88.70 | 58.12 |
| 15.00 | 0 | 13.79 | 90.30 | 58.56 |
| 20.00 | 3.45 | 17.24 | 92.98 | 58.30 |
| 25.00 | 0 | 10.35 | 94.28 | 59.03 |
| 30.00 | 0 | 10.35 | 94.28 | 57.51 |
| Sampling Proportion% | VP | He | H | A |
|---|---|---|---|---|
| 5.00 | 0.91 | 0.46 | 0.31 | 2.19 |
| 10.00 | 0.93 | 0.54 | 0.36 | 2.81 |
| 15.00 | 0.93 | 0.56 | 0.38 | 2.83 |
| 20.00 | 0.94 | 0.57 | 0.39 | 2.84 |
| 25.00 | 0.94 | 0.58 | 0.40 | 2.72 |
| 30.00 | 0.94 | 0.59 | 0.41 | 2.67 |
| Category | Group | Sample Size | Extraction Proportion | Extraction Count |
|---|---|---|---|---|
| Morphology | I | 36 | 22.22% | 8 |
| II | 16 | 25% | 4 | |
| III | 14 | 21.43% | 3 | |
| IV | 40 | 30% | 12 | |
| Molecular Markers | I | 3 | 33.33% | 1 |
| II | 42 | 11.90% | 5 | |
| III | 7 | 57.14% | 4 | |
| IV | 54 | 20.37% | 11 | |
| Total | - | 106 | 40.57% | 43 |
| Traits | Mean | Variance | Range | CV% | H’ | t-Test | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Original | Core | Original | Core | Original | Core | Original | Core | Original | Core | ||
| SW | 356.2 | 381.1 | 1.4 | 1.9 | 635.8 | 553.8 | 33.0 | 36.0 | 1.9 | 1.9 | NS |
| HY | 1.9 | 1.8 | 0.8 | 0.8 | 2.0 | 2.0 | 46.6 | 50.0 | 1.1 | 1.0 | NS |
| GV | 3.7 | 3.8 | 1.1 | 0.8 | 4.0 | 3.0 | 28.4 | 23.0 | 1.4 | 1.2 | NS |
| CH | 1.3 | 1.2 | 0.2 | 0.2 | 1.0 | 1.0 | 35.4 | 33.4 | 0.6 | 0.5 | NS |
| LN | 17.6 | 17.0 | 6.3 | 8.6 | 13.0 | 13.0 | 14.3 | 17.2 | 2.0 | 1.9 | NS |
| LC | 1.7 | 1.6 | 0.6 | 0.3 | 3.0 | 2.0 | 47.3 | 35.9 | 1.0 | 0.8 | * |
| LS | 1.4 | 1.5 | 0.3 | 0.3 | 1.0 | 1.0 | 34.6 | 34.4 | 0.7 | 0.7 | NS |
| LM | 2.9 | 2.8 | 0.7 | 0.8 | 3.0 | 3.0 | 28.0 | 31.2 | 1.1 | 1.2 | NS |
| LS | 2.2 | 2.3 | 1.3 | 1.3 | 3.0 | 3.0 | 52.9 | 49.5 | 1.1 | 1.3 | NS |
| LT | 1.7 | 1.5 | 0.5 | 0.3 | 2.0 | 2.0 | 41.1 | 39.1 | 1.0 | 0.8 | NS |
| PW | 2.6 | 2.5 | 0.3 | 0.3 | 2.0 | 2.0 | 19.8 | 23.4 | 0.7 | 0.8 | NS |
| PL | 1.2 | 1.3 | 0.2 | 0.2 | 1.0 | 1.0 | 33.7 | 35.5 | 0.5 | 0.6 | NS |
| FL | 1.4 | 1.3 | 0.3 | 0.4 | 2.0 | 2.0 | 40.2 | 46.5 | 0.8 | 0.7 | NS |
| RW | 8.4 | 8.2 | 2.0 | 3.4 | 8.1 | 8.1 | 16.9 | 22.5 | 1.8 | 1.9 | * |
| RL | 23.4 | 23.8 | 7.6 | 8.8 | 13.4 | 11.4 | 11.7 | 12.5 | 2.0 | 1.9 | NS |
| AR | 2.8 | 3.0 | 0.1 | 0.2 | 1.8 | 1.8 | 12.1 | 13.1 | 2.0 | 1.8 | NS |
| VB | 6.1 | 6.2 | 0.4 | 0.5 | 2.9 | 2.9 | 10.0 | 12.0 | 2.0 | 2.1 | NS |
| RY | 36,698 | 37,251 | 8.0 | 9.4 | 50,294 | 50,294 | 24.4 | 26.1 | 2.0 | 1.9 | NS |
| SY | 5605 | 5653 | 2.5 | 2.9 | 7535 | 7159 | 26.2 | 29.9 | 2.0 | 1.8 | NS |
| BR | 20.1 | 19.9 | 5.7 | 9.7 | 12.9 | 12.9 | 11.9 | 15.6 | 2.0 | 2.1 | * |
| SC | 15.7 | 16.5 | 13.3 | 19.8 | 37.6 | 37.6 | 23.3 | 38.2 | 1.6 | 1.7 | NS |
| K+ | 4.3 | 4.3 | 0.6 | 0.6 | 5.2 | 4.3 | 17.9 | 17.7 | 1.7 | 1.5 | NS |
| Na+ | 3.4 | 3.6 | 2.3 | 3.4 | 7.6 | 7.6 | 44.6 | 51.2 | 1.9 | 1.8 | NS |
| α-N | 5.0 | 5.1 | 1.5 | 1.8 | 5.9 | 5.9 | 24.1 | 26.4 | 2.0 | 2.0 | NS |
| RS | 1.8 | 1.7 | 0.7 | 0.5 | 3.0 | 2.0 | 45.1 | 44.0 | 1.1 | 0.5 | NS |
| RG | 2.9 | 2.9 | 0.8 | 1.2 | 3.0 | 3.0 | 32.0 | 38.5 | 1.3 | 1.9 | NS |
| SK | 2.3 | 2.4 | 0.5 | 0.6 | 2.0 | 2.0 | 30.0 | 31.3 | 1.0 | 0.8 | NS |
| FC | 1.4 | 1.3 | 0.2 | 0.2 | 1.0 | 1.0 | 35.4 | 35.9 | 0.7 | 0.7 | NS |
| FL | 2.1 | 2.2 | 0.6 | 0.5 | 2.0 | 2.0 | 36.7 | 33.7 | 1.1 | 1.2 | NS |
| Index | Original Germplasm | Core Collection |
|---|---|---|
| Na | 1.7492 | 1.7617 |
| Ne | 1.4256 | 1.4236 |
| H | 0.2614 | 0.2551 |
| I | 0.3772 | 0.3875 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Song, Y.; Li, S.; Pi, Z.; Wu, Z. Developing Chinese Sugar Beet Core Collection: Comprehensive Analysis Based on Morphology and Molecular Markers. Horticulturae 2025, 11, 990. https://doi.org/10.3390/horticulturae11080990
Li J, Song Y, Li S, Pi Z, Wu Z. Developing Chinese Sugar Beet Core Collection: Comprehensive Analysis Based on Morphology and Molecular Markers. Horticulturae. 2025; 11(8):990. https://doi.org/10.3390/horticulturae11080990
Chicago/Turabian StyleLi, Jinghao, Yue Song, Shengnan Li, Zhi Pi, and Zedong Wu. 2025. "Developing Chinese Sugar Beet Core Collection: Comprehensive Analysis Based on Morphology and Molecular Markers" Horticulturae 11, no. 8: 990. https://doi.org/10.3390/horticulturae11080990
APA StyleLi, J., Song, Y., Li, S., Pi, Z., & Wu, Z. (2025). Developing Chinese Sugar Beet Core Collection: Comprehensive Analysis Based on Morphology and Molecular Markers. Horticulturae, 11(8), 990. https://doi.org/10.3390/horticulturae11080990






