Advances in Droplet-Based Microfluidic High-Throughput Screening of Engineered Strains and Enzymes Based on Ultraviolet, Visible, and Fluorescent Spectroscopy
Abstract
1. Introduction
2. Detection and Screening Based on Ultraviolet Spectroscopy
3. Detection and Screening Based on the Visible Light Spectrum
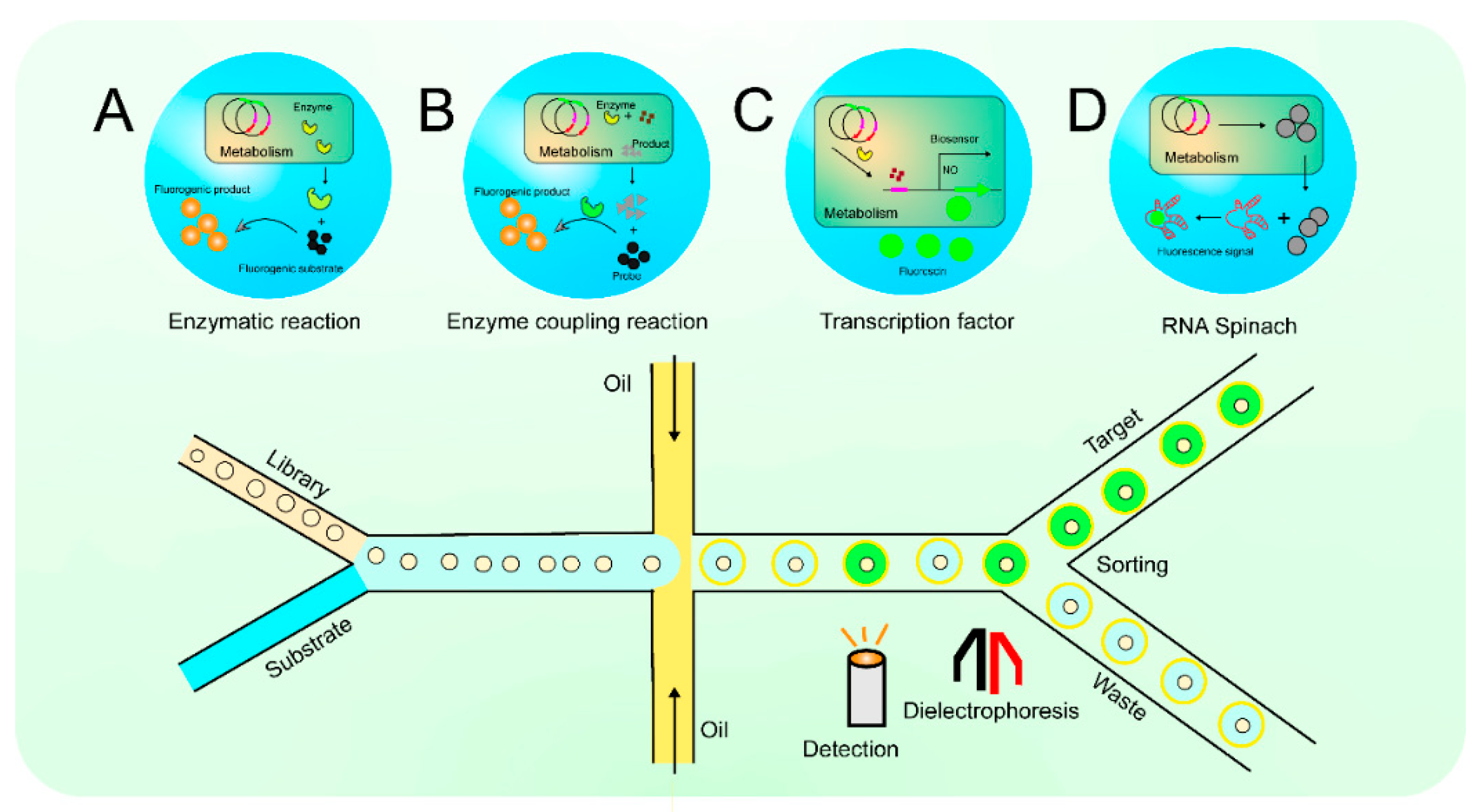
4. Detection and Screening Based on Fluorescence Spectroscopy
4.1. Detection and Screening Based on Conventional Strategies of Fluorescence Spectroscopy
4.2. Detection and Screening Based on Fluorescence Spectroscopy Using Biosensors
5. Detection Screening Based on Other Technologies
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moragues, T.; Arguijo, D.; Beneyton, T.; Modavi, C.; Simutis, K.; Abate, A.R.; Baret, J.C.; DeMello, A.J.; Densmore, D.; Griffiths, A.D. Droplet-based microfluidics. Nat. Rev. Methods Primers 2023, 3, 32. [Google Scholar] [CrossRef]
- Chiu, F.; Stavrakis, S. High-throughput droplet-based microfluidics for directed evolution of enzymes. Electrophoresis 2019, 40, 2860–2872. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yang, G.; Ma, F. Fluorescence coupling strategies in fluorescence-activated droplet sorting (FADS) for ultrahigh-throughput screening of enzymes, metabolites, and antibodies. Biotechnol. Adv. 2023, 66, 108173. [Google Scholar] [CrossRef] [PubMed]
- Muretta, J.M.; Rajasekaran, D.; Blat, Y.; Little, S.; Myers, M.; Nair, C.; Burdekin, B.; Yuen, S.L.; Jimenez, N.; Guhathakurta, P.; et al. HTS driven by fluorescence lifetime detection of FRET identifies activators and inhibitors of cardiac myosin. SLAS Discov. Adv. Life Sci. R D 2023, 28, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Sesen, M.; Alan, T.; Neild, A. Droplet control technologies for microfluidic high throughput screening (μHTS). Lab Chip 2017, 17, 2372–2394. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.; Huisman, D.H.; Vieira, H.M.; Frodyma, D.E.; Neilsen, B.K.; Chakraborty, B.; Hight, S.K.; White, M.A.; Fisher, K.W.; Lewis, R.E. A Gene Expression High-Throughput Screen (GE-HTS) for Coordinated Detection of Functionally Similar Effectors in Cancer. Cancers 2020, 12, 3143. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yu, Z.; Zheng, M.; Yang, W.; Liu, Z.; Zhou, J.; Huang, L. Artificial intelligence-accelerated high-throughput screening of antibiotic combinations on a microfluidic combinatorial droplet system. Lab Chip 2023, 23, 3961–3977. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef]
- Miansari, M.; Friend, J.R. Acoustic Nanofluidics via Room—Temperature Lithium Niobate Bonding: A Platform for Actuation and Manipulation of Nanoconfined Fluids and Particles. Adv. Funct. Mater. 2016, 26, 7861–7872. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, W.; Zhao, Q.; Yan, S. Geometric optimization of double layered microchannel with grooves array for enabling nanoparticle manipulation. Phys. Fluids 2023, 35, 062009. [Google Scholar]
- Zeng, W.; Guo, L.; Xu, S.; Chen, J.; Zhou, J. High-Throughput Screening Technology in Industrial Biotechnology. Trends Biotechnol. 2020, 38, 888–906. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.-L.; Lee, P.-Y.; Yang, C.-L.; Huang, W.-Y.; Lin, Y.-S. Recent Advances in Applications of Droplet Microfluidics. Micromachines 2015, 6, 1249–1271. [Google Scholar] [CrossRef]
- Amirifar, L.; Besanjideh, M.; Nasiri, R.; Shamloo, A.; Nasrollahi, F.; de Barros, N.R.; Davoodi, E.; Erdem, A.; Mahmoodi, M.; Hosseini, V.; et al. Droplet-based microfluidics in biomedical applications. Biofabrication 2022, 14, 022001. [Google Scholar] [CrossRef] [PubMed]
- Weitong, Q.; Guangyu, Y. Research and application progress of microdroplets high throughput screening methods. Synth. Biol. 2023, 4, 966–979. [Google Scholar]
- Sun, G.; Qu, L.; Azi, F.; Liu, Y.; Li, J.; Lv, X.; Du, G.; Chen, J.; Chen, C.H.; Liu, L. Recent progress in high-throughput droplet screening and sorting for bioanalysis. Biosens. Bioelectron. 2023, 225, 115107. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.K.; Jamali, B.; Hubbuch, J. Selective high throughput protein quantification based on UV absorption spectra. Biotechnol. Bioeng. 2013, 110, 448–460. [Google Scholar] [CrossRef]
- Duncombe, T.A.; Ponti, A.; Seebeck, F.P.; Dittrich, P.S. UV-Vis Spectra-Activated Droplet Sorting for Label-Free Chemical Identification and Collection of Droplets. Anal. Chem. 2021, 93, 13008–13013. [Google Scholar] [CrossRef]
- Medcalf, E.J.; Gantz, M.; Kaminski, T.S.; Hollfelder, F. Ultra-High-Throughput Absorbance-Activated Droplet Sorting for Enzyme Screening at Kilohertz Frequencies. Anal. Chem. 2023, 95, 4597–4604. [Google Scholar] [CrossRef]
- Baret, J.C.; Miller, O.J.; Taly, V.; Ryckelynck, M.; El-Harrak, A.; Frenz, L.; Rick, C.; Samuels, M.L.; Hutchison, J.B.; Agresti, J.J.; et al. Fluorescence-activated droplet sorting (FADS): Efficient microfluidic cell sorting based on enzymatic activity. Lab Chip 2009, 9, 1850–1858. [Google Scholar] [CrossRef]
- Wang, B.L.; Ghaderi, A.; Zhou, H.; Agresti, J.; Weitz, D.A.; Fink, G.R.; Stephanopoulos, G. Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption. Nat. Biotechnol. 2014, 32, 473–478. [Google Scholar] [CrossRef]
- Gielen, F.; Hours, R.; Emond, S.; Fischlechner, M.; Schell, U.; Hollfelder, F. Ultrahigh-throughput–directed enzyme evolution by absorbance-activated droplet sorting (AADS). Proc. Natl. Acad. Sci. USA 2016, 113, E7383–E7389. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Vestergaard, M.; Jensen, T.G.; Shen, J.; Dufva, M.; Solem, C.; Jensen, P.R. Finding the Needle in the Haystack—The Use of Microfluidic Droplet Technology to Identify Vitamin-Secreting Lactic Acid Bacteria. mBio 2017, 8, e00526-17. [Google Scholar] [CrossRef] [PubMed]
- Beneyton, T.; Thomas, S.; Griffiths, A.D.; Nicaud, J.M.; Drevelle, A.; Rossignol, T. Droplet-based microfluidic high-throughput screening of heterologous enzymes secreted by the yeast Yarrowia lipolytica. Microb. Cell Factories 2017, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Abatemarco, J.; Sarhan, M.F.; Wagner, J.M.; Lin, J.L.; Liu, L.; Hassouneh, W.; Yuan, S.F.; Alper, H.S.; Abate, A.R. RNA-aptamers-in-droplets (RAPID) high-throughput screening for secretory phenotypes. Nat. Commun. 2017, 8, 332. [Google Scholar] [CrossRef] [PubMed]
- Jankowski, P.; Samborski, A.; Ostaszewski, R.; Garstecki, P. Evaluation of droplet-based microfluidic platforms as a convenient tool for lipases and esterases assays. Prep. Biochem. Biotechnol. 2019, 49, 727–734. [Google Scholar] [CrossRef]
- Terekhov, S.S.; Smirnov, I.V.; Malakhova, M.V.; Samoilov, A.E.; Manolov, A.I.; Nazarov, A.S.; Danilov, D.V.; Dubiley, S.A.; Osterman, I.A.; Rubtsova, M.P.; et al. Ultrahigh-throughput functional profiling of microbiota communities. Proc. Natl. Acad. Sci. USA 2018, 115, 9551–9556. [Google Scholar] [CrossRef]
- Lyu, F.; Pan, M.; Patil, S.; Wang, J.-H.; Matin, A.C.; Andrews, J.R.; Tang, S.K. Phenotyping antibiotic resistance with single-cell resolution for the detection of heteroresistance. Sens. Actuators B Chem. 2018, 270, 396–404. [Google Scholar] [CrossRef]
- Cao, X.; Luo, Z.; Zeng, W.; Xu, S.; Zhao, L.; Zhou, J. Enhanced avermectin production by Streptomyces avermitilis ATCC 31267 using high-throughput screening aided by fluorescence-activated cell sorting. Appl. Microbiol. Biotechnol. 2018, 102, 703–712. [Google Scholar] [CrossRef]
- Zhang, W.; Fu, J.; Wang, Y.; Zhang, X.; Li, J. Enhanced visible-light photocatalytic activity of ZnS/BiOBr/graphene oxide ternary composite. J. Phys. Chem. Solids 2019, 127, 19–27. [Google Scholar] [CrossRef]
- Wang, Q.; Feng, L.R.; Wei, L.; Li, H.G.; Wang, L.; Zhou, Y.; Yu, X.B. Mutation Breeding of Lycopene-Producing Strain Blakeslea Trispora by a Novel Atmospheric and Room Temperature Plasma (ARTP). Appl. Biochem. Biotechnol. 2014, 174, 452–460. [Google Scholar]
- Zhou, S.; Liu, P.; Chen, J.; Du, G.; Li, H.; Zhou, J. Characterization of mutants of a tyrosine ammonia-lyase from Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 2016, 100, 10443–10452. [Google Scholar] [CrossRef]
- Mendoza, L.D.; Rodriguez, J.A.; Leclaire, J.; Buono, G.; Fotiadu, F.; Carrière, F.; Abousalham, A. An ultraviolet spectrophotometric assay for the screening of sn-2-specific lipases using 1,3-O-dioleoyl-2-O-α-eleostearoyl-sn-glycerol as substrate. J. Lipid Res. 2012, 53, 185–194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lijuan, Y.; Jiamin, W.; Lu, W.; Yi, C. Cell-based high throughput screening of a-amino acid ester hydrolase variants. Microbiology 2011, 39, 0264–0271. [Google Scholar]
- Tian, H.; Yu, B.; Ai, L.; Yu, H.; Chen, C. A high-throughput system for screening high diacetyl-producing lactic acid bacteria in fermented milk in 96-well microplates. J. Food Meas. Charact. 2020, 14, 548–556. [Google Scholar] [CrossRef]
- Xiaodan, C.; Qun, F.; Zhaolun, F. Ultraviolet detection capillary electrophoresis system based on composite microfluidic chip. Chem J Chinese U 2004, 25, 1231–1234. [Google Scholar]
- Ottevaere, H.; Van Overmeire, S.; Albero, J.; Nieradko, L.; Desmet, G.; Gorecki, C.; Thienpont, H. Plastic light coupler for absorbance detection in silicon microfluidic channels. Microfluid. Nanofluidics 2015, 18, 559–568. [Google Scholar] [CrossRef]
- Hassan, S.U.; Nightingale, A.M.; Niu, X. Micromachined optical flow cell for sensitive measurement of droplets in tubing. Biomed. Microdevices 2018, 20, 92. [Google Scholar] [CrossRef]
- Ahmadi, F.; Samlali, K.; Vo, P.Q.N.; Shih, S.C.C. An integrated droplet-digital microfluidic system for on-demand droplet creation, mixing, incubation, and sorting. Lab Chip 2019, 19, 524–535. [Google Scholar] [CrossRef]
- Maceiczyk, R.M.; Hess, D.; Chiu, F.W.Y.; Stavrakis, S.; deMello, A.J. Differential detection photothermal spectroscopy: Towards ultra-fast and sensitive label-free detection in picoliter & femtoliter droplets. Lab Chip 2017, 17, 3654–3663. [Google Scholar]
- Wagner, J.; Liu, L.; Yuan, S.-F.; Venkataraman, M.; Abate, A.; Alper, H. A comparative analysis of single cell and droplet-based FACS for improving production phenotypes: Riboflavin overproduction in Yarrowia lipolytica. Metab. Eng. 2018, 47, 346–356. [Google Scholar] [CrossRef]
- Kim, H.S.; Hsu, S.-C.; Han, S.I.; Thapa, H.R.; Guzman, A.; Browne, D.R.; Tatli, M.; Devarenne, T.P.; Stern, D.B.; Han, A. High-throughput droplet microfluidics screening platform for selecting fast-growing and high lipid-producing microalgae from a mutant library. Plant Direct 2017, 1, e00011. [Google Scholar] [CrossRef]
- Choi, J.W.; Vasamsetti, B.M.K.; Kim, K.W.; Seo, S.H.; Lee, D.H.; Chang, S.I.; Choo, J.; Kim, H.Y. Analysis of ribonuclease activity in sub-nanoliter droplets by label-free fluorescence measurements. Analyst 2017, 142, 2610–2616. [Google Scholar] [CrossRef]
- Hardiman, E.; Gibbs, M.; Reeves, R.; Bergquist, P. Directed Evolution of a Thermophilic β-glucosidase for Cellulosic Bioethanol Production. Appl. Biochem. Biotechnol. 2010, 161, 301–312. [Google Scholar] [CrossRef]
- Ma, F.; Xie, Y.; Huang, C.; Feng, Y.; Yang, G. An Improved Single Cell Ultrahigh Throughput Screening Method Based on In Vitro Compartmentalization. PLoS ONE 2014, 9, e89785. [Google Scholar] [CrossRef]
- Tu, R.; Martinez, R.; Prodanovic, R.; Klein, M.; Schwaneberg, U. A Flow Cytometry–Based Screening System for Directed Evolution of Proteases. SLAS Discov. 2011, 16, 285–294. [Google Scholar] [CrossRef]
- Agresti, J.J.; Antipov, E.; Abate, A.R.; Ahn, K.; Rowat, A.C.; Baret, J.C.; Marquez, M.; Klibanov, A.M.; Griffiths, A.D.; Weitz, D. A Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 4004–4009. [Google Scholar] [CrossRef]
- Turner, P.; Mamo, G.; Karlsson, E.N. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb. Cell Factories 2007, 6, 9. [Google Scholar] [CrossRef]
- Nakkharat, P.; Haltrich, D. Purification and characterisation of an intracellular enzyme with beta-glucosidase and beta-galactosidase activity from the thermophilic fungus Talaromyces thermophilus CBS 236.58. J. Biotechnol. 2006, 123, 304–313. [Google Scholar] [CrossRef]
- Huebner, A.; Olguin, L.F.; Bratton, D.; Whyte, G.; Huck, W.T.; de Mello, A.J.; Edel, J.B.; Abell, C.; Hollfelder, F. Development of quantitative cell-based enzyme assays in microdroplets. Anal. Chem. 2008, 80, 3890–3896. [Google Scholar] [CrossRef]
- Varadarajan, N.; Rodriguez, S.; Hwang, B.Y.; Georgiou, G.; Iverson, B.L. Highly active and selective endopeptidases with programmed substrate specificities. Nat. Chem. Biol. 2008, 4, 290–294. [Google Scholar] [CrossRef]
- Mizukami, S.; Watanabe, S.; Hori, Y.; Kikuchi, K. Covalent protein labeling based on noncatalytic beta-lactamase and a designed FRET substrate. J. Am. Chem. Soc. 2009, 131, 5016–5017. [Google Scholar] [CrossRef]
- Yang, Y.; Babiak, P.; Reymond, J.-L. Low background FRET-substrates for lipases and esterases suitable for high-throughput screening under basic (pH 11) conditions. Org. Biomol. Chem. 2006, 4, 1746–1754. [Google Scholar] [CrossRef]
- Hammar, P.; Angermayr, S.A.; Sjostrom, S.L.; van der Meer, J.; Hellingwerf, K.J.; Hudson, E.P.; Joensson, H.N. Single-cell screening of photosynthetic growth and lactate production by cyanobacteria. Biotechnol. Biofuels 2015, 8, 193. [Google Scholar] [CrossRef]
- Abalde-Cela, S.; Gould, A.; Liu, X.; Kazamia, E.; Smith, A.G.; Abell, C. High-throughput detection of ethanol-producing cyanobacteria in a microdroplet platform. J. R. Soc. Interface 2015, 12, 20150216. [Google Scholar] [CrossRef]
- Ostafe, R.; Prodanović, R.; Lloyd Ung, W.; Weitz, D.A.; Fischer, R.J.B. A high-throughput cellulase screening system based on droplet microfluidics. Biomicrofluidics 2014, 8, 041102. [Google Scholar] [CrossRef]
- Williams, T.C.; Pretorius, I.S.; Paulsen, I.T. Synthetic Evolution of Metabolic Productivity Using Biosensors. Trends Biotechnol. 2016, 34, 371–381. [Google Scholar] [CrossRef]
- Sun, Q.M.; Lu, Y.L.; Shen, X.L.; Sun, X.X.; Wang, J.; Yuan, Q.P. Fluorescence detection-based high-throughput screening systems and devices facilitate cell factories construction. Synth. Biol. 2023, 4, 947–965. [Google Scholar]
- Cheng, F.; Tang, X.L.; Kardashliev, T. Transcription Factor-Based Biosensors in High-Throughput Screening: Advances and Applications. Biotechnol. J. 2018, 13, e1700648. [Google Scholar] [CrossRef]
- Siedler, S.; Stahlhut, S.G.; Malla, S.; Maury, J.; Neves, A.R. Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab. Eng. 2014, 21, 2–8. [Google Scholar] [CrossRef]
- Tu, R.; Li, L.; Yuan, H.; He, R.; Wang, Q. Biosensor-enabled droplet microfluidic system for the rapid screening of 3-dehydroshikimic acid produced in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2020, 47, 1155–1160. [Google Scholar] [CrossRef]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, Q.; Huang, W.; Wang, K.; Zhu, X.; Xie, M. Detection of HIV-1 ribonuclease H activity in single-cell by using RNA mimics green fluorescent protein based biosensor. Sens. Actuators B Chem. 2019, 281, 439–444. [Google Scholar] [CrossRef]
- Lim, H.G.; Jang, S.; Jang, S.; Seo, S.W.; Jung, G.Y. Design and optimization of genetically encoded biosensors for high-throughput screening of chemicals. Curr. Opin. Biotechnol. 2018, 54, 18–25. [Google Scholar] [CrossRef]
- Cheng, F.; Kardashliev, T.; Pitzler, C.; Shehzad, A.; Lue, H.; Bernhagen, J.; Zhu, L.; Schwaneberg, U. A Competitive Flow Cytometry Screening System for Directed Evolution of Therapeutic Enzyme. ACS Synth. Biol. 2015, 4, 768–775. [Google Scholar] [CrossRef]
- Laohakunakorn, N.; Grasemann, L.; Lavickova, B.; Michielin, G.; Shahein, A.; Swank, Z.; Maerkl, S.J. Bottom-Up Construction of Complex Biomolecular Systems with Cell-Free Synthetic Biology. Front. Bioeng. Biotechnol. 2020, 8, 213. [Google Scholar] [CrossRef]
- Lu, Y. Cell-free synthetic biology: Engineering in an open world. Synth. Syst. Biotechnol. 2017, 2, 23–27. [Google Scholar] [CrossRef]
- Rodionov, D.A.; Vitreschak, A.G.; Mironov, A.A.; Gelfand, M.S. Comparative genomics of the methionine metabolism in Gram-positive bacteria: A variety of regulatory systems. Nucleic Acids Res. 2004, 32, 3340–3353. [Google Scholar] [CrossRef]
- Shin, J.; Noireaux, V. An E. coli cell-free expression toolbox: Application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 2012, 1, 29–41. [Google Scholar] [CrossRef]
- Tabuchi, T.; Yokobayashi, Y. High-throughput screening of cell-free riboswitches by fluorescence-activated droplet sorting. Nucleic Acids Res. 2022, 50, 3535–3550. [Google Scholar] [CrossRef]
- Vallejo, D.; Nikoomanzar, A.; Paegel, B.M.; Chaput, J.C. Fluorescence-Activated Droplet Sorting for Single-Cell Directed Evolution. ACS Synth. Biol. 2019, 8, 1430–1440. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, Y.; Xu, Q.; Sun, X.; Meng, F. Recent Advances on Sorting Methods of High-Throughput Droplet-Based Microfluidics in Enzyme Directed Evolution. Front. Chem. 2021, 6, 666867. [Google Scholar] [CrossRef]
- Gu, S.; Lu, Y.; Ding, Y.; Li, L.; Zhang, F.; Wu, Q. Droplet-based microfluidics for dose–response assay of enzyme inhibitors by electrochemical method. Anal. Chim. Acta 2013, 796, 68–74. [Google Scholar] [CrossRef]
- Gasilova, N.; Yu, Q.; Qiao, L.; Girault, H.H. On-chip spyhole mass spectrometry for droplet-based microfluidics. Angew. Chem. (Int. Ed. Engl.) 2014, 53, 4408–4412. [Google Scholar] [CrossRef]
- Pullagura, B.K.; Amarapalli, S.; Gundabala, V. Coupling electrohydrodynamics with photopolymerization for microfluidics-based generation of polyethylene glycol diacrylate (PEGDA) microparticles and hydrogels. Colloids Surf. A Physicochem. Eng. Asp. 2021, 608, 125586. [Google Scholar] [CrossRef]
- Goto, H.; Kanai, Y.; Yotsui, A.; Shimokihara, S.; Shitara, S.; Oyobiki, R.; Fujiwara, K.; Watanabe, T.; Einaga, Y.; Matsumoto, Y.; et al. Microfluidic screening system based on boron-doped diamond electrodes and dielectrophoretic sorting for directed evolution of NAD(P)-dependent oxidoreductases. Lab Chip 2020, 20, 852–861. [Google Scholar] [CrossRef]
- Norris, J.L.; Porter, N.A.; Caprioli, R.M. Mass spectrometry of intracellular and membrane proteins using cleavable detergents. Anal. Chem. 2003, 75, 6642–6647. [Google Scholar] [CrossRef]
- Heinemann, J.; Deng, K.; Shih, S.C.C.; Gao, J.; Adams, P.D.; Singh, A.K.; Northen, T.R. On-chip integration of droplet microfluidics and nanostructure-initiator mass spectrometry for enzyme screening. Lab Chip 2017, 17, 323–331. [Google Scholar] [CrossRef]
- Holland-Moritz, D.A.; Wismer, M.K.; Mann, B.F.; Farasat, I.; Devine, P.; Guetschow, E.D.; Mangion, I.; Welch, C.J.; Moore, J.C.; Sun, S.; et al. Mass Activated Droplet Sorting (MADS) Enables High-Throughput Screening of Enzymatic Reactions at Nanoliter Scale. Angew. Chem. (Int. Ed. Engl.) 2020, 59, 4470–4477. [Google Scholar] [CrossRef]
- Willner, M.R.; McMillan, K.S.; Graham, D.; Vikesland, P.J.; Zagnoni, M. Surface-Enhanced Raman Scattering Based Microfluidics for Single-Cell Analysis. Anal. Chem. 2018, 90, 12004–12010. [Google Scholar] [CrossRef]
- Wang, X.; Xin, Y.; Ren, L.; Sun, Z.; Zhu, P.; Ji, Y.; Li, C.; Xu, J.; Ma, B. Positive dielectrophoresis-based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo. Sci. Adv. 2020, 6, eabb3521. [Google Scholar] [CrossRef]
- Liu, W.W.; Zhu, Y. “Development and application of analytical detection techniques for droplet-based microfluidics”—A review. Anal. Chim. Acta 2020, 1113, 66–84. [Google Scholar] [CrossRef] [PubMed]
- Swyer, I.; Soong, R.; Dryden, M.D.M.; Fey, M.; Maas, W.E.; Simpson, A.; Wheeler, A.R. Interfacing digital microfluidics with high-field nuclear magnetic resonance spectroscopy. Lab Chip 2016, 16, 4424–4435. [Google Scholar] [CrossRef] [PubMed]
- Hale, W.; Rossetto, G.; Greenhalgh, R.; Finch, G.; Utz, M. High-resolution nuclear magnetic resonance spectroscopy in microfluidic droplets. Lab Chip 2018, 18, 3018–3024. [Google Scholar] [CrossRef]
- Choi, K.; Mudrik, J.M.; Wheeler, A.R. A guiding light: Spectroscopy on digital microfluidic devices using in-plane optical fibre waveguides. Anal. Bioanal. Chem. 2015, 407, 7467–7475. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Zhao, X.; Zhu, J.; Tu, R.; Dong, L.; Wang, L.; Dong, Z.; Wang, Q.; Du, W. Fluorescence-activated droplet sorting of lipolytic microorganisms using a compact optical system. Lab Chip 2017, 18, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.G.; Huang, M.S.; Wang, H.F.; Fang, Q. Forming a Large-Scale Droplet Array in a Microcage Array Chip for High-Throughput Screening. Anal. Chem. 2019, 91, 10757–10763. [Google Scholar] [CrossRef]
- Qiao, Y.; Hu, R.; Chen, D.; Wang, L.; Wang, Z.; Yu, H.; Fu, Y.; Li, C.; Dong, Z.; Weng, Y.; et al. Fluorescence-activated droplet sorting of PET degrading microorganisms. J. Hazard Mater. 2022, 424 Pt B, 127417. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, Y.; Li, Q.; Zhou, J.; Li, J.; Du, G. Growth-coupled evolution and high-throughput screening assisted rapid enhancement for amylase-producing Bacillus licheniformis. Bioresour. Technol. 2021, 337, 125467. [Google Scholar] [CrossRef]
- Prodanovic, R.; Ung, W.L.; Durdic, K.I.; Fischer, R.; Weitz, D.A.; Ostafe, R. A High-Throughput Screening System Based on Droplet Microfluidics for Glucose Oxidase Gene Libraries. Molecules 2020, 25, 2418. [Google Scholar] [CrossRef]
- Guo, L.; Zeng, W.; Xu, S.; Zhou, J. Fluorescence-activated droplet sorting for enhanced pyruvic acid accumulation by Candida glabrata. Bioresour. Technol. 2020, 318, 124258. [Google Scholar] [CrossRef]
- Steyer, D.J.; Kennedy, R.T. High-Throughput Nanoelectrospray Ionization-Mass Spectrometry Analysis of Microfluidic Droplet Samples. Anal. Chem. 2019, 91, 6645–6651. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, X.W.; Farasat, I.; Guetschow, E.D.; Welch, C.J.; Kennedy, R.T.; Sun, S.; Moore, J.C. Enabling Biocatalysis by High-Throughput Protein Engineering Using Droplet Microfluidics Coupled to Mass Spectrometry. ACS Omega 2018, 3, 1498–1508. [Google Scholar] [CrossRef] [PubMed]

| Spectrum | Screening Analysis Strategies | Screening Target | Specific Applications | References |
|---|---|---|---|---|
| Ultraviolet light | Addition of metabolite dyes | - | A planar microfluidic spectroscopy detection system was proposed, which could continuously analyze and determine thymol blue of the staining at 180–890 nm and realize the high-throughput detection of the corresponding stained species. | [28] |
| Ultraviolet light | Enzymatic reaction | E. coli | UV–Vis full-wavelength detection system to monitor E. coli growth at 280 nm and thiouric acid assay at 311 nm to identify strain expression ergothionease activity. | [16] |
| Visible light | Direct measurement of absorbance | Yeast | Integrated droplet digital microfluidic system to measure the growth of mutant and wild-type yeasts in ionic liquids. | [29] |
| Visible light | Enzymatic reaction | HL-60 Cell population | Differential detection photothermal interferometry combined with droplet microfluidics, relying on electronic media and mitochondrial succinate-tetrazolium reductase reaction to generate absorbance and the high-throughput analysis of the HL-60 cell population’s metabolic activity. | [30] |
| Visible light | Enzyme-coupled reaction | Glucose oxidase | Using the principle of enzyme colorimetry, glucose hydrolysis intermediate H2O2, 4-aminoantipyrine, and phenol generate red quinone imine, thereby continuously determining glucose oxidase activity with a high throughput. | [31] |
| Fluorescence | Direct measurement of target metabolite | Lactic acid bacteria | Riboflavin has a natural fluorescent signal, and high-yield mutant strains are screened for use in milk fermentation, and riboflavin reaches 2.81 mg/L. | [32] |
| Fluorescence | Addition of metabolite dyes | Microalgae strain | The fluorescent stain BODIPY was added to stain the lipids, and microalgae strains with a 2.75-fold increase in lipid yield were screened. | [33] |
| Fluorescence | Embedding metal chelating agents | - | Add EtBr to bind to RNA to determine ribonuclease activity: the higher the activity, the lower the fluorescence signal. | [34] |
| Fluorescence | Enzymatic reaction | Environmental microorganisms | Fluorescein dibutyrate was introduced as a fluorescent substrate, and 11 lipase-producing strains were screened from environmental microorganisms using FADS. | [35] |
| Fluorescence | Enzymatic reaction | E. coli | A method was developed to rapidly form a large-scale droplet array using microcage array chips, which improved the operability of droplets and introduced fluorescent substrates to screen strains expressing esterase AFEST from mixed bacteria. | [36] |
| Fluorescence | Enzymatic reaction | Viable bacteria in wastewater | The fluorescent substrate diphenyl dibenzoate can be degraded by PETase to produce a fluorescent signal, which can screen for strains which express PETase efficiently. | [37] |
| Fluorescence | Enzyme-coupled reaction | Bacillus licheniformus | According to the modified 3,5-dinitrosalicylic acid (DNS) method, starch substrates are decomposed into glucose; DNS and glucose undergo redox reactions to produce fluorescent substances, and strains with α-amylase expression increased by 67% were screened. | [38] |
| Fluorescence | Enzyme-coupled reaction | Yeast | The glucose in the microdroplets was decomposed via oxidase to produce H2O2; H2O2 reacted with vanadium bromoperoxidase to produce BrO−, and BrO- reacted with aminophenoxyfluorescein to produce a strong fluorescence, and an enzyme coupling strategy was established to screen out glucose oxidase mutants 2.1 times higher than those of wild-type Kcat. | [39] |
| Fluorescence | Protein-based biosensors | Candida glabrum | A biosensor-expressing pH-sensitive fluorescent protein was constructed, and the ankyrin gene was introduced for expression so that the fluorescent protein was anchored on the surface of Candida glabricis to show the accumulation of pyruvate, and the strain with a 73.6% increase in pyruvate yield, reaching 48.6 g/L, was screened. | [40] |
| Fluorescence | Transcription factor-based biosensors | E. coli | A biosensor in response to 3-dehydroshikimic acid (3-DHS) was constructed, and 3-DHS was positively regulated, expressing a fluorescent protein, and screening mutant strains with high yields of 3-DHS, with a yield of 2.46 g/L at 24 h. | [41] |
| Fluorescence | Nucleic acid-based biosensors (RNA Spinach) | Saccharomyces cerevisiae | Innovated on the basis of RNA Spinach, a universal sensing technology which provides small molecule metabolites by altering RNA sequences, and screened mutant strains with 28-fold and 3-fold higher yields of tyrosine and recombinant protein streptomycin, respectively. | [24] |
| Non-spectral | Coupled mass spectrometry analysis | Transaminase | Combined with droplet microfluidics, a high-throughput and stable droplet analysis system was constructed to detect the reaction products of intradroplet transaminases, quantify enzyme activity, and evaluate enzyme mutants. | [42] |
| Non-spectral | Coupled mass spectrometry analysis | E. coli | Transaminases can convert methyl 4-methyl-3-oxovalerate into the corresponding amine, and the amine is then spontaneously hydrolyzed to β-leucine, which is detected using the microfluidic-mass spectrometry analysis system and screened out a variety of mutants of transaminases. | [43] |
| Raman spectroscopy | Coupled surface-enhanced Raman scattering | Prostate cancer cell population | Surface-enhanced Raman scattering combined with microfluidic system improved the sensitivity of the detection system, and the glycan expression and cell variability of prostate cancer cells were analyzed at high throughput using wheat germ lectin-modified nanometal particles. | [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Wang, B.; Luo, Q.; Zeng, R.; Zhang, J.; Cheng, J. Advances in Droplet-Based Microfluidic High-Throughput Screening of Engineered Strains and Enzymes Based on Ultraviolet, Visible, and Fluorescent Spectroscopy. Fermentation 2024, 10, 33. https://doi.org/10.3390/fermentation10010033
Hu S, Wang B, Luo Q, Zeng R, Zhang J, Cheng J. Advances in Droplet-Based Microfluidic High-Throughput Screening of Engineered Strains and Enzymes Based on Ultraviolet, Visible, and Fluorescent Spectroscopy. Fermentation. 2024; 10(1):33. https://doi.org/10.3390/fermentation10010033
Chicago/Turabian StyleHu, Shunyang, Bangxu Wang, Qing Luo, Rumei Zeng, Jiamin Zhang, and Jie Cheng. 2024. "Advances in Droplet-Based Microfluidic High-Throughput Screening of Engineered Strains and Enzymes Based on Ultraviolet, Visible, and Fluorescent Spectroscopy" Fermentation 10, no. 1: 33. https://doi.org/10.3390/fermentation10010033
APA StyleHu, S., Wang, B., Luo, Q., Zeng, R., Zhang, J., & Cheng, J. (2024). Advances in Droplet-Based Microfluidic High-Throughput Screening of Engineered Strains and Enzymes Based on Ultraviolet, Visible, and Fluorescent Spectroscopy. Fermentation, 10(1), 33. https://doi.org/10.3390/fermentation10010033





