Abstract
Watermelon (Citrullus lanatus), a vital economic crop, is severely threatened by Fusarium wilt (FW), which is caused by the soil-borne fungal pathogen Fusarium oxysporum f. sp. niveum (Fon). To elucidate the molecular mechanisms underlying FW resistance in watermelon, we tracked the infection process via microscopy, identifying three critical time points (1, 6, and 8 days post-inoculation) corresponding to spore germination, hyphal invasion of the xylem vascular system, and symptom onset, respectively. Transcriptional profiling at these stages revealed six disease-resistance-associated gene modules through differential expression analysis, expression pattern clustering, weighted gene co-expression network analysis, and functional enrichment. These modules exhibited strong correlations with distinct infection phases. Protein–protein interaction networks identified 35 hub genes, including receptor-like kinases; WRKY and ethylene-responsive factor transcription factors; and genes involved in cell wall reinforcement, hormone signaling, defense metabolism/detoxification, programmed cell death regulation, and antimicrobial compound biosynthesis. Differential expressions of these genes across infection stages likely underpin the observed phenotypic disparities. Five hub regulatory genes were identified by quantitative real-time PCR in the SRgreen and SRblack modules, namely, Cla97C01G014990 (WRKY transcription factor 42), Cla97C02G042360 (calcium-transporting ATPase), Cla97C08G155710 (AIG2), Cla97C09G170380 (ethylene-responsive factor 1B-like), and Cla97C06G121810 (receptor kinase, putative). These genes mediate early rapid defense responses via SRgreen and sustain long-term resistance through SRblack. By validating the expression patterns of hub genes, the study elucidated the watermelon resistance response and provided insights into transcriptional regulation during different stages of Fon–watermelon interactions. Additionally, it identified candidate genes that could enhance watermelon resistance to wilt disease.
1. Introduction
Watermelon (Citrullus lanatus) is a globally important cucurbit crop, with China dominating its production and consumption, generating more than 60 million metric tons annually [1]. A critical constraint to watermelon cultivation is Fusarium wilt (FW), caused by the soil-borne pathogen Fusarium oxysporum f. sp. niveum (Fon), which reduces yields by 30–80% through vascular system colonization and water transport disruption [2]. Fon persists as chlamydospores for more than a decade in soil, germinating upon detecting host root exudates to develop infection hyphae that penetrate root tissues [3]. Comparative studies using confocal laser scanning microscopy revealed that both resistant and susceptible watermelon varieties permit initial root colonization by Fon, but defense mechanisms in resistant varieties restrict pathogen progression beyond cortical layers, preventing vascular invasion [4,5]. In susceptible plants, xylem occlusion by fungal hyphae, gums, and tyloses triggers hydraulic failure and wilting, underscoring the critical role of transcriptional reprogramming in vascular defense.
Recent genetic mapping efforts have identified key quantitative trait loci (QTLs) underlying Fon resistance in watermelon. Fon-1, a major QTL, was mapped to chromosome 1 in a recombinant inbred line (RIL) population derived from the cultivated variety 97103 and wild accession PI296341-FR, and this QTL explained 48.1% of the phenotypic variance (LOD = 13.2). This locus contains candidate genes, including a receptor kinase, glucan endo-1,3-β-glucosidase precursors, and three acidic chitinases, suggesting roles in pathogen recognition and cell wall remodeling [6]. A QTL mapping analysis was performed using F2:3 and RIL populations derived from the Fon race 1-resistant donor Citrullus amarus USVL246-FR2 and susceptible parent USVL114. This study identified a major QTL (qFon1-9) on chromosome 9 of USVL246-FR2 that confers resistance to Fon race 1 and represents an uncharacterized genetic resource for enhancing FW resistance in watermelon. Notably, seven receptor-like kinase (RLK) genes mapped within the 1.5-LOD confidence interval of qFon1-9 emerged as strong candidates for broad-spectrum resistance. These RLKs might function as pattern recognition receptors to perceive conserved pathogen-associated molecular patterns (PAMPs), thereby activating basal defense responses independent of pathogen race specialization [7].
Transcription factors (TFs) are essential DNA-binding proteins that regulate gene expression by modulating transcriptional dynamics through interactions with cis-regulatory elements in the promoters of stress-responsive genes. These proteins act as central regulators of plant defenses by controlling the expression of stress-induced genes critical for activating host defense mechanisms [8]. Among plant TF families, the WRKY superfamily is a prominent regulator of immunity. Its members are characterized by a conserved WRKY domain that mediates DNA recognition, and they are widely distributed across plant species [9]. WRKY TFs govern diverse immune responses, functioning as both activators and repressors of defense pathways. They are integral to the regulation of PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI). In cotton, group IIc WRKY TFs enhanced resistance to F. oxysporum f. sp. vasinfectum by activating the GhMKK2–GhNTF6 signaling cascade, which increases the GhMYC2-mediated expression of flavonoid biosynthesis genes. This pathway increased the levels of antimicrobial secondary metabolites, thereby restricting pathogen colonization [10]. PdpapWRKY28 overexpression significantly suppressed reactive oxygen species (ROS) accumulation in Populus davidiana and enhanced antioxidant enzyme activity, thereby conferring robust tolerance to environmental stresses [11]. In addition to WRKY TFs, APETALA2/ethylene-responsive factor (AP2/ERF) TFs also play central roles in regulating plant responses to diverse biotic and abiotic stresses, including salinity, drought, temperature extremes, and pathogenic infections [12]. AP2/ERF family members modulate defense responses through intricate molecular mechanisms, such as the transcriptional activation of stress-responsive genes and crosstalk with phytohormone signaling pathways. MPK-mediated phosphorylation of Glycine max ERF113 (GmERF113) enhances its transcriptional activity, thereby increasing the expression of defense-related genes and augmenting immunity against the oomycete pathogen Phytophthora sojae [13]. In Gossypium hirsutum, GbERF1 overexpression significantly increased lignin biosynthesis gene transcription, resulting in increased lignin deposition and enhanced resistance to the wilt pathogen Verticillium dahliae [14]. The Arabidopsis ERF protein ORA59 directly binds to the PDF1.2 promoter to activate jasmonic acid/ethylene (JA/ET)-dependent defense responses, thereby improving resistance to the necrotrophic fungus Botrytis cinerea [15].
Weighted gene co-expression network analysis (WGCNA), a method for identifying trait-related modules [16], has been widely applied to dissect hub genes associated with disease resistance [17,18,19]. In this study, we employed WGCNA to characterize genome-wide transcriptomic responses in two watermelon varieties, namely, the FW-resistant variety PI 296341 (R) and FW-susceptible variety W1-1 (S), following infection with Fon. This study performed a comprehensive analysis of compatible (susceptible interaction) and incompatible (resistant interaction) molecular mechanisms between Fon and watermelon, thereby generating genome-wide expression profiles of disease resistance-related genes. By integrating co-expression networks and pathogen challenge dynamics, our work provides a systems-level framework for unraveling genetic determinants of FW resistance.
Expression pattern analysis of selected hub genes was conducted to elucidate their potential roles in disease resistance mechanisms. These findings provide critical molecular insights into plant–pathogen interactions, establishing a robust framework for further exploration of Fusarium oxysporum defense mechanisms in host plants.
2. Materials and Methods
2.1. Plant Materials
FW-resistant variety PI 296341 (R) and FW-susceptible variety W1-1 (S) were obtained from the molecular genetic breeding research group for watermelons and melons of Northeast Agricultural University, China. Seeds were germinated and grown under controlled conditions (light/dark = 16 h/8 h, light intensity of 120 μmol m−2 s−1, 28 °C, 70% relative humidity).
2.2. Fungal Transformation with GFP
The Fon (Fusarium oxysporum f. sp. niveum race 1) isolate was laboratory-preserved and maintained on potato dextrose agar (PDA) at 25 °C. Fon was transformed as previously described [20]. Briefly, Agrobacterium tumefaciens strain AGL1 (Coolaber Science & Technology Co., Ltd., Beijing, China) harboring pCAMBIA1303-GFP was resuscitated in lysogeny broth (LB) medium supplemented with 50 μg/mL kanamycin (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and 50 μg/mL rifampicin (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) under standard incubation conditions (28 °C, 180 rpm). AGL1 (OD600 = 0.15) was mixed with Fon microconidia (1 × 106 conidia/mL) in equal volumes (100 μL each). The mixture (200 μL) was plated onto nitrocellulose membranes (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and incubated in a co-cultivation medium supplemented with 5 mM glucose at 25°C for 48 h. Membranes were then transferred to PDA containing 120 μg/mL hygromycin B (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and 300 μg/mL cefotaxime (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) for 7–10 days at 25 °C to permit transformant growth. Hygromycin-resistant transformants were isolated and maintained on PDA supplemented with hygromycin B (120 μg/mL).
2.3. Inoculation of Watermelon Plants
Fungal conidia were produced by culturing Fon in a 250 mL conical flask containing 150 mL of potato dextrose broth at 28 °C on a rotary shaker (150 rpm) for 5–7 days. The conidial suspension was adjusted to a final concentration of 1 × 106 conidia/mL using sterile distilled water for subsequent plant inoculation. Fifteen-day-old watermelon seedlings grown in a controlled growth chamber (16 h/8 h light/dark photoperiod, 28 °C, 70% relative humidity) were carefully removed from vermiculite substrate and gently rinsed with tap water to remove residual particles. Seedling roots were then immersed in freshly prepared spore suspensions (1 × 106 conidia/mL) for 20 min. Inoculated seedlings were replanted into fresh vermiculite, whereas control plants were subjected to identical procedures except they were immersed in sterile distilled water instead of the spore suspension.
2.4. Microscopic Analysis
Watermelon roots were sampled at 24 h intervals up to 8 days post-inoculation (dpi). At each time point, five plants were harvested, rinsed to remove the growth medium, and dissected to isolate the taproot and lower hypocotyl. Root segments adjacent to the root base were excised and sectioned into 5 mm pieces, whereas hypocotyl segments (0–1 cm above the main root) were hand-sectioned into 2–3 mm pieces. To determine the critical time points of Fon infection in watermelon roots, the colonization process of Fon-GFP was observed in watermelon roots at different time points, with five biological replicates per time point. Samples were mounted in water droplets on glass slides under coverslips for microscopic examination using a confocal laser scanning microscope (TCS SP8, Leica, Berlin, Germany) with a 10× objective. Roots treated with non-GFP-labeled Fon served as control samples to ensure that the observed fluorescence was attributable solely to the colonization of Fon-GFP. Fluorescence was excited at 488 nm, and emitted light was recorded between 520 and 540 nm.
2.5. RNA Sequencing (RNA-Seq) and Data Analysis
Root samples were collected from Fon-inoculated plants (S and R) and water-treated controls at 0, 1, 6, and 8 dpi. To explore nonspecific immune responses in the plants, they were exposed to killed pathogens (autoclaved at 121 °C for 5 min) at 1 dpi. In total, 48 samples (10–12 roots per time point) were rapidly frozen in liquid nitrogen for RNA extraction and subsequent RNA-seq. Raw sequencing data were processed using fastp v0.20.1 to trim low-quality bases and remove adapter sequences [21], yielding clean reads. Clean reads were then aligned to the reference genome (http://cucurbitgenomics.org/organism/21, accessed on 19 January 2024) using HISAT2 v2.1.1 [22]. StringTie was subsequently employed to assemble transcript isoforms from mapped reads for each sample, generating gene expression quantification matrices [23].
2.6. Data Integration and Network Construction
Differentially expressed genes (DEGs, log2 fold change > 1 and false discovery rate < 0.05) were clustered and visualized using the ClusterGVis package (https://github.com/junjunlab/ClusterGVis, accessed on 24 March 2024). Co-expression networks were constructed using the R package WGCNA (version 1.73). Subsequently, Cytoscape v3.9.1 was employed to visualize intramodule gene networks and highlight biological interactions among hub genes [24]. Networks were filtered to retain the top 100 edges with the highest weights. Hub genes were prioritized by degree, with the top 10 genes with the highest degrees designated as critical nodes and visualized as red vertices in the network.
2.7. Quantitative Real-Time PCR
qRT-PCR was used to verify the expression of select DEGs. Root samples were collected from Fon-inoculated plants (S and R) and water-treated controls at 0, 1, 6, and 8 dpi. Total RNA was extracted from watermelon root samples using the Spin Column Plant Total RNA Purification Kit (Sangon Biotech, Shanghai, China). Approximately 1 µg of the total RNA was used as a template to synthesize the cDNA with HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China). qRT-PCR was performed using ChamQ™ Universal SYBR® qPCR Master Mix (Vazyme, Nanjing, China) and a LightCycler® 480 II Real-time PCR instrument (Roche, Basel, Switzerland). The relative mRNA level of each candidate gene was evaluated against watermelon ClACTIN (Gene Accession Number Cla97C02G038590) as a reference gene. The qPrimerDB website (https://biodb.swu.edu.cn/qprimerdb/, accessed on 1 April 2024) was used to design qRT-PCR-specific primers (Table S1). The gene expression level was calculated using the 2−ΔΔCT method with three biological replicates.
2.8. Statistical Analysis
Data came from three biological replications and were analyzed using Graphpad software (version 10.4.0) for statistical analysis. Statistically significant differences were assessed using one-way ANOVA, followed by a post hoc Tukey test at a significance level of p < 0.05.
3. Results
3.1. Analysis by Confocal Laser Scanning Microscopy
Following inoculation with Fon, S exhibited pronounced leaf drooping and chlorosis at 8 dpi (Figure 1A,B). In contrast, R displayed no discernible phenotypic alterations under identical infection conditions (Figure 1C,D). Within 1 dpi, most Fon-GFP spores germinated on the root surface, forming initial hyphae (green fluorescence; Figure 1E). By 2 dpi, the fungal hyphae developed densely across the root surface, indicating robust colonization (Figure 1F). Between 4 and 5 dpi, Fon-GFP hyphae penetrated the intercellular spaces along the edges of root cells (Figure 1G and Figure S1). At 6 dpi, Fon-GFP hyphae entered the root vascular system, as evidenced by their presence within xylem vessels (Figure 1H,I). By 8 dpi, extensive colonization of the root vascular system by Fon-GFP was observed, coinciding with the onset of wilt symptoms in watermelon seedlings (Figure 1B,J).
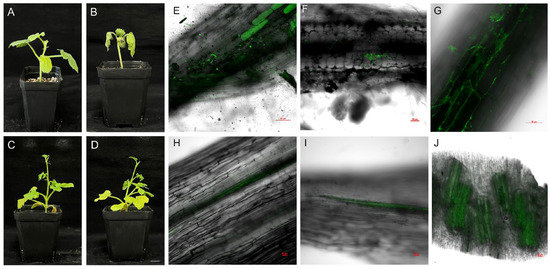
Figure 1.
Phenotypes of watermelon seedlings (S: susceptible, FW-susceptible watermelon variety W1-1, and R: resistant, FW-resistant watermelon variety PI 296341) after 8 days of Fon (Fusarium oxysporum f. sp. niveum race 1) and water treatment and confocal laser scanning microscopic analysis of Fon-GFP colonization in S. (A) S seedlings after 8 days of water treatment. (B) S seedlings after 8 days of Fon inoculation. (C) R seedlings after 8 days of water treatment. (D) R seedlings after 8 days of Fon inoculation. Scale bar = 1.5 cm. (E) At 1 dpi, Fon-GFP attached to the surface of watermelon roots began to germinate. (F) At 2 dpi, Fon-GFP on the surface of watermelon roots began to exhibit dense hyphal proliferation. (G) Between 4 and 5 dpi, Fon-GFP hyphae penetrated the intercellular spaces along the edges of root cells. (H,I) Longitudinal section of the root. At 6 dpi, hyphae were observed to extend into the xylem vessels. (J) In transverse sections of the root, extensive colonization of hyphae in the xylem vessels was observed at 8 dpi. Fluorescence was excited at 488 nm and emitted light between 520 and 540 nm. Scale bar = 50 µm.
Combining observations of the infection process with phenotypic responses in S inoculated with Fon-GFP, we identified 1, 6, and 8 dpi as critical time points. Spore germination occurred at 1 dpi, initiating the formation of hyphae, which likely marks the early recognition phase of plant defense against fungal pathogens (Figure 1E). By 6 dpi, hyphae had invaded the root vascular system, as evidenced by their presence within xylem vessels (Figure 1H,I), suggesting a transition to necrotrophic growth and triggering a moderate defense response in the host. Extensive colonization of the vascular system at 8 dpi coincided with the onset of wilt symptoms in watermelon seedlings (Figure 1B,J), indicating an advanced stage of infection in which the pathogen disrupts cellular integrity. These key time points (1, 6, and 8 dpi) were pivotal for subsequent experiments aimed at elucidating the molecular mechanisms underlying plant–pathogen interactions during these critical phases of infection.
3.2. Identification of DEGs
To identify DEGs associated with Fon infection, mRNA libraries were constructed and divided into 16 groups, which comprised samples from two genotypes of infected plants and water-treated controls at 0, 1, 6, and 8 dpi, with three biological replicates per group (n = 3). In total, 2,397,660,805 raw reads were generated, and after quality control, 2,357,983,183 clean reads were obtained for downstream analysis (Table S2).
Comparative analysis of pathogen-treated and water-treated samples revealed distinct transcriptional patterns. In cultivar S, 1, 1, 1197, and 1250 upregulated DEGs were identified at 0, 1, 6, and 8 dpi, whereas 13, 18, 1561, and 1713 downregulated DEGs, respectively, were identified at these time points (Figure 2A). In cultivar R, pathogen-challenged plants exhibited 0, 219, 200, and 754 upregulated DEGs and 0, 365, 31, and 1171 downregulated DEGs at 0, 1, 6, and 8 dpi, respectively (Figure 2B).
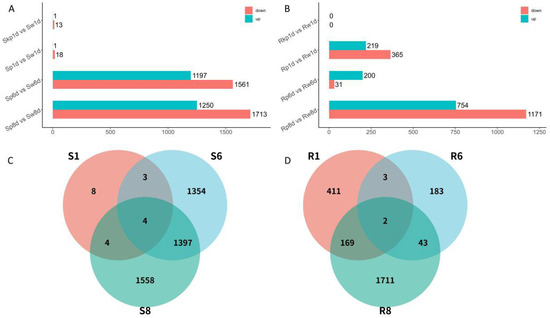
Figure 2.
Graphical representation of gene expression changes following inoculation with Fusarium oxysporum f.sp. niveum (Fon). (A) DEGs between different treatments at each time point in susceptible watermelon (S). (B) DEGs between different treatments at each time point in resistant watermelon (R). (C) Venn diagram of overlapped genes in S. (D) Venn diagram of overlapped genes in R. R: FW-resistant watermelon variety PI 296341; S: FW-susceptible watermelon variety W1-1; w: water treatment; p: pathogen treatment (Fusarium oxysporum f. sp. niveum race 1); kp: killed pathogen treatment; S1: Sp1d vs. Sw1d; S6: Sp6d vs. Sw6d; S8: Sp8d vs. Sw8d; R1: Rp1d vs. Rw1d; R6: Rp6d vs. Rw6d; R8: Rp8d vs. Rw8d.
Given the minimal differential expression observed between water-treated and killed pathogen-treated samples at 1 dpi in both cultivars, data from killed pathogen-treated samples at this time point were excluded from subsequent analyses. A comparative overlap analysis was conducted across three critical infection stages (1, 6, and 8 dpi) in R and S. In R, 411, 183, and 1711 DEGs were uniquely expressed at 1, 6, and 8 dpi, respectively. Conversely, cultivar S exhibited 8, 1354, and 1558 uniquely expressed DEGs, respectively, at these time points (Figure 2C). Notably, cultivar R displayed the temporal conservation of two DEGs across all stages, whereas cultivar S retained four persistently expressed DEGs (Figure 2D).
3.3. Determination of Key Clusters
To systematically characterize cultivar-specific defense responses, DEGs upregulated by pathogen exposure in the S and R cultivars were clustered using the ClusterGVis package (Version 0.1.2), yielding 12 distinct clusters in each cultivar. Subsequent functional annotation prioritized defense-associated clusters by integrating expression dynamics with functional enrichment analyses of Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. In R, 1058 upregulated DEGs were grouped into 12 clusters, five of which (R1, R5, R7, R9, R12) exhibited potential associations with disease resistance. DEGs in R1 (contained 59 genes) and R5 (contained 96 genes) were upregulated at 6 dpi, whereas those in R7 (contained 74 genes) displayed sustained upregulation at both 6 and 8 dpi. Genes in R9 (contained 132 genes) and R12 (contained 91 genes) were specifically activated at 8 and 1 dpi, respectively. Functional enrichment analysis revealed that these clusters were significantly associated with immune system processes, defense responses, phenylpropanoid biosynthesis, responses to biotic stimuli, glutathione metabolism, salicylic acid (SA) signaling, and NOD-like receptor pathways (Figure 3). In S, four clusters (S5, S6, S7, S10) displayed putative roles in disease resistance. Genes in S5 (contained 106 genes) were upregulated at 6 and 8 dpi, those in S6 (contained 136 genes) were upregulated at 8 dpi, and those in S7 (contained 123 genes) and S10 (contained 213 genes) were upregulated at 6 dpi. The enriched functions included immune system processes, responses to biotic stimuli, glutathione metabolism, defense responses, plant–pathogen interactions, hormone signaling, and phenylpropanoid biosynthesis (Figure 4). Merging these key clusters yielded 941 DEGs involved in various aspects of disease resistance.
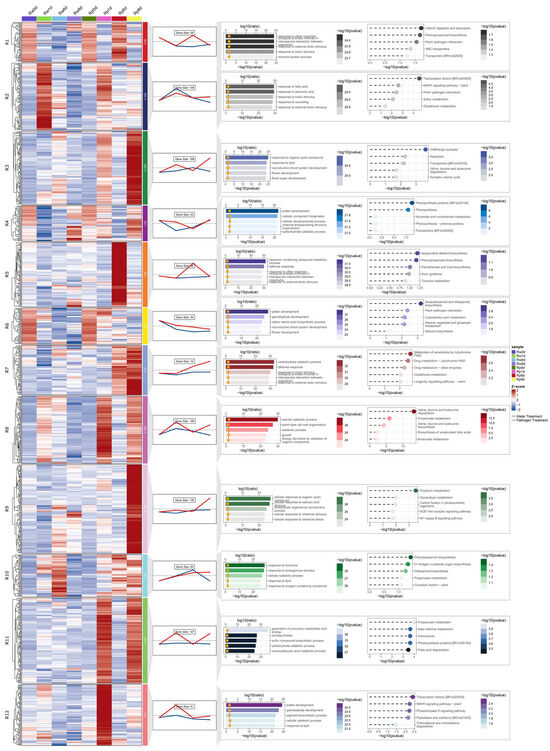
Figure 3.
Co-expression cluster identification and functional annotation of DEGs in resistant (R) watermelon upregulated upon inoculation with Fusarium oxysporum f.sp. niveum (Fon). The Rw0d data are the same as the Rp0d data. From left to right are expression calorimetric maps, expression trend maps and the number of genes contained, GO enrichment results, and KEGG pathway enrichment results. R: FW-resistant watermelon variety PI 296341, w: water treatment, p: pathogen treatment (Fusarium oxysporum f. sp. niveum race 1). In expression calorimetric maps, red indicates upregulation and blue indicates downregulation, with white representing baseline expression (z-score normalized), and R1–R12 denote the 12 gene clusters obtained. In expression trend maps, the red line represents data from the pathogen treatment, the blue line represents data from the water treatment, and trend lines denote mean expression values per cluster. In the GO enrichment results, the top 5 enriched terms in Biological Process (BP) are shown. In the KEGG pathway enrichment results, the top 5 most significant terms from the KEGG enrichment analysis are shown.
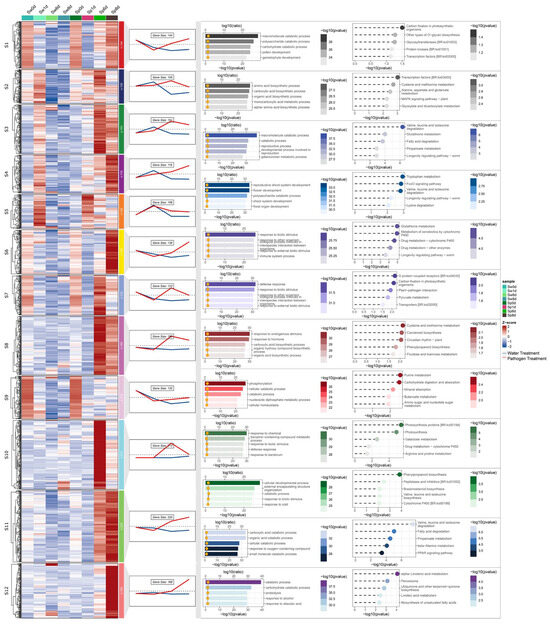
Figure 4.
Co-expression cluster identification and functional annotation of DEGs in susceptible (S) watermelon upregulated upon inoculation with Fusarium oxysporum f.sp. niveum (Fon). The Sw0d data are the same as the Sp0d data. From left to right are expression calorimetric maps, expression trend maps and the number of genes contained, GO enrichment results, and KEGG pathway enrichment results. S: FW-susceptible watermelon variety W1-1, w: water treatment, p: pathogen treatment (Fusarium oxysporum f. sp. niveum race 1). In expression calorimetric maps, red indicates upregulation and blue indicates downregulation, with white representing baseline expression (z-score normalized), and S1–S12 denote the 12 gene clusters obtained. In expression trend maps, the red line represents data from the pathogen treatment, the blue line represents data from the water treatment, and trend lines denote mean expression values per cluster. In the GO enrichment results, the top 5 enriched terms in Biological Process (BP) are shown. In the KEGG pathway enrichment results, the top 5 most significant terms from the KEGG enrichment analysis are shown.
3.4. Determination of Key Modules Associated with Fon Resistance Through Co-Expression Network Analysis
To systematically characterize the expression divergence of defense-related DEGs between S and R, WGCNA was performed on 941 defense-associated DEGs from key clusters (R1, R5, R7, R9, R12, S5, S6, S7, S10). A soft threshold power of β = 17 was selected to construct hierarchical clustering trees based on scale-free topology criteria (Figure S2). Following this, genes were partitioned into 10 distinct co-expression modules using dynamic tree cutting (mergeCutHeight = 0.25; Figure 5A). To delineate key modules linked to disease resistance, we analyzed correlations between co-expression modules and phenotypic traits associated with specific infection stages. In R, three modules demonstrated robust associations with pathogen responses. Specifically, SRgreen exhibited a significant positive correlation with Rp1d (r2 = 0.85, p = 3 × 10−12), SRyellow exhibited a significant positive correlation with Rp6d (r2 = 0.64, p = 7 × 10−6), and SRblue exhibited a significant positive correlation with Rp8d (r2 = 0.83, p = 2 ×10−11). In S, two modules displayed strong pathogen-responsive patterns, with SRbrown being positively correlated with Sp8d (r2 = 0.66, p = 2 × 10−6) and SRturquoise being positively correlated with Sp6d (r2 = 0.93, p = 2 × 10−18). SRred and SRblack exhibited conserved expression trajectories in both S and R, suggesting shared regulatory mechanisms underlying basal pathogen responses (Figure 5B). Conversely, SRmagenta displayed divergent transcriptional dynamics between the two genotypes, with antagonistic expression patterns in S versus R, indicative of the cultivar-specific modulation of defense-related pathways. The SRpurple and SRpink modules exhibited strong correlations with the water treatment group, and thus, they were excluded from further analysis.
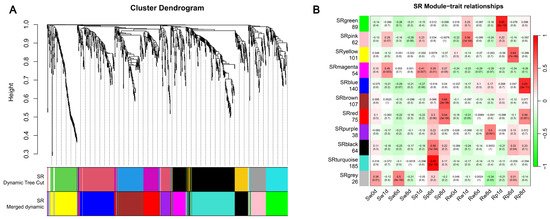
Figure 5.
Co-expression network analysis of DEGs and the module–trait relationship in response to Fusarium oxysporum f.sp. niveum (Fon) infection of watermelon. (A) Cluster dendrogram of different genes in co-expression modules. Dynamic tree cut (minimum module size = 30 genes, merge cut height = 0.25). Color bars below the dendrogram represent assigned modules. SR denotes the data used in WGCNA from susceptible (S) and resistant (R) watermelon. (B) Heat map of the correlation between templates and groups. Each row represents a module eigengene, and each column represents a different characteristic. Each cell contains the corresponding correlation and p-value, and the table is color coded by correlation according to the color legend. The number below each module name denotes the number of genes within the module. Excluding the gray module, a total of 10 gene modules were identified.
3.5. GO and KEGG Enrichment Analyses of Key Modules
GO and KEGG pathway enrichment analyses were performed to delineate the biological significance of the identified modules. The SRgreen module exhibited significant enrichment for plant-specific signaling pathways, including the mitogen-activated protein kinase (MAPK) signaling pathway and plant and plant hormone signal transduction (Figure 6A). SRyellow was associated with metabolic reprogramming, particularly the amine metabolic process and tryptophan biosynthetic process (Figure S3). SRblue displayed enrichment in stress-responsive functions, such as cytokinin metabolic process and responses to toxic substances (Figure S4). In the S cultivar, SRbrown was associated with structural and interactive pathways, including cellulose biosynthetic processes and plant–pathogen interactions (Figure S5), whereas SRturquoise was linked to defense-related processes, including external encapsulating structure organization and phenylpropanoid biosynthesis (Figure S6). The conserved modules SRred and SRblack demonstrated significant enrichment for pathways associated with stress adaptation and secondary metabolism. SRred was specifically linked to cellular responses to abiotic stimuli and beta-alanine metabolism (Figure S7), whereas SRblack encoded functions critical to phytohormone dynamics and structural defense, including brassinosteroid biosynthesis and phenylpropanoid biosynthesis (Figure 6B). In stark contrast, the divergent SRmagenta module exhibited a unique enrichment of glutamine metabolic processes, reflecting genotype-specific metabolic reprogramming during pathogen challenge (Figure S8). Based on module correlation and GO and KEGG enrichment analyses, six key modules, namely, SRgreen, SRblue, SRbrown, SRturquoise, SRblack, and SRmagenta, were identified as being associated with disease resistance responses.
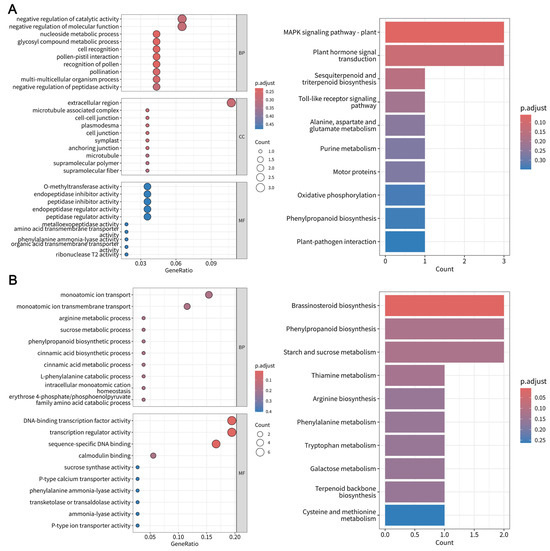
Figure 6.
Functional enrichment analysis of key co-expression modules in pathogen-responsive genes in the Fusarium oxysporum f.sp. niveum–watermelon pathosystem. (A) GO and KEGG pathway enrichment analyses of SRgreen module genes. (B) GO and KEGG pathway enrichment analyses of SRblack module genes.
3.6. Identification of Hub Genes Involved in FW Resistance Through WGCNA
To identify hub genes within the defense-related modules, gene co-expression networks were constructed and analyzed using Cytoscape (Figure 7). Networks were filtered to retain the top 100 edges with the highest weights. Hub genes were prioritized according to the degree, with the top 10 highest-degree genes designated as critical nodes and visualized as red vertices in the network (Table S3). Based on the annotated functions in the reference genome, 35 hub genes were selected (Table 1).
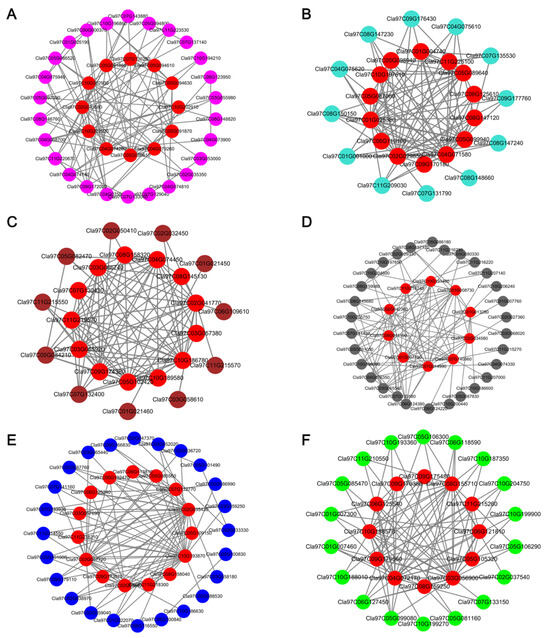
Figure 7.
Identification of hub genes in the protein–protein interaction (PPI) networks following inoculation of watermelon with Fusarium oxysporum f.sp. niveum. (A) PPI network of SRmagenta. (B) PPI network of SRturquoise. (C) PPI network of SRbrown. (D) PPI network of SRblack. (E) PPI network of SRblue. (F) PPI network of SRgreen.

Table 1.
Hub genes associated with watermelon resistance to Fusarium oxysporum f.sp. niveum.
The SRblack module encompassed key regulatory and metabolic components, including WRKY TF 42, calcium-transporting ATPase, 1-deoxy-d-xylulose-5-phosphate synthase 2a, and benzyl alcohol O-benzoyltransferase-like. The SRgreen module featured signaling and hormone-related elements: basic-leucine zipper (bZIP) TF family, ethylene-responsive TF 1B-like, putative receptor kinase, expansin-like B1, patatin, and gibberellin receptor GID1B-like.
SRblue contained genes associated with cell wall dynamics and defense signaling, notably cellulose synthase-like protein H1, SA-binding protein 2-like, gibberellin 2-beta-dioxygenase 8, cellulose synthase-like protein B3, xylose isomerase, and detoxification-related proteins. SRbrown is enriched for structural remodeling and stress-responsive factors, including mannan endo-1,4-beta-mannosidase 1-like, receptor-like protein kinase 4, NINJA family transcriptional repressor, and bZIP TF 53. The SRturquoise module comprised defense execution machinery: metacaspase-9, FAD-binding berberine family protein, peroxidase, MADS-box TF, endo-1,4-beta-xylanase A-like, NAC domain-containing protein, thaumatin-like protein, and pectinesterase inhibitor. Conversely, SRmagenta encoded glutathione S-transferase and MADS-box TFs.
3.7. qRT-PCR Validation of RNA-Seq Data
To validate the reliability of RNA-seq data and differential expression profiles, 15 hub genes were selected for analysis by qRT-PCR (Figure 8). These genes represented diverse expression patterns observed in the transcriptomic dataset. Among them, in the SRblack module, Cla97C01G014990 (WRKY TF 42) and Cla97C02G042360 (calcium-transporting ATPase) were upregulated at 6 and 8 dpi. By contrast, Cla97C08G155710 (AIG2), Cla97C09G170380 (ethylene-responsive TF 1B-like), and Cla97C06G121810 (receptor kinase, putative) in the SRgreen module were upregulated at 1 dpi.
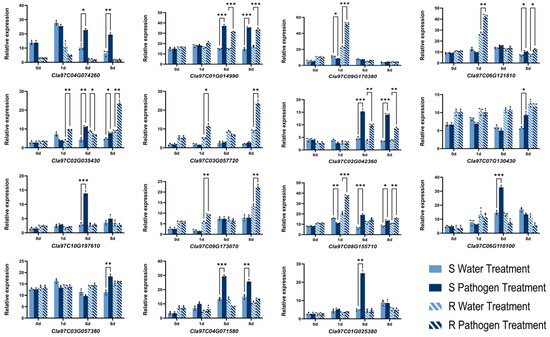
Figure 8.
qRT-PCR analysis of 15 hub genes following inoculation of resistant (R) and susceptible (S) watermelon with Fusarium oxysporum f.sp. niveum. Error bars indicate the mean ± standard deviation, and the three data points in each column indicate three biological replicates. * p < 0.05, ** p < 0.01, *** p < 0.001.
4. Discussion
The interaction between Fon and watermelon involves distinct infection phases, including a biotrophic stage during which the pathogen obtains nutrients from living host cells and a necrotrophic stage characterized by cell death and tissue collapse. Our time-course microscopic observations revealed that Fon-GFP spores initiated germination and root colonization at 1 dpi, followed by hyphal penetration into xylem vessels at 6 dpi, culminating in wilt symptoms at 8 dpi (Figure 1). This progression aligns with the biotrophy-to-necrotrophy transition described in previous studies [4,5,25,26,27], establishing 1, 6, and 8 dpi as critical infection milestones for dissecting host defense mechanisms.
Transcriptomic profiling has emerged as a cornerstone for dissecting the molecular underpinnings of plant–F. oxysporum interactions, particularly in unraveling the dynamic interplay between host defense mechanisms and pathogen virulence strategies [11,28,29]. Differential expression analysis revealed distinct transcriptional strategies between R and S during Fon infection. In S, the majority of DEGs were observed at 6 and 8 dpi, coinciding with xylem colonization and symptom onset. Conversely, the R genotype exhibited high numbers of DEGs at all three critical infection stages (1, 6, and 8 dpi), suggesting an earlier and more dynamic defense response. This early transcriptional activation (1 dpi) in R likely reflects rapid pathogen recognition and signaling, which might suppress Fon development during the biotrophic phase [5]. Venn diagram analysis further highlighted stage-specific defense mechanisms (Figure 2). In R, the minimal overlap of DEGs across time points indicated distinct transcriptional programs tailored to each infection phase [early recognition (1 dpi), xylem defense (6 dpi), and necrosis-associated resistance (8 dpi)]. As the infection process progresses, F. oxysporum secretes different proteins to facilitate its progression [30]. This phased response likely enhances the ability to counter Fon progression at multiple stages [31]. By contrast, the S genotype displayed significant overlap in DEGs between 6 and 8 dpi, suggesting a delayed and generalized defensive response. The lack of early (1 dpi) DEGs in S might have permitted Fon to proliferate unchecked during the biotrophic phase, leading to uncontrolled xylem invasion and subsequent necrotrophic expansion [4].
The application of ClusterGVis for transcriptomic clustering and functional enrichment analysis provided a systematic framework to identify biologically meaningful gene sets underlying host–pathogen interactions [32]. Expression clustering analysis revealed distinct transcriptional strategies in the R and S cultivars during Fon infection. Using ClusterGVis, we identified nine clusters (R1, R5, R7, R9, R12, S5, S6, S7, S10) enriched for upregulated defense-related genes in Fon-treated samples. Notably, R8, R11, and R12 exhibited pronounced upregulation on 1 dpi in R, whereas S lacked significant upregulation at this stage, highlighting a critical temporal divergence in defense initiation.
The R8 cluster was enriched for pathways related to valine, leucine, and isoleucine degradation and cellular catabolic processes. Concurrently, upregulated genes in cluster R11 were involved in the generation of precursor metabolites and energy, sulfur compound biosynthetic processes, and carbohydrate catabolic processes. This metabolic rewiring likely provides precursors and energy for defense responses [33]. Cluster R12, activated at 1 dpi, was enriched for transcription factors (e.g., WRKY) and the MAPK signaling pathway. The MAPK cascade is a conserved regulator of PTI and ETI in plants [34]. The early upregulation of MAPK components in R aligned with rapid pathogen recognition and signal amplification, which are critical for deploying localized defense barriers (e.g., callose deposition, ROS bursts) to restrict Fon proliferation [35,36]. Contrasting these findings, S displayed minimal early transcriptional activation (1 dpi), with significant upregulation delayed until 6 and 8 dpi. This delay likely permitted Fon to establish biotrophic colonization unimpeded, leading to unchecked xylem invasion and eventual necrosis.
Integrating our findings, the SRblue module coordinated late-stage defense (8 dpi) against Fon through synergistic cell wall reinforcement and hormonal regulation. Hub genes, such as Cla97C02G035430 (cellulose synthase-like H1) and Cla97C02G035420 (cellulose synthase-like B3), likely enhance structural resistance by promoting cellulose deposition in papillae, analogous to barley, in which silencing a cellulose synthase-like gene (HvCslD2) increased fungal penetration because of reduced epidermal cellulose content [37]. Additionally, Cla97C03G057720 (SA-binding protein 2-like) is implicated in SA transport, which is crucial for priming defenses, as demonstrated in Arabidopsis, where exogenous SA treatment increased PR1 expression and reduced F. oxysporum-induced necrosis [38]. The SRturquoise module established a robust immune barrier during late infection (6 dpi) by coordinating ROS signaling, programmed cell death (PCD), and antimicrobial metabolic pathways. For instance, Cla97C04G071580 (metacaspase-9) regulates post-mortem cellular clearance, similar to Arabidopsis AtMC9, which facilitates the autolysis of xylem vessel contents post-vacuolar rupture, thereby restricting F. oxysporum spread by eliminating compromised host tissues [39]. Furthermore, Cla97C10G197610 (thaumatin-like protein) exhibits direct antifungal activity, akin to ClTLP27 in watermelon, which inhibits Fo mycelial growth through conserved cysteine-rich domains, thereby disrupting fungal cell wall integrity [40]. The SRbrown module, which was positively correlated with late-stage Fon infection (8 dpi), might orchestrate immune responses against Fon through the synergistic regulation of JA signaling. Cla97C07G130430 (NINJA-family protein AFP2-like) potentially suppresses JA hyperproduction by recruiting TOPLESS/TPR corepressors to JAZ–MYC2 complexes, as demonstrated in Arabidopsis, in which NINJA-mediated repression fine-tunes defense–growth trade-offs [41]. The SRmagenta module, which was associated with the responses of the S cultivar to Fon infection, might coordinate oxidative stress management and transcriptional reprogramming to mitigate pathogenic impact. Cla97C04G074260 (glutathione S-transferase U8-like) and Cla97C04G079260 (glutathione S-transferase) potentially enhance ROS detoxification, as demonstrated in Lilium regale, in which LrGSTU5 overexpression upregulated PR1b, chitinase, and antioxidant enzymes (GST, SOD, APX), thereby reducing superoxide anion accumulation and improving Fo resistance [42].
The SRgreen module coordinated early transcriptional defense responses in watermelon during F, oxysporum (Fo) infection, potentially through synergistic interactions of its constituent genes. Cla97C08G155710 (protein AIG2) can fine-tune and balance SA and tryptophan-derived secondary metabolite chemical defense systems in response to nonpathogenic and pathogenic microbes [43]. Cla97C09G170380 (ethylene-responsive TF 1B-like) might enhance peroxidase activity and ROS scavenging, as demonstrated by PdPapERF109 overexpression in Populus, which improved F. oxysporum resistance by reducing oxidative damage [44]. Cla97C05G105320 (bZIP TF) likely upregulates antioxidant enzymes (e.g., SOD, CAT) and pathogenesis-related genes, as supported by studies demonstrating that LrbZIP1 in Lilium amplified defense responses against F. oxysporum through stress signaling pathways [45]. Cla97C06G121810 (receptor kinase, putative) could initiate PTI by suppressing Fo-induced ROS bursts, paralleling findings in which VfLRR–RLK1 in Vernicia enhanced root architecture and Fo resistance via ROS detoxification [46]. Cla97C09G175480 (expansin-like B1) might regulate structural defenses by modifying cell wall dynamics, as sugarcane Expansin genes were found to limit fungal entry by modulating stomatal apertures [47]. Cla97C08G159250 (patatin) might balance localized cell death and defense through oxylipin metabolism, akin to PLP2 in Arabidopsis, which controls lipid signaling to restrict pathogen spread [48]. Cla97C11G215260 (gibberellin receptor GID1B-like) potentially prioritizes defense over growth by stabilizing DELLA proteins, consistent with SA-mediated GID1 degradation mechanisms that reallocate resources to promote Fo resistance [49].
The SRblack module, which was positively correlated with both the S and R genotypes during late-stage Fon infection, likely orchestrates multifaceted defense mechanisms through coordinated hormonal signaling, calcium dynamics, and metabolic reprogramming. Cla97C07G140960 (cytochrome P450) might enhance SA/ET-dependent defenses, as demonstrated in cucumber, in which CYP82D47 overexpression increased PR1, PR2, and EIN3 expression, improving resistance to F. oxysporum and powdery mildew [50]. Cla97C02G042360 (calcium-transporting ATPase) potentially regulates PCD by modulating intracellular calcium fluxes, akin to NbCA1 in Nicotiana, which delays pathogen-induced cell death to balance immunity and tissue integrity [51]. Cla97C11G216240 (dynamin-related protein 4C) might fine-tune callose deposition by regulating the vesicle trafficking of callose synthases, as observed in Arabidopsis, in which DRP2B restricts excessive callose accumulation to optimize pathogen containment [52]. Cla97C02G034580 (benzyl alcohol O-benzoyltransferase-like) could contribute to SA biosynthesis via peroxisomal β-oxidation, similar to HSR201 in tobacco, which cooperates with NtCNL and NtCHD to produce SA precursors critical for defense priming [53].
The SRgreen and SRblack modules represent distinct sources of early- and late-phase resistance, respectively. In the SRblack module, Cla97C01G014990 (WRKY TF 42) and Cla97C02G042360 (calcium-transporting ATPase) exhibited high node degree values, suggesting that sustained resistance is mediated through WRKY-dependent transcriptional regulation and calcium signaling pathways. By contrast, the SRgreen module harbored the high-degree genes Cla97C08G155710 (AIG2), Cla97C09G170380 (ethylene-responsive TF 1B-like), and Cla97C06G121810 (receptor kinase, putative), which likely recognize Fon via receptor kinases and rapidly initiate defense responses through AIG2-mediated signaling and ethylene-responsive hormonal pathways, enabling early-stage pathogen resistance.
5. Conclusions
In this study, we identified 1, 6, and 8 dpi as key infection stages of Fon. Through the analysis of differential expression profiles, WGCNA, and functional enrichment, we revealed six gene modules closely associated with disease resistance, which contribute to resistance at different infection stages. Furthermore, PPI network analysis combined with RT-qPCR identified five hub genes in the SRgreen and SRblack modules, including Cla97C01G014990, Cla97C02G042360, Cla97C08G155710, Cla97C09G170380, and Cla97C06G121810. The identified candidate genes provide rapid resistance during early infection and long-term resistance during later infection stages. This study screens for hub genes related to disease resistance in the Fon–watermelon interaction at different stages, offering novel disease-resistant genes for breeding watermelon resistant to Fusarium wilt.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11060625/s1, Figure S1. Confocal laser scanning microscopic analysis of Fon-GFP colonization in S (FW-susceptible watermelon variety W1-1). Between 4 and 5 dpi, Fon-GFP hyphae penetrated the intercellular spaces along the edges of root cells and single frames; Figure S2: Analysis of network topology for various soft-thresholding powers. The soft threshold was determined by the scale-free network index; R2 was set to 0. 8, and the best soft threshold was 17; Figure S3: GO and KEGG pathway enrichment analysis of SRyellow module genes; Figure S4: GO and KEGG pathway enrichment analysis of SRblue module genes; Figure S5: GO and KEGG pathway enrichment analysis of SRbrown module genes; Figure S6: GO and KEGG pathway enrichment analysis of SRturquoise module genes; Figure S7: GO and KEGG pathway enrichment analysis of SRred module genes; Figure S8: GO and KEGG pathway enrichment analysis of SRmagenta module genes. Table S1: qRT-PCR-specific primers; Table S2: Quality control (QC) analysis of transcriptome data; Table S3: The top 10 hub genes (highest degree) in the modules.
Author Contributions
Conceptualization, P.G.; data curation, C.Z.; formal analysis, X.F.; funding acquisition, P.G.; investigation, C.Z.; methodology, X.F.; project administration, X.F.; resources, Z.L., S.L. and Z.S.; software, C.Z.; supervision, F.L.; validation, J.Z. and X.W.; visualization, X.F.; writing—original draft preparation, C.Z.; writing—review and editing, P.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science and Technology-based Small and Medium-sized Enterprises Innovation Capacity Enhancement Project of Shandong Province (grant number 2023TSGC0804), the Weifang City Science and Technology-based Small and Medium-sized Enterprises Innovation Capacity Enhancement Project (grant number 2023TS1072), the National Nature Science Foundation of China (grant number U21A20229), the “Young Leading Talents” support program of Northeast Agricultural University (grant number NEAU2023QNLJ-005), the major development program of Heilongjiang Province (grant number GA23B007), and the Natural Science Foundation of Heilongjiang Province (grant number ZL2024C011).
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
We appreciate all the people who have collaborated on this project.
Conflicts of Interest
Authors Zhao Liu, Shusen Liu and Zhengfeng Song were employed by the Shandong Engineering Research Center for Watermelon and Melon Breeding Co., Ltd.
References
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org (accessed on 9 May 2025).
- Martyn, R.D. Resistance to Races 0, 1, and 2 of Fusarium Wilt of Watermelon in Citrullus sp. PI-296341-FR. Hortscience 1991, 26, 429. [Google Scholar] [CrossRef]
- Yvonne Couteaudier, C.A. Survival and Inoculum Potential of Conidia and Chlamydospores of Fusarium oxysporum f. sp. Lini in Soil. Dev. Agric. Manag. For. Ecol. 1991, 23, 551–556. [Google Scholar] [CrossRef]
- Lu, G.; Guo, S.; Zhang, H.; Geng, L.; Martyn, R.D.; Xu, Y. Colonization of Fusarium Wilt-Resistant and Susceptible Watermelon Roots by a Green-Fluorescent-Protein-Tagged Isolate of Fusarium oxysporum f. sp. Niveum. J. Phytopathol. 2014, 162, 228–237. [Google Scholar] [CrossRef]
- Zhang, M.; Xu, J.H.; Liu, G.; Yao, X.F.; Li, P.F.; Yang, X.P. Characterization of the Watermelon Seedling Infection Process by Fusarium oxysporum f. sp. Niveum. Plant Pathol. 2015, 64, 1076–1084. [Google Scholar] [CrossRef]
- Ren, Y.; Jiao, D.; Gong, G.; Zhang, H.; Guo, S.; Zhang, J.; Xu, Y. Genetic Analysis and Chromosome Mapping of Resistance to Fusarium oxysporum f. sp. Niveum (FON) Race 1 and Race 2 in Watermelon (Citrullus lanatus L.). Mol. Breed. 2015, 35, 183. [Google Scholar] [CrossRef]
- Branham, S.E.; Levi, A.; Wechter, W.P. QTL Mapping Identifies Novel Source of Resistance to Fusarium Wilt Race 1 in Citrullus amarus. Plant Dis. 2019, 103, 984–989. [Google Scholar] [CrossRef]
- Xu, X.P.; Chen, C.H.; Fan, B.F.; Chen, Z.X. Physical and Functional Interactions between Pathogen-Induced Arabidopsis WRKY18, WRKY40, and WRKY60 Transcription Factors. Plant Cell 2006, 18, 1310–1326. [Google Scholar] [CrossRef]
- Agarwal, P.; Reddy, M.P.; Chikara, J. WRKY: Its Structure, Evolutionary Relationship, DNA-Binding Selectivity, Role in Stress Tolerance and Development of Plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef]
- Wang, L.; Guo, D.; Zhao, G.; Wang, J.; Zhang, S.; Wang, C.; Guo, X. Group IIc WRKY Transcription Factors Regulate Cotton Resistance to Fusarium oxysporum by Promoting GhMKK2-Mediated Flavonoid Biosynthesis. New Phytol. 2022, 236, 249–265. [Google Scholar] [CrossRef]
- Diao, J.; Wang, J.; Zhang, P.; Hao, X.; Wang, Y.; Liang, L.; Zhang, Y.; Ma, W.; Ma, L. Transcriptome Analysis Reveals the Important Role of WRKY28 in Fusarium oxysporum Resistance. Front. Plant Sci. 2021, 12, 720679. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the Soybean GmERF3 Gene, an AP2/ERF Type Transcription Factor for Increased Tolerances to Salt, Drought, and Diseases in Transgenic Tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jiang, L.; Du, B.; Ning, B.; Ding, X.; Zhang, C.; Song, B.; Liu, S.; Zhao, M.; Zhao, Y.; et al. GmMKK4-Activated GmMPK6 Stimulates GmERF113 to Trigger Resistance to Phytophthora sojae in Soybean. Plant J. 2022, 111, 473–495. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Jin, L.; Miao, Y.; He, X.; Hu, Q.; Guo, K.; Zhu, L.; Zhang, X. An Ethylene Response-Related Factor, GbERF1-like, from Gossypium barbadense Improves Resistance to Verticillium dahliae via Activating Lignin Synthesis. Plant Mol. Biol. 2016, 91, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-J.; Zhang, J.; Lin, Z.; Yu, P.; Lu, M.; Li, N. The AP2/ERF Transcription Factor ORA59 Regulates Ethylene-Induced Phytoalexin Synthesis through Modulation of an Acyltransferase Gene Expression. J. Cell. Physiol. 2024, 239, e30935. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Xu, X.; Lu, X.; Tang, Z.; Zhang, X.; Lei, F.; Hou, L.; Li, M. Combined Analysis of Carotenoid Metabolites and the Transcriptome to Reveal the Molecular Mechanism Underlying Fruit Colouration in Zucchini (Cucurbita pepo L.). Food Chem. Mol. Sci. 2021, 2, 100021. [Google Scholar] [CrossRef]
- Anees, M.; Gao, L.; Umer, M.J.; Yuan, P.; Zhu, H.; Lu, X.; He, N.; Gong, C.; Kaseb, M.O.; Zhao, S.; et al. Identification of Key Gene Networks Associated With Cell Wall Components Leading to Flesh Firmness in Watermelon. Front. Plant Sci. 2021, 12, 630243. [Google Scholar] [CrossRef]
- Shen, Q.; Wu, X.; Tao, Y.; Yan, G.; Wang, X.; Cao, S.; Wang, C.; He, W. Mining Candidate Genes Related to Heavy Metals in Mature Melon (Cucumis melo L.) Peel and Pulp Using WGCNA. Genes 2022, 13, 1767. [Google Scholar] [CrossRef]
- Mullins, E.D.; Chen, X.; Romaine, P.; Raina, R.; Geiser, D.M.; Kang, S. Agrobacterium-Mediated Transformation of Fusarium oxysporum: An Efficient Tool for Insertional Mutagenesis and Gene Transfer. Phytopathology 2001, 91, 173–180. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast One-Pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.T.; Franz, M.; Kazi, F.; Donaldson, S.L.; Morris, Q.; Bader, G.D. Cytoscape Web: An Interactive Web-Based Network Browser. Bioinformatics 2010, 26, 2347–2348. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Shao, J.; Wang, Y.; Li, W.; Guo, D.; Yan, B.; Xia, Y.; Peng, M. Analysis of Banana Transcriptome and Global Gene Expression Profiles in Banana Roots in Response to Infection by Race 1 and Tropical Race 4 of Fusarium oxysporum f. sp. Cubense. BMC Genom. 2013, 14, 851. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, Z.; Zhang, C.; Song, J.; Wang, Q.; Luan, F.; Gao, P. Evaluation of Differential miRNA Expression between Fusarium Wilt-Resistant and -Susceptible Watermelon Varieties. Sci. Hortic. 2024, 332, 113189. [Google Scholar] [CrossRef]
- Sun, Y.; Yang, H.; Li, J. Transcriptome Analysis Reveals the Response Mechanism of Frl-Mediated Resistance to Fusarium oxysporum f. sp. Radicis-Lycopersici (FORL) Infection in Tomato. Int. J. Mol. Sci. 2022, 23, 7078. [Google Scholar] [CrossRef]
- Kaushal, M.; Mahuku, G.; Swennen, R. Comparative Transcriptome and Expression Profiling of Resistant and Susceptible Banana Cultivars during Infection by Fusarium oxysporum. Int. J. Mol. Sci. 2021, 22, 3002. [Google Scholar] [CrossRef]
- Lu, G.; Guo, S.; Zhang, H.; Geng, L.; Song, F.; Fei, Z.; Xu, Y. Transcriptional Profiling of Watermelon during Its Incompatible Interaction with Fusarium oxysporum f. sp. Niveum. Eur. J. Plant Pathol. 2011, 131, 585–601. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Williams, A.H.; Garg, G.; Buck, S.-A.G.; Singh, K.B. Transcriptome Analysis of the Fungal Pathogen Fusarium oxysporum f. sp. Medicaginis during Colonisation of Resistant and Susceptible Medicago truncatula Hosts Identifies Differential Pathogenicity Profiles and Novel Candidate Effectors. BMC Genom. 2016, 17, 860. [Google Scholar] [CrossRef]
- Luo, J.; Zhang, A.; Tan, K.; Yang, S.; Ma, X.; Bai, X.; Hou, Y.; Bai, J. Study on the Interaction Mechanism between Crocus Sativus and Fusarium oxysporum Based on Dual RNA-Seq. Plant Cell Rep. 2023, 42, 91–106. [Google Scholar] [CrossRef]
- Kuang, W.; Huang, J.; Yang, Y.; Liao, Y.; Zhou, Z.; Liu, Q.; Wu, H. Identification of Markers Correlating with Mitochondrial Function in Myocardial Infarction by Bioinformatics. PLoS ONE 2024, 19, e0316463. [Google Scholar] [CrossRef] [PubMed]
- Manzo, D.; Ferriello, F.; Puopolo, G.; Zoina, A.; D’Esposito, D.; Tardella, L.; Ferrarini, A.; Ercolano, M.R. Fusarium oxysporum f. sp. Radicis-Lycopersici Induces Distinct Transcriptome Reprogramming in Resistant and Susceptible Isogenic Tomato Lines. BMC Plant Biol. 2016, 16, 53. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wei, L.; Liu, T.; Ma, J.; Huang, K.; Guo, H.; Huang, Y.; Zhang, L.; Zhao, J.; Tsuda, K.; et al. Suppression of ETI by PTI Priming to Balance Plant Growth and Defense through an MPK3/MPK6-WRKYs-PP2Cs Module. Mol. Plant 2023, 16, 903–918. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Djamei, A.; Bitton, F.; Hirt, H. A Major Role of the MEKK1-MKK1/2-MPK4 Pathway in ROS Signalling. Mol. Plant 2009, 2, 120–137. [Google Scholar] [CrossRef]
- Shubchynskyy, V.; Boniecka, J.; Schweighofer, A.; Simulis, J.; Kvederaviciute, K.; Stumpe, M.; Mauch, F.; Balazadeh, S.; Mueller-Roeber, B.; Boutrot, F.; et al. Protein Phosphatase AP2C1 Negatively Regulates Basal Resistance and Defense Responses to Pseudomonas syringae. J. Exp. Bot. 2017, 68, 1169–1183. [Google Scholar] [CrossRef][Green Version]
- Douchkov, D.; Lueck, S.; Hensel, G.; Kumlehn, J.; Rajaraman, J.; Johrde, A.; Doblin, M.S.; Beahan, C.T.; Kopischke, M.; Fuchs, R.; et al. The Barley (Hordeum vulgare) Cellulose Synthase-like D2 Gene (HvCslD2) Mediates Penetration Resistance to Host-Adapted and Nonhost Isolates of the Powdery Mildew Fungus. New Phytol. 2016, 212, 421–433. [Google Scholar] [CrossRef]
- Edgar, C.I.; McGrath, K.C.; Dombrecht, B.; Manners, J.M.; Maclean, D.C.; Schenk, P.M.; Kazan, K. Salicylic Acid Mediates Resistance to the Vascular Wilt Pathogen Fusarium oxysporum in the Model Host Arabidopsis thaliana. Austral. Plant Pathol. 2006, 35, 581–591. [Google Scholar] [CrossRef]
- Bollhoner, B.; Zhang, B.; Stael, S.; Denance, N.; Overmyer, K.; Goffner, D.; Van Breusegem, F.; Tuominen, H. Post mortem Function of AtMC9 in Xylem Vessel Elements. New Phytol. 2013, 200, 498–510. [Google Scholar] [CrossRef]
- Zhang, M.; Xu, J.; Liu, G.; Yang, X. Antifungal Properties of a Thaumatin-like Protein from Watermelon. Acta Physiol. Plant. 2018, 40, 186. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Vanden Bossche, R.; Sewell, J.; Gil, E.; et al. NINJA Connects the Co-Repressor TOPLESS to Jasmonate Signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Han, Q.; Chen, R.; Yang, Y.; Cui, X.; Ge, F.; Chen, C.; Liu, D. A Glutathione S-Transferase Gene from Lilium regale Wilson Confers Transgenic Tobacco Resistance to Fusarium oxysporum. Sci. Hortic. 2016, 198, 370–378. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L.; Jander, G.; Bhawal, R.; Zhang, S.; Liu, Z.; Oakley, A.; Hua, J. AIG2A and AIG2B Limit the Activation of Salicylic Acid-Regulated Defenses by Tryptophan-Derived Secondary Metabolism in Arabidopsis. Plant Cell 2022, 34, 4641–4660. [Google Scholar] [CrossRef] [PubMed]
- Diao, J.; Li, M.; Zhang, P.; Zong, C.; Ma, W.; Ma, L. Overexpression of the PdpapERF109 Gene Enhances Resistance of Populus davidiana x P. alba Var. Pyramidalis to Fusarium oxysporum Infection. J. For. Res. 2022, 33, 1925–1937. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, D.; Zheng, W.; He, H.; Ji, B.; Han, Q.; Ge, F.; Chen, C. A bZIP Transcription Factor, LrbZIP1, Is Involved in Lilium regale Wilson Defense Responses against Fusarium oxysporum f. sp. Lilii. Genes Genom. 2014, 36, 789–798. [Google Scholar] [CrossRef]
- Yue, Z.-L.; Tian, Z.-J.; Zhang, J.-W.; Zhang, S.-W.; Li, Y.-D.; Wu, Z.-M. Overexpression of Lectin Receptor-Like Kinase 1 in Tomato Confers Resistance to Fusarium oxysporum f. sp. Radicis-Lycopersici. Front. Plant Sci. 2022, 13, 836269. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, Z.; Li, X.; Cheng, Q.; Li, R. Two Sugarcane Expansin Protein-Coding Genes Contribute to Stomatal Aperture Associated with Structural Resistance to Sugarcane Smut. J. Fungi 2024, 10, 631. [Google Scholar] [CrossRef]
- La Camera, S.; Balague, C.; Goebel, C.; Geoffroy, P.; Legrand, M.; Feussner, I.; Roby, D.; Heitz, T. The Arabidopsis Patatin-Like Protein 2 (PLP2) Plays an Essential Role in Cell Death Execution and Differentially Affects Biosynthesis of Oxylipins and Resistance to Pathogens. Mol. Plant-Microbe Interact. 2009, 22, 469–481. [Google Scholar] [CrossRef]
- Yu, X.; Cui, X.; Wu, C.; Shi, S.; Yan, S. Salicylic Acid Inhibits Gibberellin Signaling through Receptor Interactions. Mol. Plant 2022, 15, 1759–1771. [Google Scholar] [CrossRef]
- Wang, H.; Li, P.; Wang, Y.; Chi, C.; Jin, X.; Ding, G. Overexpression of Cucumber CYP82D47 Enhances Resistance to Powdery Mildew and Fusarium oxysporum f. sp. Cucumerinum. Funct. Integr. Genom. 2024, 24, 14. [Google Scholar] [CrossRef]
- Zhu, X.; Caplan, J.; Mamillapalli, P.; Czymmek, K.; Dinesh-Kumar, S.P. Function of Endoplasmic Reticulum Calcium ATPase in Innate Immunity-Mediated Programmed Cell Death. Embo J. 2010, 29, 1007–1018. [Google Scholar] [CrossRef]
- Leslie, M.E.; Rogers, S.W.; Heese, A. Increased Callose Deposition in Plants Lacking DYNAMIN-RELATED PROTEIN 2B Is Dependent upon POWDERY MILDEW RESISTANT 4. Plant Signal. Behav. 2016, 11, e1244594. [Google Scholar] [CrossRef]
- Kotera, Y.; Komori, H.; Tasaki, K.; Takagi, K.; Imano, S.; Katou, S. The Peroxisomal β-Oxidative Pathway and Benzyl Alcohol O-Benzoyltransferase HSR201 Cooperatively Contribute to the Biosynthesis of Salicylic Acid. Plant Cell Physiol. 2023, 64, 758–770. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).