Cloning, Expression, and Characterization of Family A DNA Polymerase from Massilia aurea
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Construction of a Plasmid Encoding Mau DNA Polymerase
2.3. Purification of Mau DNA Polymerase
2.4. DNA Substrates
2.5. A DNA Polymerase Activity Assay
2.6. An Exonuclease Activity Assay
2.7. Thermal Stability Analysis
2.8. Data Analysis
2.9. The Estimation of DNA-Binding Affinity
3. Results
3.1. Expression and Purification of Mau DNA Polymerase
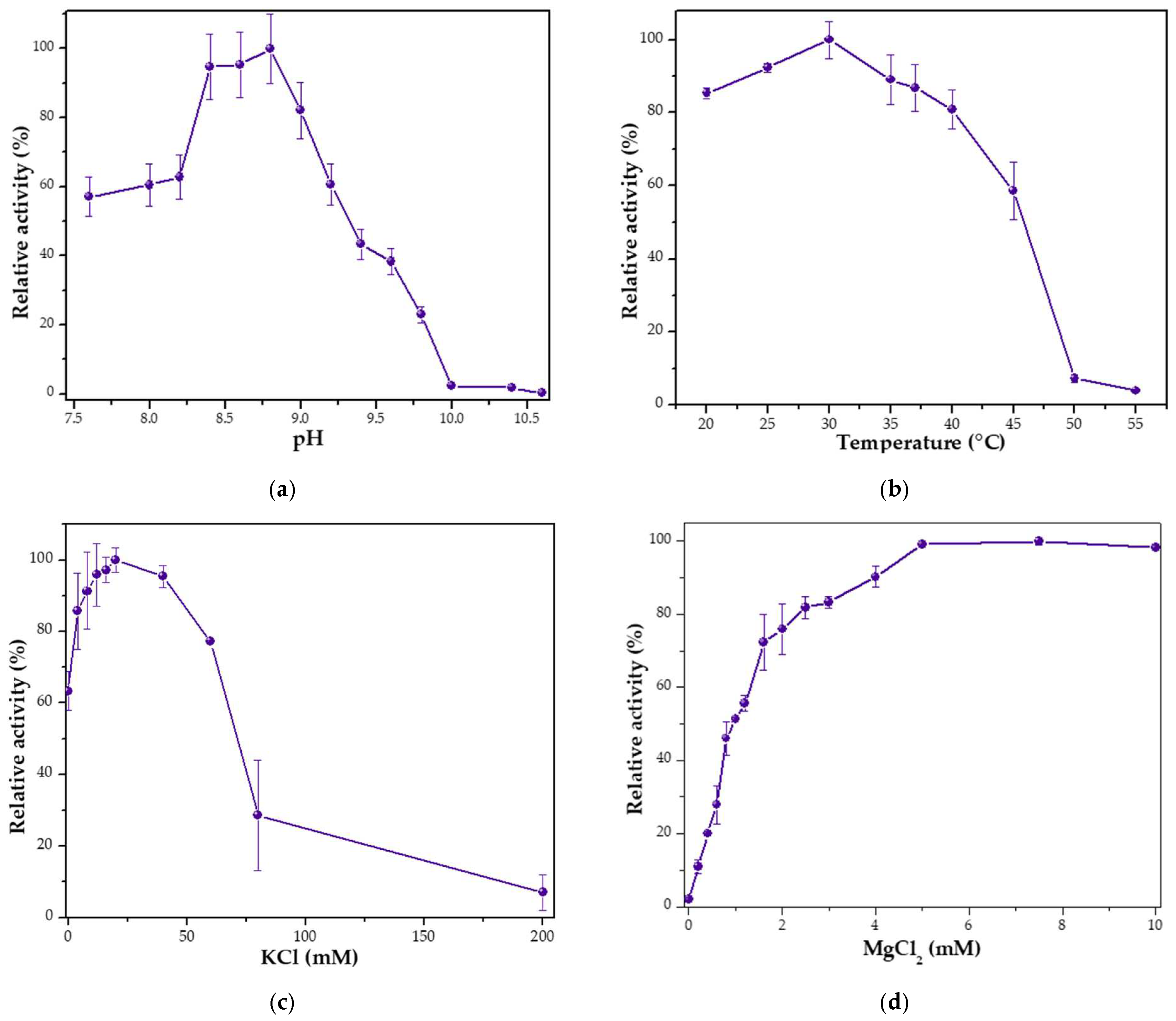
3.2. Characterization of Mau DNA Polymerase
3.3. Thermal Stability of Mau DNA Polymerase
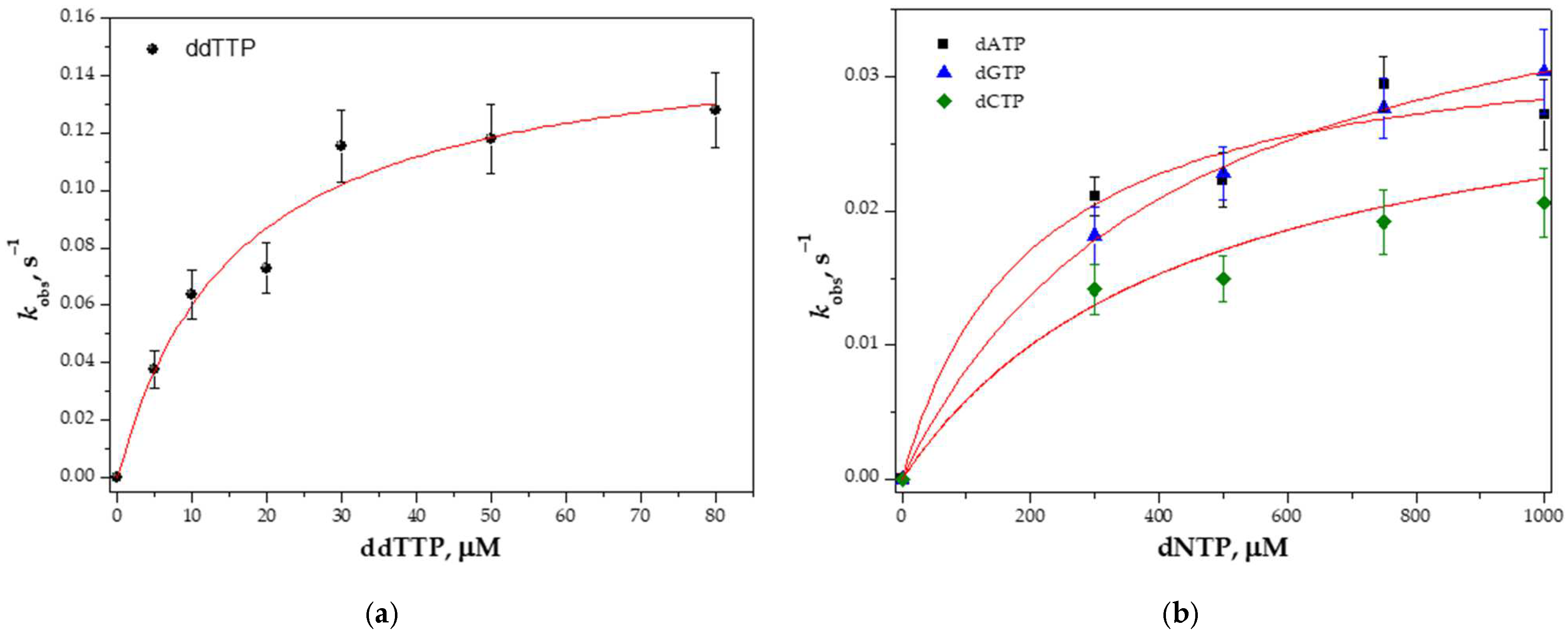
3.4. Kinetic Analysis of Mau DNA Polymerase Activity
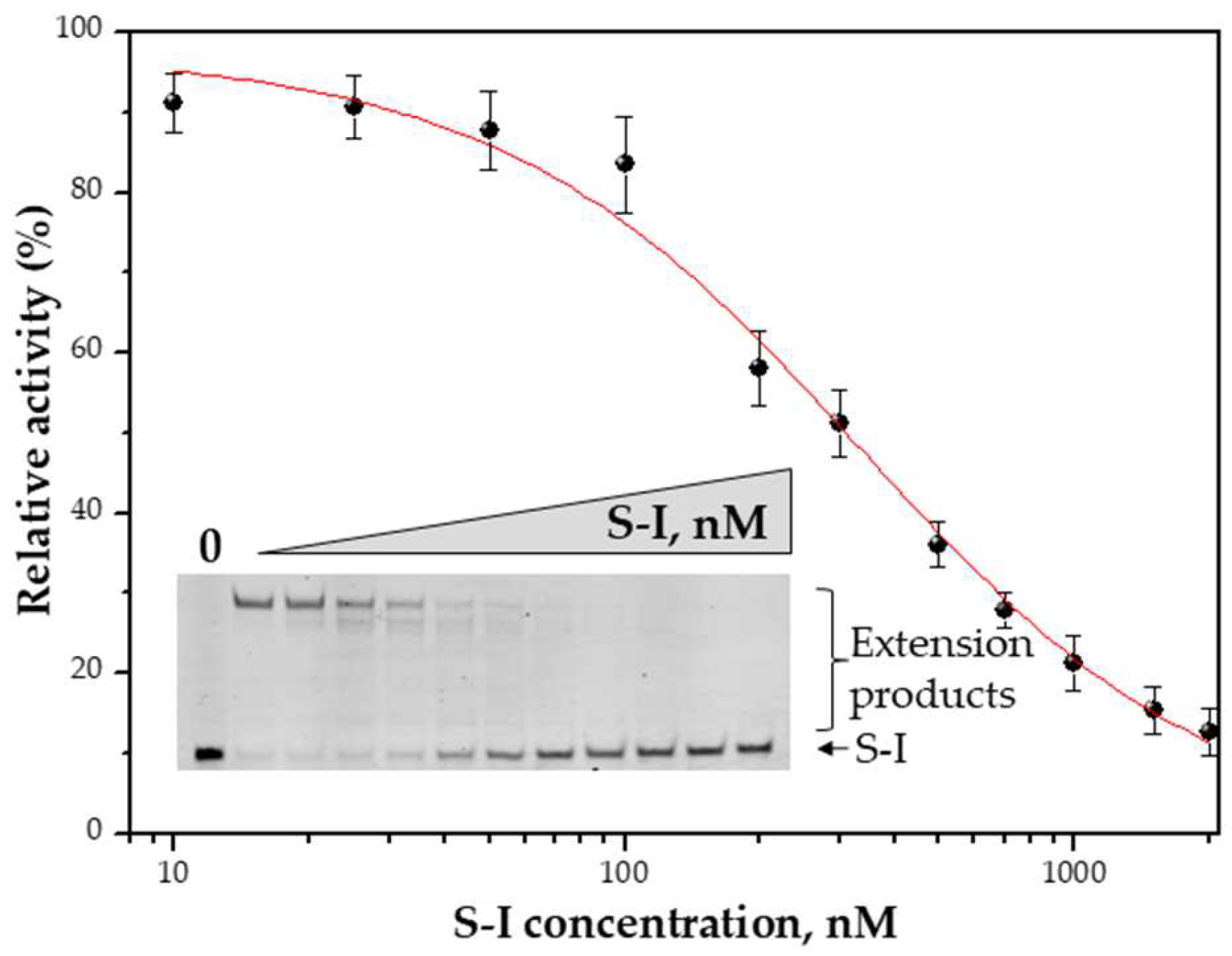
3.5. Primer-Template Binding
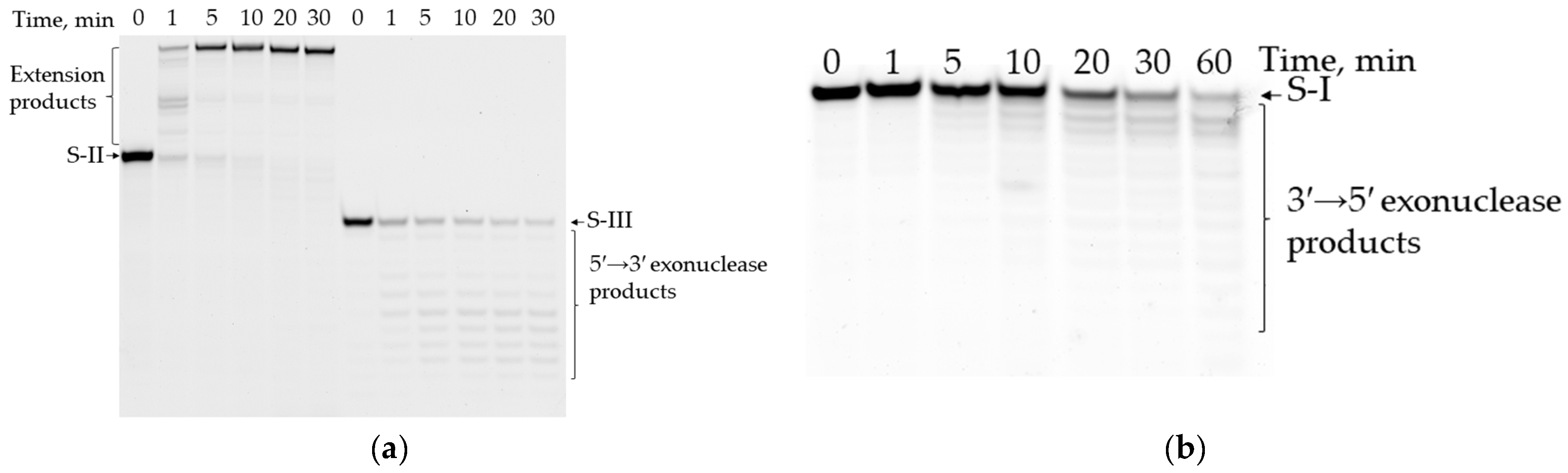
3.6. Exonuclease Activity of Mau DNA Polymerase
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johansson, E.; Dixon, N. Replicative DNA Polymerases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012799. [Google Scholar] [CrossRef]
- Jain, R.; Aggarwal, A.K.; Rechkoblit, O. Eukaryotic DNA Polymerases. Curr. Opin. Struct. Biol. 2018, 53, 77–87. [Google Scholar] [CrossRef]
- Lange, S.S.; Takata, K.; Wood, R.D. DNA Polymerases and Cancer. Nat. Rev. Cancer 2011, 11, 96–110. [Google Scholar] [CrossRef]
- Henrikus, S.S.; van Oijen, A.M.; Robinson, A. Specialised DNA Polymerases in Escherichia coli: Roles within Multiple Pathways. Curr. Genet. 2018, 64, 1189–1196. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Structural and Molecular Kinetic Features of Activities of DNA Polymerases. Int. J. Mol. Sci. 2022, 23, 6373. [Google Scholar] [CrossRef]
- Delagoutte, E. DNA Polymerases: Mechanistic Insight from Biochemical and Biophysical Studies. Front. Biosci. 2012, 17, 509. [Google Scholar] [CrossRef]
- Ling, J.A.; Frevert, Z.; Washington, M.T. Recent Advances in Understanding the Structures of Translesion Synthesis DNA Polymerases. Genes 2022, 13, 915. [Google Scholar] [CrossRef]
- Aschenbrenner, J.; Marx, A. DNA Polymerases and Biotechnological Applications. Curr. Opin. Biotechnol. 2017, 48, 187–195. [Google Scholar] [CrossRef]
- Ishino, S.; Ishino, Y. DNA Polymerases as Useful Reagents for Biotechnology—The History of Developmental Research in the Field. Front. Microbiol. 2014, 5, 465. [Google Scholar] [CrossRef]
- Houlihan, G.; Arangundy-Franklin, S.; Holliger, P. Engineering and Application of Polymerases for Synthetic Genetics. Curr. Opin. Biotechnol. 2017, 48, 168–179. [Google Scholar] [CrossRef]
- Terpe, K. Overview of Thermostable DNA Polymerases for Classical PCR Applications: From Molecular and Biochemical Fundamentals to Commercial Systems. Appl. Microbiol. Biotechnol. 2013, 97, 10243–10254. [Google Scholar] [CrossRef]
- Zhang, L.; Kang, M.; Xu, J.; Huang, Y. Archaeal DNA Polymerases in Biotechnology. Appl. Microbiol. Biotechnol. 2015, 99, 6585–6597. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR Past, Present and Future. Biotechniques 2020, 69, 317–325. [Google Scholar] [CrossRef]
- Coulther, T.A.; Stern, H.R.; Beuning, P.J. Engineering Polymerases for New Functions. Trends Biotechnol. 2019, 37, 1091–1103. [Google Scholar] [CrossRef]
- Karunanathie, H.; Kee, P.S.; Ng, S.F.; Kennedy, M.A.; Chua, E.W. PCR Enhancers: Types, Mechanisms, and Applications in Long-Range PCR. Biochimie 2022, 197, 130–143. [Google Scholar] [CrossRef]
- Wang, Y.; Prosen, D.E.; Mei, L.; Sullivan, J.C.; Finney, M.; Vander Horn, P.B. A Novel Strategy to Engineer DNA Polymerases for Enhanced Processivity and Improved Performance in Vitro. Nucleic Acids Res. 2004, 32, 1197–1207. [Google Scholar] [CrossRef]
- Śpibida, M.; Krawczyk, B.; Olszewski, M.; Kur, J. Modified DNA Polymerases for PCR Troubleshooting. J. Appl. Genet. 2017, 58, 133–142. [Google Scholar] [CrossRef]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR Inhibition in QPCR, DPCR and MPS—Mechanisms and Solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef]
- Dietrich, J.; Schmitt, P.; Zieger, M.; Preve, B.; Rolland, J.L.; Chaabihi, H.; Gueguen, Y. PCR Performance of the Highly Thermostable Proof-Reading B-Type DNA Polymerase from Pyrococcus Abyssi. FEMS Microbiol. Lett. 2002, 217, 89–94. [Google Scholar] [CrossRef]
- Cline, J.; Braman, J.C.; Hogrefe, H.H. PCR Fidelity of Pfu DNA Polymerase and Other Thermostable DNA Polymerases. Nucleic Acids Res. 1996, 24, 3546–3551. [Google Scholar] [CrossRef]
- Bulygin, A.A.; Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Comparative Analysis of Family A DNA-Polymerases as a Searching Tool for Enzymes with New Properties. Mol. Biol. 2023, 57, 182–192. [Google Scholar] [CrossRef]
- Stenesh, J.; McGowan, G.R. DNA Polymerase from Mesophilic and Thermophilic Bacteria. Biochim. Biophys. Acta Nucleic Acids Protein Synth. 1977, 475, 32–41. [Google Scholar] [CrossRef]
- Gallego, V.; Sánchez-Porro, C.; García, M.T.; Ventosa, A. Massilia Aurea Sp. Nov., Isolated from Drinking Water. Int. J. Syst. Evol. Microbiol. 2006, 56, 2449–2453. [Google Scholar] [CrossRef]
- Malboeuf, C.M.; Isaacs, S.J.; Tran, N.H.; Kim, B. Thermal Effects on Reverse Transcription: Improvement of Accuracy and Processivity in CDNA Synthesis. Biotechniques 2001, 30, 1074–1084. [Google Scholar] [CrossRef]
- Johnson, K.A. Rapid Quench Kinetic Analysis of Polymerases, Adenosinetriphosphatases, and Enzyme Intermediates. Methods Enzymol. 1995, 249, 38–61. [Google Scholar] [CrossRef]
- Weiss, K.K.; Chen, R.; Skasko, M.; Reynolds, H.M.; Lee, K.; Bambara, R.A.; Mansky, L.M.; Kim, B. A Role for DNTP Binding of Human Immunodeficiency Virus Type 1 Reverse Transcriptase in Viral Mutagenesis. Biochemistry 2004, 43, 4490–4500. [Google Scholar] [CrossRef]
- Skasko, M.; Weiss, K.K.; Reynolds, H.M.; Jamburuthugoda, V.; Lee, K.; Kim, B. Mechanistic Differences in RNA-Dependent DNA Polymerization and Fidelity between Murine Leukemia Virus and HIV-1 Reverse Transcriptases. J. Biol. Chem. 2005, 280, 12190–12200. [Google Scholar] [CrossRef]
- Elshawadfy, A.M.; Keith, B.J.; Ee Ooi, H.; Kinsman, T.; Heslop, P.; Connolly, B.A. DNA Polymerase Hybrids Derived from the Family-B Enzymes of Pyrococcus Furiosus and Thermococcus Kodakarensis: Improving Performance in the Polymerase Chain Reaction. Front. Microbiol. 2014, 5, 224. [Google Scholar] [CrossRef]
- Oscorbin, I.P.; Belousova, E.A.; Boyarskikh, U.A.; Zakabunin, A.I.; Khrapov, E.A.; Filipenko, M.L. Derivatives of Bst-like Gss-Polymerase with Improved Processivity and Inhibitor Tolerance. Nucleic Acids Res. 2017, 45, 9595–9610. [Google Scholar] [CrossRef]
- Rigby, P.W.J.; Dieckmann, M.; Rhodes, C.; Berg, P. Labeling Deoxyribonucleic Acid to High Specific Activity in Vitro by Nick Translation with DNA Polymerase I. J. Mol. Biol. 1977, 113, 237–251. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Downie, J.A.; Ito, J. Primary Structure of the DNA Polymerase I Gene of an α-Proteobacterium, Rhizobium Leguminosarum, and Comparison with Other Family A DNA Polymerases. Curr. Microbiol. 1999, 38, 355–359. [Google Scholar] [CrossRef]
- Sellmann, E.; Schröder, K.L.; Knoblich, I.M.; Westermann, P. Purification and Characterization of DNA Polymerases from Bacillus Species. J. Bacteriol. 1992, 174, 4350–4355. [Google Scholar] [CrossRef]
- Setlow, P. DNA Polymerase I from Escherichia coli. Methods Enzymol. 1974, 29, 3–12. [Google Scholar] [CrossRef]
- McClure, W.; Jovin, T. The Steady State Kinetic Parameters and Non-Processivity of Escherichia coli Deoxyribonucleic Acid Polymerase I. J. Biol. Chem. 1975, 250, 4073–4080. [Google Scholar] [CrossRef]
- Lowe, L.G.; Guengerich, F.P. Steady-State and Pre-Steady-State Kinetic Analysis of DNTP Insertion Opposite 8-Oxo-7,8-Dihydroguanine by Escherichia coli Polymerases I Exo- and II Exo-. Biochemistry 1996, 35, 9840–9849. [Google Scholar] [CrossRef]
- Driscoll, M.D.; Rentergent, J.; Hay, S. A Quantitative Fluorescence-Based Steady-State Assay of DNA Polymerase. FEBS J. 2014, 281, 2042–2050. [Google Scholar] [CrossRef]
- Polesky, A.H.; Steitz, T.A.; Grindley, N.D.; Joyce, C.M. Identification of Residues Critical for the Polymerase Activity of the Klenow Fragment of DNA Polymerase I from Escherichia coli. J. Biol. Chem. 1990, 265, 14579–14591. [Google Scholar] [CrossRef]
- Spratt, T. Structure of the Hydrogen Bonding Complex of O6-Methylguanine with Cytosine and Thymine during DNA Replication. Nucleic Acids Res. 1997, 25, 3354–3361. [Google Scholar] [CrossRef]
- Gillin, F.D.; Nossal, N.G. T4 DNA Polymerase Has a Lower Apparent Km for Deoxynucleoside Triphosphates Complementary Rather than Noncomplementary to the Template. Biochem. Biophys. Res. Commun. 1975, 64, 457–464. [Google Scholar] [CrossRef]
- Wu, P.; Nossal, N.; Benkovic, S.J. Kinetic Characterization of a Bacteriophage T4 Antimutator DNA Polymerase. Biochemistry 1998, 37, 14748–14755. [Google Scholar] [CrossRef]
- Kong, H.; Kucera, R.B.; Jack, W.E. Characterization of a DNA Polymerase from the Hyperthermophile Archaea Thermococcus Litoralis. Vent DNA Polymerase, Steady State Kinetics, Thermal Stability, Processivity, Strand Displacement, and Exonuclease Activities. J. Biol. Chem. 1993, 268, 1965–1975. [Google Scholar] [CrossRef]
- Tang, F.; Liu, S.; Li, Q.-Y.; Yuan, J.; Li, L.; Wang, Y.; Yuan, B.-F.; Feng, Y.-Q. Location Analysis of 8-Oxo-7,8-Dihydroguanine in DNA by Polymerase-Mediated Differential Coding. Chem. Sci. 2019, 10, 4272–4281. [Google Scholar] [CrossRef]
- Patel, S.S.; Wong, I.; Johnson, K.A. Pre-Steady-State Kinetic Analysis of Processive DNA Replication Including Complete Characterization of an Exonuclease-Deficient Mutant. Biochemistry 1991, 30, 511–525. [Google Scholar] [CrossRef]
- Boosalis, M.S.; Petruska, J.; Goodman, M.F. DNA Polymerase Insertion Fidelity. Gel Assay for Site-Specific Kinetics. J. Biol. Chem. 1987, 262, 14689–14696. [Google Scholar] [CrossRef]
- Takata, K.; Shimizu, T.; Iwai, S.; Wood, R.D. Human DNA Polymerase N (POLN) Is a Low Fidelity Enzyme Capable of Error-Free Bypass of 5S-Thymine Glycol. J. Biol. Chem. 2006, 281, 23445–23455. [Google Scholar] [CrossRef]

| Short Name | Sequence |
|---|---|
| S-I | FAM-5′CGGCCCCCAGATGAGTCGAGCAGC3′ 3′GCCGGGGGTCTACTCAGCTCGTCGATAGTTCCGTTTTACAGAGG5′ |
| S-II | FAM-5′CGGCCCCCAGATGAGTCGAGCAGC3′5′TCAAGGCAAAATGTCTCC3′ 3′GCCGGGGGTCTACTCAGCTCGTCGATAGTTCCGTTTTACAGAGG5′ |
| S-III | 5′CGGCCCCCAGATGAGTCGAGCAGC3′5′TCAAGGCAAAATGTCTCC3′-FAM 3′GCCGGGGGTCTACTCAGCTCGTCGATAGTTCCGTTTTACAGAGG5′ |
| Template-trap | 5′CGGCCCCCAGATGAGTCGAGCAGCTATCAAGGCAAAATGTCTCC3′ |
| dNTP | kcat (s−1) | Kd,appdNTP (µM) | kcat/Kd,appdNTP (µM−1·s−1) |
|---|---|---|---|
| dTTP (corr.) | 0.20 ± 0.07 | 16 ± 4 | (1.0 ± 0.5) × 10−2 |
| dATP (incorr.) | 0.034 ± 0.004 | 200 ± 100 | (1.0 ± 0.5) × 10−4 |
| dCTP (incorr.) | 0.030 ± 0.004 | 500 ± 100 | (0.6 ± 0.2) × 10−4 |
| dGTP (incorr.) | 0.044 ± 0.003 | 430 ± 50 | (1.0 ± 0.1) × 10−4 |
| DNA Polymerase | Kd,appDNA, nM | Kd,appdNTP, µM | kcat, s−1 | Ref. |
|---|---|---|---|---|
| Pol I (E. coli) | 5 – | 1–2 12–147 | 8.3 – | [33] [34] |
| KF (E. coli) | – – 8 – | 1.1 2.8–8.0 2.3–3.2 0.014–0.042 | 0.37 0.93–1.55 2.0–2.8 – | [35] [36] [37] [38] |
| T4 (E. coli phage T4) | – 70 | 6–17 20 | – – | [39] [40] |
| Taq (T. aquaticus) | 1.0–1.8 – | (corr.) 14–17 (incorr.) 6–12 0.99 | – – | [41] [36] |
| Tth (T. thermophiles) | – – | (corr.) 2.5 (incorr.) 230–280 0.022 | – – | [42] [38] |
| Bsu (B. subtilis) | – | (corr.) 1.8 (incorr.) 180 | – | [42] |
| T7 | 17.8 | 18 | 0.24 | [43] |
| Pol ɑ (Drosophila) | – | 3.7 | 2.2 | [44] |
| Pol η (Human) | – | 7.8–8.2 | – | [45] |
| Mau (M. aurea) | <280 | (corr.) 16 (incorr.) 200–500 | (corr.) 0.2 (incorr.) 0.03–0.044 | this study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuznetsova, A.A.; Bedritskikh, K.S.; Bulygin, A.A.; Kuznetsov, N.A. Cloning, Expression, and Characterization of Family A DNA Polymerase from Massilia aurea. Fermentation 2023, 9, 650. https://doi.org/10.3390/fermentation9070650
Kuznetsova AA, Bedritskikh KS, Bulygin AA, Kuznetsov NA. Cloning, Expression, and Characterization of Family A DNA Polymerase from Massilia aurea. Fermentation. 2023; 9(7):650. https://doi.org/10.3390/fermentation9070650
Chicago/Turabian StyleKuznetsova, Aleksandra A., Ksenia S. Bedritskikh, Anatoly A. Bulygin, and Nikita A. Kuznetsov. 2023. "Cloning, Expression, and Characterization of Family A DNA Polymerase from Massilia aurea" Fermentation 9, no. 7: 650. https://doi.org/10.3390/fermentation9070650
APA StyleKuznetsova, A. A., Bedritskikh, K. S., Bulygin, A. A., & Kuznetsov, N. A. (2023). Cloning, Expression, and Characterization of Family A DNA Polymerase from Massilia aurea. Fermentation, 9(7), 650. https://doi.org/10.3390/fermentation9070650






