Induction and Characterisation of Lignocellulolytic Activities from Novel Deep-Sea Fungal Secretomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates Used
2.2. Cultivation of Fungi and Supernatant Harvesting
2.3. Xylanase Activity Measurements
2.4. Qualitative Plate-Based Assays for Ligninolytic Enzyme Activity
Polyphenol-Oxidase/Peroxidase-like Activity
2.5. Exo-Glycoside Hydrolase and Feruloyl Esterase Activity Measurements
2.6. Electrophoresis and Zymography
2.7. Statistics
3. Results and Discussion
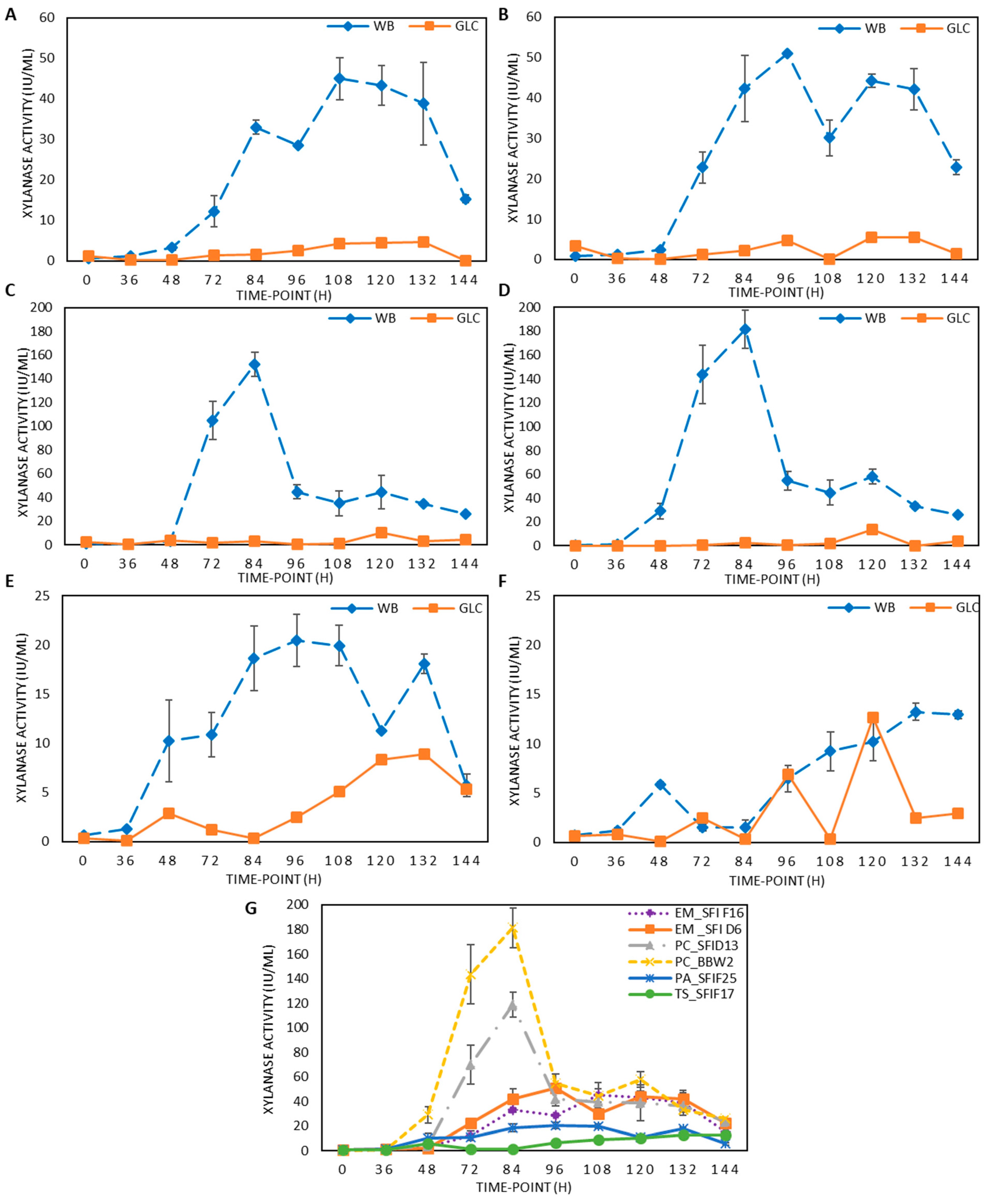
3.1. Xylanase Induction by WB during LSF, and the Qualitative Assessment of Lignin-Degrading Activities
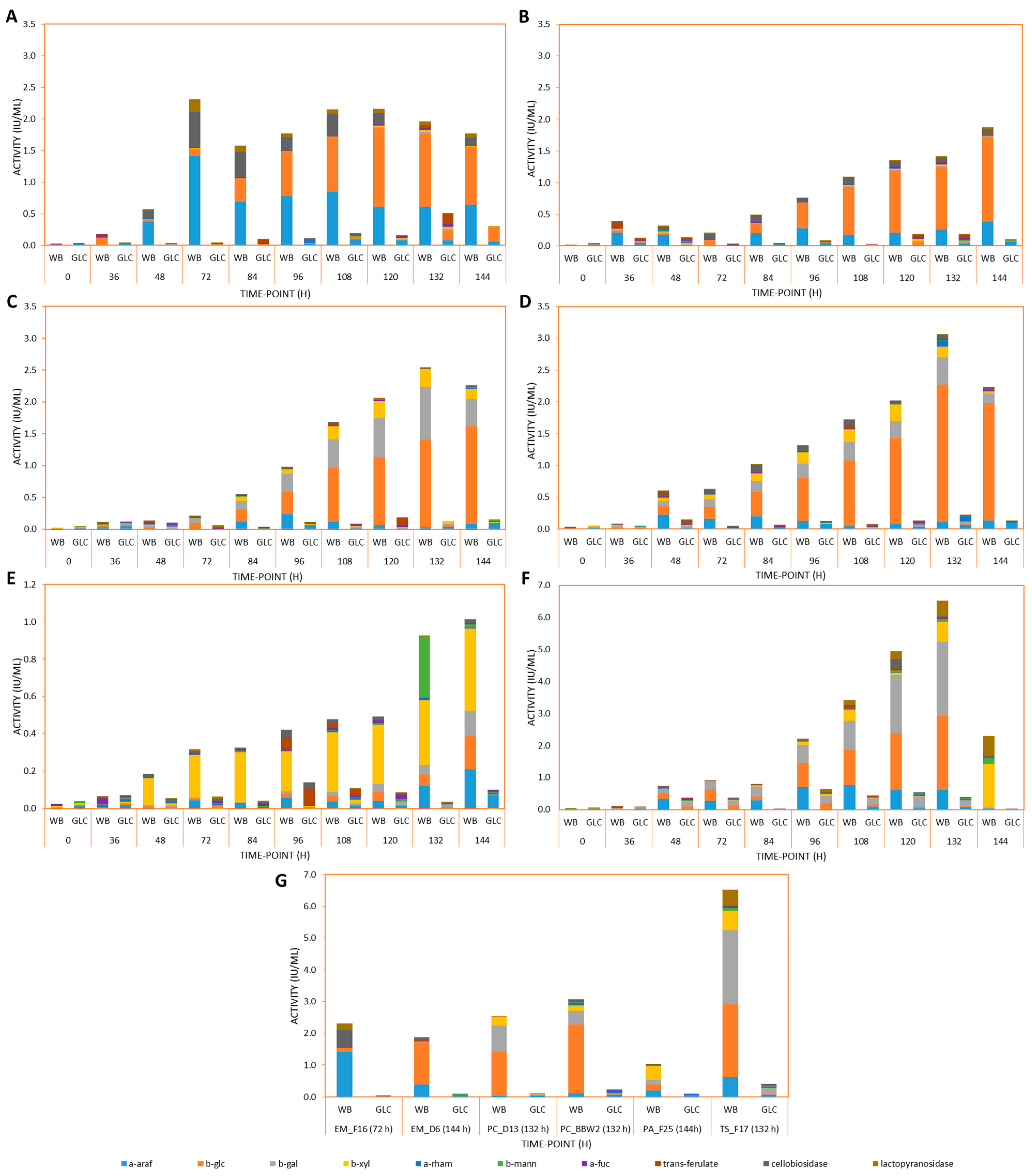
3.2. Exo-Acting Glycoside Hydrolase and Feruloyl Esterase Induction by WB during LSF
3.3. Effect of Temperature and pH on Xylanase Activity in the Fungal Secretomes
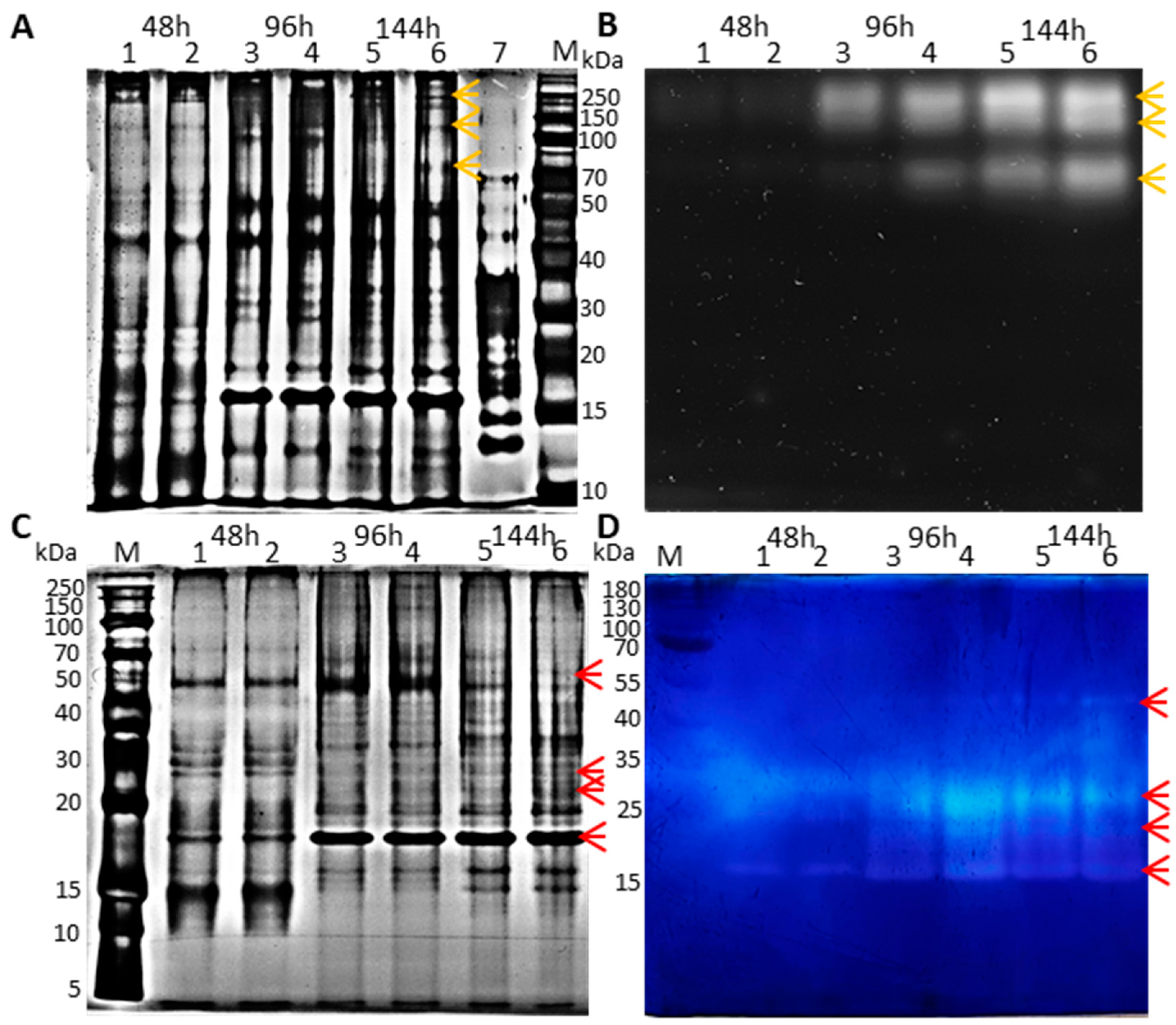
3.4. Electrophoretic Separation of Proteins in the Crude Secretomes and Zymogram Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Birolli, W.G.; Lima, R.N.; Porto, A.L.M. Applications of Marine-Derived Microorganisms and Their Enzymes in Biocatalysis and Biotransformation, the Underexplored Potentials. Front. Microbiol. 2019, 10, 1453. [Google Scholar] [CrossRef] [PubMed]
- Chávez, R.; Fierro, F.; García-Rico, R.O.; Vaca, I. Filamentous Fungi from Extreme Environments as a Promising Source of Novel Bioactive Secondary Metabolites. Front. Microbiol. 2015, 6, 903. [Google Scholar] [CrossRef]
- Marchese, P.; Garzoli, L.; Young, R.; Allcock, L.; Barry, F.; Tuohy, M.; Murphy, M. Fungi Populate Deep-Sea Coral Gardens as Well as Marine Sediments in the Irish Atlantic Ocean. Environ. Microbiol. 2021, 23, 4168–4184. [Google Scholar] [CrossRef] [PubMed]
- Coleine, C.; Stajich, J.E.; Selbmann, L. Fungi Are Key Players in Extreme Ecosystems. Trends Ecol. Evol. 2022, 37, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Nagano, Y.; Nagahama, T. Fungal Diversity in Deep-Sea Extreme Environments. Fungal Ecol. 2012, 5, 463–471. [Google Scholar] [CrossRef]
- Rédou, V.; Navarri, M.; Meslet-Cladière, L.; Barbier, G.; Burgaud, G. Species Richness and Adaptation of Marine Fungi from Deep-Subseafloor Sediments. Appl. Environ. Microbiol. 2015, 81, 3571–3583. [Google Scholar] [CrossRef]
- Batista-García, R.A.; Sutton, T.; Jackson, S.A.; Tovar-Herrera, O.E.; Balcázar-López, E.; del Rayo Sánchez-Carbente, M.; Sánchez-Reyes, A.; Dobson, A.D.W.; Folch-Mallol, J.L. Characterization of Lignocellulolytic Activities from Fungi Isolated from the Deep-Sea Sponge Stelletta normani. PLoS ONE 2017, 12, e0173750. [Google Scholar] [CrossRef]
- Hu, X.Y.; Li, X.M.; Wang, B.G.; Meng, L.H. Tanzawaic Acid Derivatives: Fungal Polyketides from the Deep-Sea Coral-Derived Endozoic Penicillium steckii AS-324. J. Nat. Prod. 2022, 85, 1398–1406. [Google Scholar] [CrossRef]
- Cheng, Z.; Liu, W.; Fan, R.; Han, S.; Li, Y.; Cui, X.; Zhang, J.; Wu, Y.; Lv, X.; Zhang, Y.; et al. Terpenoids from the Deep-Sea-Derived Fungus Penicillium thomii YPGA3 and Their Bioactivities. Mar. Drugs 2020, 18, 164. [Google Scholar] [CrossRef]
- Limbadri, S.; Luo, X.; Lin, X.; Liao, S.; Wang, J.; Zhou, X.; Yang, B.; Liu, Y. Bioactive Novel Indole Alkaloids and Steroids from Deep Sea-Derived Fungus Aspergillus fumigatus SCSIO 41012. Molecules 2018, 23, 2379. [Google Scholar] [CrossRef]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose Degradation: An Overview of Fungi and Fungal Enzymes Involved in Lignocellulose Degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Abdeshahian, P.; Kadier, A.; Rai, P.K.; Silva, S.S. Lignocellulose as a Renewable Carbon Source for Microbial Synthesis of Different Enzymes. In Lignocellulosic Biorefining Technologies; Ingle, A.P., Chandel, A.K., da Silva, S.S., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2020; pp. 185–202. [Google Scholar]
- Howard, R.L.; Abotsi, E.; Van Rensburg, E.L.J.; Howard, S. Lignocellulose Biotechnology: Issues of Bioconversion and Enzyme Production. Afr. J. Biotechnol. 2003, 2, 702–733. [Google Scholar] [CrossRef]
- Smith, M.D. An Abbreviated Historical and Structural Introduction to Lignocellulose. In Understanding Lignocellulose: Synergistic Computational and Analytic Methods; American Chemical Society: Washington, DC, USA, 2019; Volume 1338, pp. 1–15. [Google Scholar] [CrossRef]
- Dowd, B.; McDonnell, D.; Tuohy, M.G. Current Progress in Optimising Sustainable Energy Recovery From Cattle Paunch Contents, a Slaughterhouse Waste Product. Front. Sustain. Food Syst. 2022, 6, 722424. [Google Scholar] [CrossRef]
- Saini, J.K.; Saini, R.; Tewari, L. Lignocellulosic Agriculture Wastes as Biomass Feedstocks for Second-Generation Bioethanol Production: Concepts and Recent Developments. 3 Biotech 2015, 5, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Raghukumar, C.; Muraleedharan, U.; Gaud, V.R.; Mishra, R. Xylanases of Marine Fungi of Potential Use for Biobleaching of Paper Pulp. J. Ind. Microbiol. Biotechnol. 2004, 31, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Damare, S.; Raghukumar, C.; Muraleedharan, U.D.; Raghukumar, S. Deep-Sea Fungi as a Source of Alkaline and Cold-Tolerant Proteases. Enzyme Microb. Technol. 2006, 39, 172–181. [Google Scholar] [CrossRef]
- National Oceanic and Atmospheric Administration-Ocean Acidification. Available online: https://www.noaa.gov/education/resource-collections/ocean-coasts/ocean-acidification (accessed on 15 June 2023).
- Hagestad, O.C.; Hou, L.; Andersen, J.H.; Hansen, E.H.; Altermark, B.; Li, C.; Kuhnert, E.; Cox, R.J.; Crous, P.W.; Spatafora, J.W.; et al. Genomic Characterization of Three Marine Fungi, Including Emericellopsis atlantica Sp. Nov. with Signatures of a Generalist Lifestyle and Marine Biomass Degradation. IMA Fungus 2021, 12, 21. [Google Scholar] [CrossRef]
- Ruginescu, R.; Enache, M.; Popescu, O.; Gomoiu, I.; Cojoc, R.; Batrinescu-Moteau, C.; Maria, G.; Dumbravician, M.; Neagu, S. Characterization of Some Salt-Tolerant Bacterial Hydrolases with Potential Utility in Cultural Heritage Bio-Cleaning. Microorganisms 2022, 10, 644. [Google Scholar] [CrossRef]
- Terrone, C.C.; de Freitas, C.; Terrasan, C.R.F.; de Almeida, A.F.; Carmona, E.C. Agroindustrial Biomass for Xylanase Production by Penicillium chrysogenum: Purification, Biochemical Properties and Hydrolysis of Hemicelluloses. Electron. J. Biotechnol. 2018, 33, 39–45. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, J.; Wang, R.; Liu, J.; Zhang, Y.; Liu, L.; Wang, F.; Yuan, H. Cooperation of Hydrolysis Modes among Xylanases Reveals the Mechanism of Hemicellulose Hydrolysis by Penicillium chrysogenum P33. Microb. Cell Fact. 2019, 18, 159. [Google Scholar] [CrossRef]
- Haas, H.; Herfurth, E.; Stöffler, G.; Redl, B. Purification, Characterization and Partial Amino Acid Sequences of a Xylanase Produced by Penicillium chrysogenum. Biochim. Biophys. Acta Gen. Subj. 1992, 1117, 279–286. [Google Scholar] [CrossRef]
- Ullah, S.F.; Souza, A.A.; Hamann, P.R.V.; Ticona, A.R.P.; Oliveira, G.M.; Barbosa, J.A.R.G.; Freitas, S.M.; Noronha, E.F. Structural and Functional Characterisation of Xylanase Purified from Penicillium chrysogenum Produced in Response to Raw Agricultural Waste. Int. J. Biol. Macromol. 2019, 127, 385–395. [Google Scholar] [CrossRef]
- Leshchenko, E.V.; Antonov, A.S.; Borkunov, G.V.; Hauschild, J.; Zhuravleva, O.I.; Khudyakova, Y.V.; Menshov, A.S.; Popov, R.S.; Kim, N.Y.; Graefen, M.; et al. New Bioactive β-Resorcylic Acid Derivatives from the Alga-Derived Fungus Penicillium antarcticum KMM 4685. Mar. Drugs 2023, 21, 178. [Google Scholar] [CrossRef] [PubMed]
- Leshchenko, E.V.; Antonov, A.S.; Dyshlovoy, S.A.; Berdyshev, D.V.; Hauschild, J.; Zhuravleva, O.I.; Borkunov, G.V.; Menshov, A.S.; Kirichuk, N.N.; Popov, R.S.; et al. Meroantarctines A-C, Meroterpenoids with Rearranged Skeletons from the Alga-Derived Fungus Penicillium antarcticum KMM 4685 with Potent p-Glycoprotein Inhibitory Activity. J. Nat. Prod. 2022, 85, 2746–2752. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Zhang, H.; Cui, H.; Davari, M.D.; Wei, B.; Wang, W.; Yuan, Q. Efficient Enzyme-Catalyzed Production of Diosgenin: Inspired by the Biotransformation Mechanisms of Steroid Saponins in: Talaromyces stollii CLY-6. Green Chem. 2021, 23, 5896–5910. [Google Scholar] [CrossRef]
- Waters, D.M.; Murray, P.G.; Ryan, L.A.; Arendt, E.K.; Tuohy, M.G. Talaromyces Emersonii Thermostable Enzyme Systems and Their Applications in Wheat Baking Systems. J. Agric. Food Chem. 2010, 58, 7415–7422. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Pointing, S.B. Qualitative Methods for the Determination of Lignocellulolytic Enzyme Production by Tropical Fungi. Fungal Divers. 1999, 2, 17–33. [Google Scholar]
- Pan, D.; Wilson, K.A.; Tan-Wilson, A. Transfer Zymography. Methods Mol. Biol. 2017, 1626, 253–269. [Google Scholar] [CrossRef]
- McCarthy, T.; Tuohy, M.G. A Multi-Step Chromatographic Strategy to Purify Three Fungal Endo-β-Glucanases. Methods Mol. Biol. 2011, 681, 497–524. [Google Scholar] [CrossRef]
- Grudkowska, M.; Lisik, P.; Rybka, K. Two-dimensional zymography in detection of proteolytic enzymes in wheat leaves. Acta Physiol. Plant. 2013, 35, 3477–3482. [Google Scholar] [CrossRef]
- Minic, Z.; Jouanin, L. Plant Glycoside Hydrolases Involved in Cell Wall Polysaccharide Degradation. Plant Physiol. Biochem. 2006, 44, 435–449. [Google Scholar] [CrossRef]
- Olfa, E.; Mondher, M.; Issam, S.; Ferid, L.; Nejib, M.M. Induction, Properties and Application of Xylanase Activity from Sclerotinia sclerotiorum S2 Fungus. J. Food Biochem. 2007, 31, 96–107. [Google Scholar] [CrossRef]
- Gaffney, M.; Doyle, S.; Murphy, R. Optimization of Xylanase Production by Thermomyces lanuginosus in Solid State Fermentation. Biosci. Biotechnol. Biochem. 2009, 73, 2640–2644. [Google Scholar] [CrossRef]
- Kumar, B.A.; Amit, K.; Alok, K.; Dharm, D. Wheat Bran Fermentation for the Production of Cellulase and Xylanase by Aspergillus niger NFCCI 4113. Res. J. Biotechnol. 2018, 13, 11–18. [Google Scholar]
- Barbieri, G.S.; Bento, H.B.S.; de Oliveira, F.; Picheli, F.P.; Dias, L.M.; Masarin, F.; Santos-Ebinuma, V.C. Xylanase Production by Talaromyces amestolkiae Valuing Agroindustrial Byproducts. BioTech 2022, 11, 15. [Google Scholar] [CrossRef]
- Shuster, L. Repression and De-Repression of Enzyme Synthesis as a Possible Explanation of some Aspects of Drug Action. Nature 1961, 189, 314–315. [Google Scholar] [CrossRef]
- Purkarthofer, H.; Sinner, M.; Steiner, W. Cellulase-free xylanase from Optimization of production in Thermomyces lanuginosus: Submerged and solid-state culture. Enzyme Microb. Technol. 1993, 15, 677–682. [Google Scholar] [CrossRef]
- Mach-Aigner, A.R.; Pucher, M.E.; Mach, R.L. D-Xylose as a Repressor or Inducer of Xylanase Expression in Hypocrea jecorina (Trichoderma reesei). Appl. Environ. Microbiol. 2010, 76, 1770–1776. [Google Scholar] [CrossRef]
- Daly, P.; Peng, M.; Di Falco, M.; Lipzen, A.; Wang, M.; Ng, V.; Grigoriev, I.V.; Tsang, A.; Mäkelä, M.R.; de Vries, R.P. Glucose-Mediated Repression of Plant Biomass Utilization in the White-Rot Fungus Dichomitus squalens. Appl. Environ. Microbiol. 2019, 85, e01828-19. [Google Scholar] [CrossRef] [PubMed]
- Khanahmadi, M.; Arezi, I.; Amiri, M.S.; Miranzadeh, M. Bioprocessing of agro-industrial residues for optimization of xylanase production by solid- state fermentation in flask and tray bioreactor. Biocatal. Agric. Biotechnol. 2018, 13, 272–282. [Google Scholar] [CrossRef]
- Okafor, U.A.; Emezue, T.N.; Okochi, V.I.; Onyegeme-Okerenta, B.M.; Nwodo-Chinedu, S. Xylanase Production by Penicillium chrysogenum (PCL501) Fermented on Cellulosic Wastes. Afr. J. Biochem. Res. 2007, 1, 48–053. [Google Scholar]
- Bajaj, B.K.; Sharma, M.; Sharma, S. Alkalistable Endo-β-1,4-Xylanase Production from a Newly Isolated Alkalitolerant Penicillium Sp. SS1 Using Agro-Residues. 3 Biotech 2011, 1, 83–90. [Google Scholar] [CrossRef][Green Version]
- Hou, Y.-H.; Wang, T.-H.; Long, H.; Zhu, H.-Y. Novel Cold-adaptive Penicillium Strain FS010 Secreting Thermo-labile Xylanase Isolated from Yellow Sea. Acta Biochim. Biophys. Sin. 2006, 38, 142–149. [Google Scholar] [CrossRef]
- Orencio-Trejo, M.; Torres-Granados, J.; Rangel-Lara, A.; Beltrán-Guerrero, E.; García-Aguilar, S.; Moss-Acosta, C.; Valenzuela-Soto, H.; De la Torre-Zavala, S.; Gastelum-Arellanez, A.; Martinez, A.; et al. Cellulase and Xylanase Production by the Mexican Strain Talaromyces stollii LV186 and Its Application in the Saccharification of Pretreated Corn and Sorghum Stover. Bioenergy Res. 2016, 9, 1034–1045. [Google Scholar] [CrossRef]
- Zuccaro, A.; Summerbell, R.C.; Gams, W.; Schroers, H.J.; Mitchell, J.I. A New Acremonium Species Associated with Fucus Spp., and Its Affinity with a Phylogenetically Distinct Marine Emericellopsis Clade. Stud. Mycol. 2004, 50, 283–297. [Google Scholar]
- Petruccioli, M.; Fenice, M.; Piccioni, P. Distribution and Typology of Glucose Oxidase Activity in the Genus Penicillium. Lett. Appl. Microbiol. 1993, 17, 285–288. [Google Scholar] [CrossRef]
- Hao, D.C.; Song, S.M.; Cheng, Y.; Qin, Z.Q.; Ge, G.B.; An, B.L.; Xiao, P.G. Functional and Transcriptomic Characterization of a Dye-Decolorizing Fungus from Taxus Rhizosphere. Pol. J. Microbiol. 2018, 67, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Zhang, H.; Cui, H.; Cheng, J.; Wang, W.; Wei, B.; Liu, F.; Liang, H.; Shen, X.; Yuan, Q. A Novel α-L-Rhamnosidase Renders Efficient and Clean Production of Icaritin. J. Clean. Prod. 2022, 341, 130903. [Google Scholar] [CrossRef]
- Gil, J. Estudio de Las β-Glucosidasas Del Complejo Celulítico de Talaromyces amestolkiae: Caracterización y Aplicaciones Biotecnológicas. Ph.D. Thesis, Universidad Complutense, Madrid, Spain, 2015. [Google Scholar]
- Nieto-Domínguez, M.; de Eugenio, L.I.; Barriuso, J.; Prieto, A.; Fernández de Toro, B.; Canales-Mayordomo, Á.; Martínez, M.J. Novel PH-Stable Glycoside Hydrolase Family 3 β-Xylosidase from Talaromyces amestolkiae: An Enzyme Displaying Regioselective Transxylosylation. Appl. Environ. Microbiol. 2015, 81, 6380–6392. [Google Scholar] [CrossRef][Green Version]
- Subramaniyan, S.; Prema, P. Biotechnology of Microbial Xylanases: Enzymology, Molecular Biology, and Application. Crit. Rev. Biotechnol. 2002, 22, 33–64. [Google Scholar] [CrossRef] [PubMed]
- dos Reis, L.; Schneider, W.D.H.; Fontana, R.C.; Camassola, M.; Dillon, A.J.P. Cellulase and Xylanase Expression in Response to Different pH Levels of Penicillium echinulatum S1M29 Medium. Bioenergy Res. 2014, 7, 60–67. [Google Scholar] [CrossRef]
- Sunkar, B.; Kannoju, B.; Bhukya, B. Optimized Production of Xylanase by Penicillium purpurogenum and Ultrasound Impact on Enzyme Kinetics for the Production of Monomeric Sugars From Pretreated Corn Cobs. Front. Microbiol. 2020, 11, 772. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, N.; Shendye, A.; Rao, M. Molecular and Biotechnological Aspects of Xylanases. FEMS Microbiol. Rev. 1999, 23, 411–456. [Google Scholar] [CrossRef]
- Bates, N.R.; Best, M.H.P.; Neely, K.; Garley, R.; Dickson, A.G.; Johnson, R.J. Detecting Anthropogenic Carbon Dioxide Uptake and Ocean Acidification in the North Atlantic Ocean. Biogeosciences 2012, 9, 2509–2522. [Google Scholar] [CrossRef]
- Arif, A.R.; Natsir, H.; Rohani, H.; Karim, A. Effect of pH fermentation on production bioethanol from jackfruit seeds (Artocarpus heterophyllus) through separate fermentation hydrolysis method. J. Phys. Conf. Ser. 2018, 979, 012015. [Google Scholar] [CrossRef]
- Corradini, F.A.S.; Milessi, T.S.; Gonçalves, V.M.; Ruller, R.; Sargo, C.R.; Lopes, L.A.; Zangirolami, T.C.; Tardioli, P.W.; Giordano, R.C.; Giordano, R.L.C. High stabilization and hyperactivation of a Recombinant β-Xylosidase through Immobilization Strategies. Enzyme Microb. Technol. 2021, 145, 109725. [Google Scholar] [CrossRef] [PubMed]
- Kuvarina, A.E.; Gavryushina, I.A.; Kulko, A.B.; Ivanov, I.A.; Rogozhin, E.A.; Georgieva, M.L.; Sadykova, V.S. The Emericellipsins a-e from an Alkalophilic Fungus Emericellopsis alkalina Show Potent Activity against Multidrug-Resistant Pathogenic Fungi. J. Fungi 2021, 7, 153. [Google Scholar] [CrossRef]
- Kuvarina, A.E.; Sukonnikov, M.A.; Rogozhin, E.A.; Serebryakova, M.V.; Timofeeva, A.V.; Georgieva, M.L.; Sadykova, V.S. Formation of Various Antimicrobial Peptide Emericellipsin Isoforms in Emericellopsos alkalina under Different Cultivation Conditions. Appl. Biochem. Microbiol. 2023, 59, 160–167. [Google Scholar] [CrossRef]
- Kuvarina, A.E.; Gavryushina, I.A.; Sykonnikov, M.A.; Efimenko, T.A.; Markelova, N.N.; Bilanenko, E.N.; Bondarenko, S.A.; Kokaeva, L.Y.; Timofeeva, A.V.; Serebryakova, M.V.; et al. Exploring Peptaibol’s Profile, Antifungal, and Antitumor Activity of Emericellipsin A of Emericellopsis Species from Soda and Saline Soils. Molecules 2022, 27, 1736. [Google Scholar] [CrossRef]
- Rogozhin, E.A.; Sadykova, V.S.; Baranova, A.A.; Vasilchenko, A.S.; Lushpa, V.A.; Mineev, K.S.; Georgieva, M.L.; Kul’ko, A.B.; Krasheninnikov, M.E.; Lyundup, A.V.; et al. A Novel Lipopeptaibol Emericellipsin a with Antimicrobial and Antitumor Activity Produced by the Extremophilic Fungus Emericellopsis alkalina. Molecules 2018, 23, 2785. [Google Scholar] [CrossRef] [PubMed]
- Abraúl, M.; Alves, A.; Hilário, S.; Melo, T.; Conde, T.; Domingues, M.R.; Rey, F. Evaluation of Lipid Extracts from the Marine Fungi Emericellopsis cladophorae and Zalerion maritima as a Source of Anti-Inflammatory, Antioxidant and Antibacterial Compounds. Mar. Drugs 2023, 21, 199. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Dufossé, L.; Deshmukh, S.K. Antibacterial Metabolites from an Unexplored Strain of Marine Fungi Emericellopsis minima and Determination of the Probable Mode of Action against Staphylococcus aureus and Methicillin-Resistant S. aureus. Biotechnol. Appl. Biochem. 2023, 70, 120–129. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Paço, A.; Escada, L.F.; Albuquerque, M.S.F.; Pinto, C.A.; Saraiva, J.; Duarte, A.S.; Rocha-Santos, T.A.P.; Esteves, A.C.; Alves, A. Unveiling Biological Activities of Marine Fungi: The Effect of Sea Salt. Appl. Sci. 2021, 11, 6008. [Google Scholar] [CrossRef]
- Gomoiu, I.; Cojoc, R.; Ruginescu, R.; Neagu, S.; Enache, M.; Maria, G.; Dumbrăvician, M.; Olteanu, I.; Rădvan, R.; Ratoiu, L.C.; et al. Brackish and Hypersaline Lakes as Potential Reservoir for Enzymes Involved in Decomposition of Organic Materials on Frescoes. Fermentation 2022, 8, 462. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Hilário, S.; Van de Peer, Y.; Esteves, A.C.; Alves, A. Genomic and Metabolomic Analyses of the Marine Fungus Emericellopsis cladophorae: Insights into Saltwater Adaptability Mechanisms and Its Biosynthetic Potential. J. Fungi 2022, 8, 31. [Google Scholar] [CrossRef]
- Tuohy, M.G.; Laffey, C.D.; Coughlan, M.P. Characterization of the Individual Components of the Xylanolytic Enzyme System of Talaromyces emersonii. Bioresour. Technol. 1994, 50, 37–42. [Google Scholar] [CrossRef]
- Liao, H.; Xu, C.; Tan, S.; Wei, Z.; Ling, N.; Yu, G.; Raza, W.; Zhang, R.; Shen, Q.; Xu, Y. Production and Characterization of Acidophilic Xylanolytic Enzymes from Penicillium oxalicum GZ-2. Bioresour. Technol. 2012, 123, 117–124. [Google Scholar] [CrossRef]
- Ang, S.K.; Shaza, E.M.; Adibah, Y.A.; Suraini, A.A.; Madihah, M.S. Production of Cellulases and Xylanase by Aspergillus fumigatus SK1 Using Untreated Oil Palm Trunk through Solid State Fermentation. Process Biochem. 2013, 48, 1293–1302. [Google Scholar] [CrossRef]
- Miao, Y.; Li, J.; Xiao, Z.; Shen, Q.; Zhang, R. Characterization and Identification of the Xylanolytic Enzymes from Aspergillus fumigatus Z5. BMC Microbiol. 2015, 15, 126. [Google Scholar] [CrossRef] [PubMed]
- Chipeta, Z.A.; du Preez, J.C.; Szakacs, J.; Christopher, L. Xylanase Production by Fungal Strains on Spent Sulphite Liquor. Appl. Microbiol. Biotechnol. 2005, 69, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.; Khare, S.K. Induction of Xylanase in Thermophilic Fungi Scytalidium thermophilum and Sporotrichum Thermophile. Braz. Arch. Biol. Technol. 2012, 55, 21–27. [Google Scholar] [CrossRef][Green Version]
- Lin, C.; Shen, Z.; Zhu, T.; Qin, W. Newly Isolated Penicillium ramulosum N1 Is Excellent for Producing Protease-Resistant Acidophilic Xylanase. J. Mol. Microbiol. Biotechnol. 2015, 25, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Badhan, A.K.; Chadha, B.S.; Sonia, K.G.; Saini, H.S.; Bhat, M.K. Functionally Diverse Multiple Xylanases of Thermophilic Fungus Myceliophthora Sp. IMI 387099. Enzyme Microb Technol. 2004, 35, 460–466. [Google Scholar] [CrossRef]
- Sachslehner, A.; Nidetzky, B.; Kulbe, K.D.; Haltrich, D. Induction of Mannanase, Xylanase, and Endoglucanase Activities in Sclerotium rolfsii. Appl. Environ. Microbiol. 1998, 64, 594–600. [Google Scholar] [CrossRef]
- Bachmann, S.L.; Mccarthy, A.J. Purification and Cooperative Activity of Enzymes Constituting the Xylan-Degrading System of Thermomonospora fusca. Appl. Environ. Microbiol. 1991, 57, 2121–2130. [Google Scholar] [CrossRef]
- Álvarez-Cervantes, J.; Domínguez-Hernández, E.M.; Mercado-Flores, Y.; O’Donovan, A.; Díaz-Godínez, G. Mycosphere Essay 10: Properties and Characteristics of Microbial Xylanases. Mycosphere 2016, 7, 1600–1619. [Google Scholar] [CrossRef]
- Hong, P.Y.; Iakiviak, M.; Dodd, D.; Zhang, M.; Mackie, R.I.; Cann, I. Two New Xylanases with Different Substrate Specificities from the Human Gut Bacterium Bacteroides intestinalis DSM 17393. Appl. Environ. Microbiol. 2014, 80, 2084–2093. [Google Scholar] [CrossRef]
- Knob, A.; Terrasan, C.R.F.; Carmona, E.C. β-Xylosidases from Filamentous Fungi: An Overview. World J. Microbiol. Biotechnol. 2010, 26, 389–407. [Google Scholar] [CrossRef]
- Mamo, G.; Thunnissen, M.; Hatti-Kaul, R.; Mattiasson, B. An Alkaline Active Xylanase: Insights into Mechanisms of High pH Catalytic Adaptation. Biochimie 2009, 91, 1187–1196. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, J.; Liu, J.; Wang, R.; Liu, L.; Wang, F.; Yuan, H. The Composition of Accessory Enzymes of Penicillium chrysogenum P33 Revealed by Secretome and Synergistic Effects with Commercial Cellulase on Lignocellulose Hydrolysis. Bioresour. Technol. 2018, 257, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Tuohy, M.G.; Murray, G.P.; Gilleran, C.T.; Collins, C.T.; Reen, F.J.; McLoughlin, L.P.; Lydon, A.G.S.; Maloney, A.P.; Heneghan, M.N.; O’Donoghue, A.G.; et al. Talaromyces emersonii Xylanase. European Patent No. 11157751.6, 7 September 2011. [Google Scholar]
- Guerfali, M.; Gargouri, A.; Belghith, H. Talaromyces thermophilus β-d-Xylosidase: Purification, Characterization and Xylobiose Synthesis. Appl. Biochem. Biotechnol. 2008, 150, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Dutta, T.; Sarkar, T.S.; Ghosh, S. Novel Xylanases from Simplicillium obclavatum MTCC 9604: Comparative Analysis of Production, Purification and Characterization of Enzyme from Submerged and Solid State Fermentation. SpringerPlus 2013, 2, 382. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.; Grinyer, J.; Nevalainen, H. Secretome of the Coprophilous Fungus Doratomyces stemonitis C8, Isolated from Koala Feces. Appl. Environ. Microbiol. 2011, 77, 3793–3801. [Google Scholar] [CrossRef]
- Sethupathy, S.; Morales, G.M.; Li, Y.; Wang, Y.; Jiang, J.; Sun, J.; Zhu, D. Harnessing Microbial Wealth for Lignocellulose Biomass Valorization through Secretomics: A Review. Biotechnol. Biofuels 2021, 14, 154. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dowd, B.; Tuohy, M.G. Induction and Characterisation of Lignocellulolytic Activities from Novel Deep-Sea Fungal Secretomes. Fermentation 2023, 9, 780. https://doi.org/10.3390/fermentation9090780
Dowd B, Tuohy MG. Induction and Characterisation of Lignocellulolytic Activities from Novel Deep-Sea Fungal Secretomes. Fermentation. 2023; 9(9):780. https://doi.org/10.3390/fermentation9090780
Chicago/Turabian StyleDowd, Bronwyn, and Maria G. Tuohy. 2023. "Induction and Characterisation of Lignocellulolytic Activities from Novel Deep-Sea Fungal Secretomes" Fermentation 9, no. 9: 780. https://doi.org/10.3390/fermentation9090780
APA StyleDowd, B., & Tuohy, M. G. (2023). Induction and Characterisation of Lignocellulolytic Activities from Novel Deep-Sea Fungal Secretomes. Fermentation, 9(9), 780. https://doi.org/10.3390/fermentation9090780




