The Microbial Community in a Substrate of Solid-State Fermentation by Lentinula edodes: A Preliminary Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Experiment Settings
2.2. Microbial Incubation and Respiration Measurement
2.3. Profiling of Microbial Community Structure and Composition
2.4. Data Analysis
3. Results
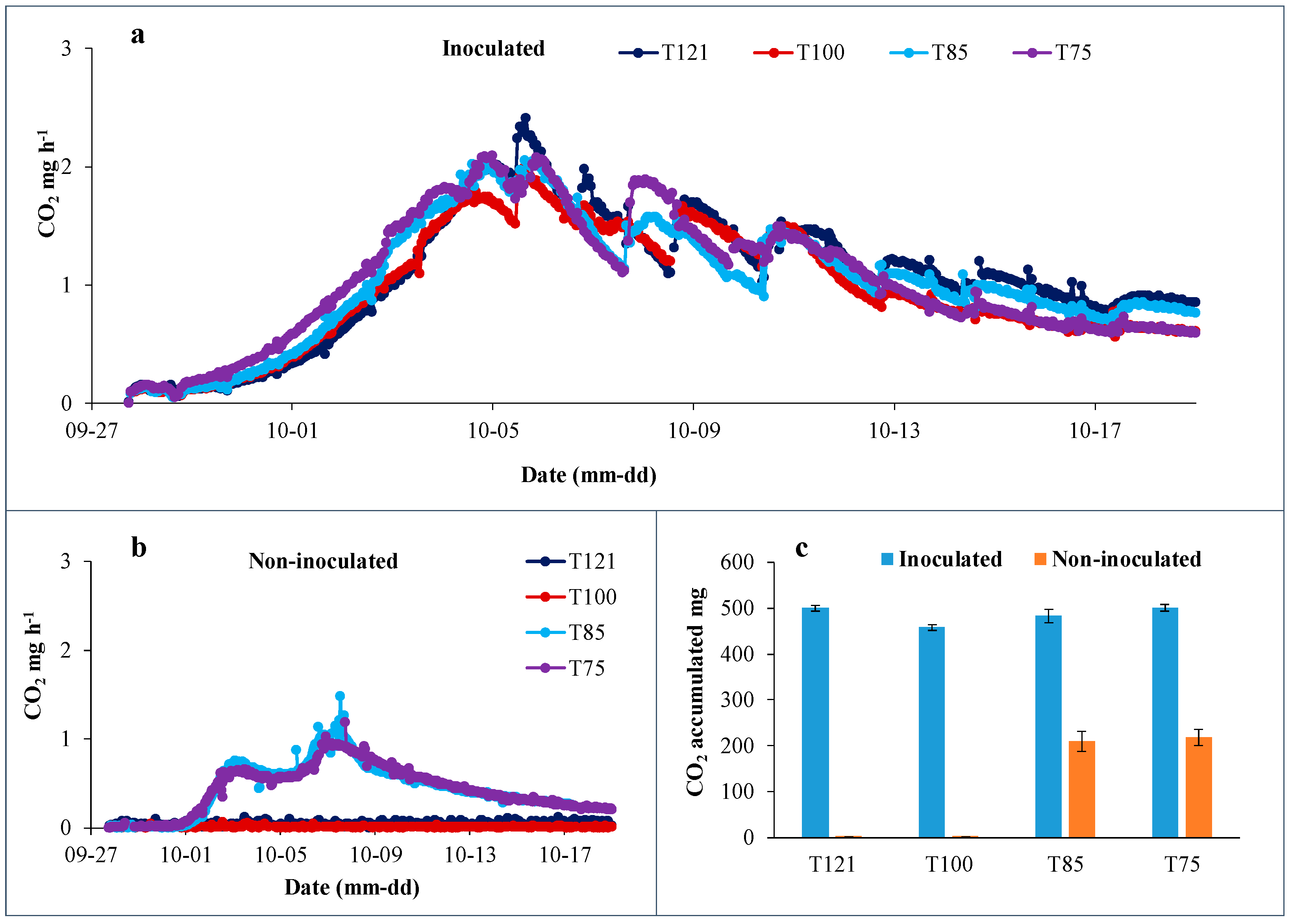
3.1. Microbial Activities in Shiitake Substrates
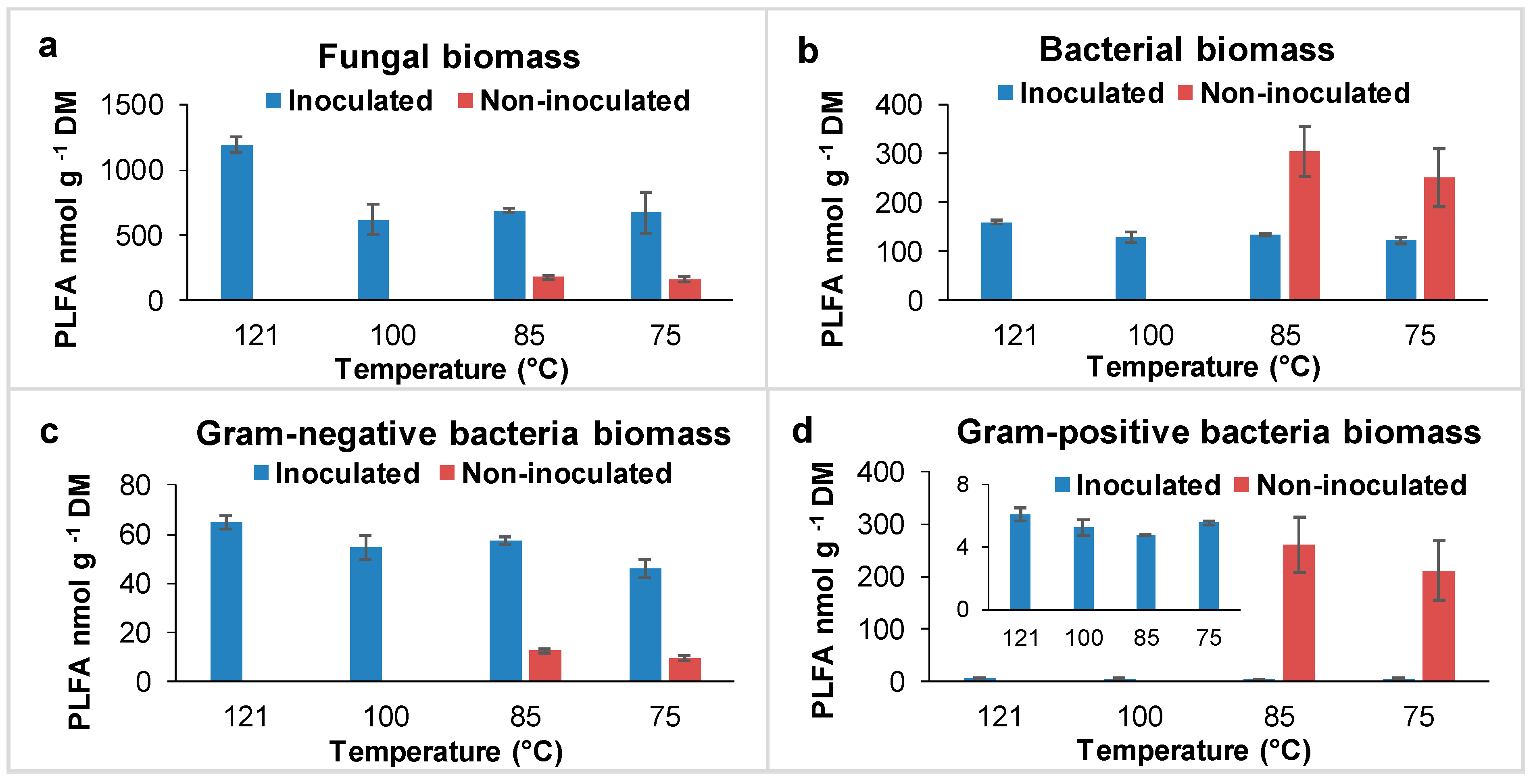
3.2. Microbial Community Structure Profile Based on PFLA Studies
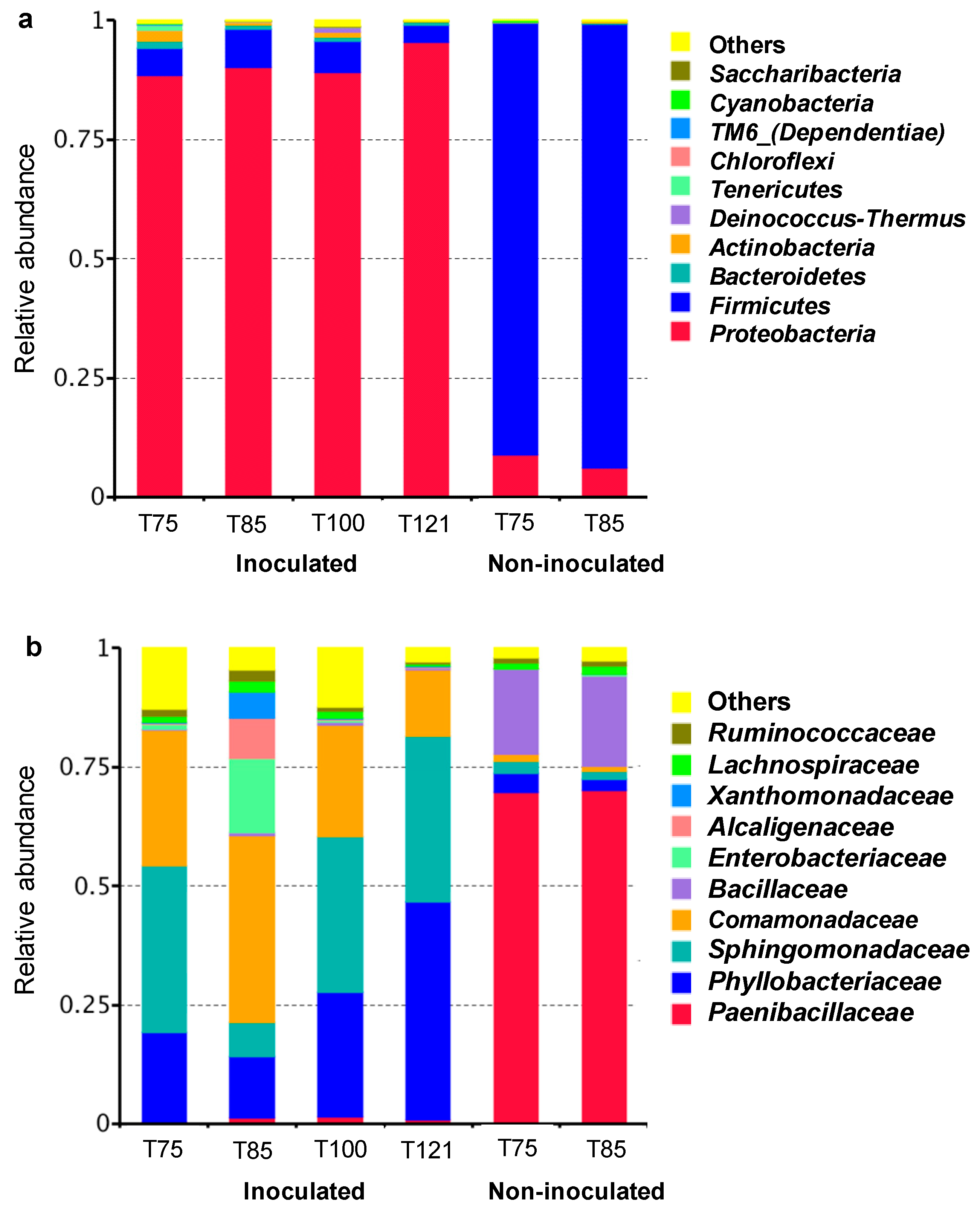
3.3. Microbial Community Composition Based on DNA Sequencing
4. Discussion
4.1. The Microbial Community after Different Heat Treatments
4.2. Coexisting Strains of Bacteria in Shiitake Substrates
4.3. Implication for Biorefinery Development
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, Y.; Ge, X.; Liu, Z.; Li, Y. Integration of shiitake cultivation and solid-state anaerobic digestion for utilization of wood biomass. Bioresour. Technol. 2015, 182, 128–135. [Google Scholar] [CrossRef]
- Xiong, S.J.; Martín, C.; Eilertsen, L.; Wei, M.; Myronycheva, O.; Larsson, S.; Lestander, T.; Atterhem, L.; Jönsson, L. Energy-efficient substrate pasteurisation for combined production of shiitake mushroom (Lentinula edodes) and bioethanol. Bioresour. Technol. 2019, 274, 65–72. [Google Scholar] [CrossRef]
- Wan, C.X.; Li, Y.B. Fungal pretreatment of lignocellulosic biomass. Biotechnol. Adv. 2012, 30, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Xiong, S.J.; Chen, F.; Geladi, P.; Eilertsen, L.; Myronycheva, O.; Lestander, T.A.; Thyrel, M. Energy smart hot-air pasteurisation as effective as energy intense autoclaving for fungal preprocessing of lignocellulose feedstock for bioethanol fuel production. Renew. Energy 2020, 155, 237–247. [Google Scholar] [CrossRef]
- Fellows, P.J. Food Processing Technology Principles and Practice. In Food Science, Technology and Nutrition, 5th ed.; Woodhead Publishing: Cambridge, UK, 2017; pp. 343–383. [Google Scholar]
- Vieira, F.R.; Pecchia, J.A. An exploration into the bacterial community under different pasteurization conditions during substrate preparation (compostingphase II) for Agaricus bisporus cultivation. Microb. Ecol. 2018, 75, 318–330. [Google Scholar] [CrossRef]
- Carrasco, J.; García-Delgado, C.; Lavega, R.; Tello, M.L.; De Toro, M.; Barba-Vicente, V.; Rodríguez-Cruz, M.S.; Sánchez-Martín, M.J.; Pérez, M.; Preston, G.M. Holistic assessment of the microbiome dynamics in the substrates used for commercial champignon (Agaricus bisporus) cultivation. Microb. Biotechnol. 2020, 13, 1933–1947. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.L.; Sun, B.; Zhang, J.Y.; Zhang, Y.T.; Gu, L.K.; Bao, L.J.; Liu, S.X. Metagenomic analysis revealed the succession of microbiota and metabolic function in corncob composting for preparation of cultivation medium for Pleurotus ostreatus. Bioresour. Technol. 2020, 306, 123156. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhou, R.; Ma, Y.; Fang, Y.; Xie, G.P.; Gao, X.Z.; Xiao, Y.Z.; Liu, J.J.; Fang, Z.M. Development of a consortium-based microbial agent beneficial to composting of distilled grain waste for Pleurotus ostreatus cultivation. Biotechnol. Biofuels 2021, 14, 242. [Google Scholar] [CrossRef]
- Lee, C.K.; Haque, M.A.; Choi, B.R.; Lee, H.Y.; Eun, C. Molecular diversity of endobacterial communities in edible part of king oyster mushroom (Pleurotus eryngii) based on 16S rRNA. Korean J. Microbiol. 2015, 51, 148–155. [Google Scholar] [CrossRef]
- Chen, L.; Yan, M.; Qian, X.; Yang, Z.; Xu, Y.; Wang, T.; Cao, J.; Sun, S. Bacterial community composition in the growth process of Pleurotus eryngii and growth-promoting abilities of isolated bacteria. Front. Microbiol. 2022, 13, 787628. [Google Scholar] [CrossRef]
- Carrasco, J.; Preston, G.M. Growing edible mushrooms: A conversation between bacteria and fungi. Environ. Microbiol. 2020, 22, 858–872. [Google Scholar] [CrossRef] [PubMed]
- Torres-Ruiz, E.; Sánchez, J.E.; Guillén-Navarro, G.K.; Ramos-Pérez, D.G.; Royse, D.J. Microbial promoters of mycelial growth: Fruiting and production of Pleurotus ostreatus. Sydowia 2016, 68, 151–161. [Google Scholar]
- Kertesz, M.A.; Thai, M. Compost bacteria and fungi that influence growth and development of Agaricus bisporus and other commercial mushrooms. Appl. Microbiol. Biotechnol. 2018, 102, 1639–1650. [Google Scholar] [CrossRef]
- Nordgren, A. Apparatus for the continuous, long-term monitoring of soil respiration rate in large number of samples. Soil Biol. Biochem. 1988, 20, 955–957. [Google Scholar] [CrossRef]
- Frostegård, Å.; Bååth, E. The use of phospholipid analysis to estimate bacterial and fungal biomass in soil. Biol. Fertil. Soils 1996, 22, 59–65. [Google Scholar] [CrossRef]
- Steger, K.; Jarvis, A.; Smårs, S.; Sundh, I. Comparison of signature lipid methods to determine microbial community structure in compost. J. Microbiol. Methods 2003, 55, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Kroppenstedt, R.M. Fatty acid menaquinone analysis of actinomycetes and related organisms. In Chemical Methods in Bacterial Systematics; Goodfellow, M., Minnikin, D.E., Eds.; Academic Press: New York, NY, USA, 1985; pp. 173–199. [Google Scholar]
- Willers, C.; Jansen van Rensburg, P.; Claassens, S. Phospholipid fatty acid profiling of microbial communities—A review of interpretations and recent applications. J. Appl. Microbiol. 2015, 119, 1207–1218. [Google Scholar] [CrossRef]
- Chung, I.M.; Kim, S.Y.; Han, J.G.; Kong, W.S.; Jung, M.Y.; Kim, S.H. Fatty acids and stable isotope ratios in shiitake mushrooms (Lentinula edodes) indicate the origin of the cultivation substrate used: A preliminary case study in Korea. Foods 2020, 9, 1210. [Google Scholar] [CrossRef]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 2011, 43, 1621–1625. [Google Scholar] [CrossRef]
- Silva, J.P.; Ticona, A.R.P.; Hamann, P.R.V.; Quirino, B.F.; Noronha, E.F. Deconstruction of lignin: From enzymes to microorganisms. Molecules 2021, 26, 2299. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, P.; Selvankumar, T.; Kamala-Kannan, S.; Mythili, R.; Sengottaiyan, A.; Govarthanan, M.; Senthilkumar, B.; Selvam, K. Production and purification of laccase by Bacillus spp. using millet husks and its pesticide degradation application. 3 Biotech 2019, 9, 396. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, C. Cultivation of Pleurotus ostreatus and other edible mushrooms. Appl. Microbiol. Biotechnol. 2010, 85, 1321–1337. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, X.; Lou, K.; Adams, M. Isolation, characterization and insecticidal activity of an endophyte of drunken horse grass: Achnatherum inebrians. J. Insect Sci. 2013, 13, 1. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Eisen, J.A. Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina. PLoS ONE 2020, 15, e0236135. [Google Scholar] [CrossRef]
- Mantelin, S.; Saux, M.F.; Zakhia, F.; Bena, G.; Bonneau, S.; Jeder, H.; de Lajudie, P.; Cleyet-Marel, J.C. Emended description of the genus Phyllobacterium and description of four novel species associated with plant roots: Phyllobacterium bourgognense sp. nov., Phyllobacterium ifriqiyense sp. nov., Phyllobacterium leguminum sp. nov. and Phyllobacterium brassicacearum sp. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 827–839. [Google Scholar]
- Holguin, G.; Guzman, M.A.; Bashan, Y. Two new nitrogen-fixing bacteria from the rhizosphere of mangrove trees: Their isolation, identification and in vitro interaction with rhizosphere Staphylococcus sp. FEMS Microbiol. Lett. 1992, 101, 207–216. [Google Scholar] [CrossRef]
- Rojas, A.; Holguin, G.; Glick, B.R.; Bashan, Y. Synergism between Phyllobacterium sp. (N2-fixer) and Bacillus licheniformis (P-solubilizer), both from a semiarid mangrove rhizosphere. FEMS Microbiol. Ecol. 2001, 35, 181–187. [Google Scholar] [CrossRef]
- Makino, A.; Nakai, R.; Yoneda, Y.; Toyama, T.; Tanaka, Y.; Meng, X.Y.; Mori, K.; Ike, M.; Morikawa, M.; Kamagata, Y.; et al. Isolation of aquatic plant growth-promoting bacteria for the floating plant duckweed (Lemna minor). Microorganisms 2022, 10, 1564. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Almeida, C.; Silva Pereira, C.; Gonzalez-Menendez, V.; Bills, G.; Pascual, J.; Sánchez-Hidalgo, M.; Kehraus, S.; Genilloud, O. Unveiling concealed functions of endosymbiotic bacteria harbored in the ascomycete Stachylidium bicolor. Appl. Environ. Microbiol. 2018, 84, e00660-18. [Google Scholar] [CrossRef]
- Jiao, Y.S.; Yan, H.; Ji, Z.J.; Liu, Y.H.; Sui, X.H.; Zhang, X.X.; Wang, E.T.; Chen, W.X.; Chen, W.F. Phyllobacterium sophorae sp. nov.; a symbiotic bacterium isolated from root nodules of Sophora flavescens. Int. J. Syst. Evol. Microbiol. 2015, 65, 399–406. [Google Scholar] [CrossRef]
- Larcher, M.; Muller, B.; Mantelin, S.; Rapior, S.; Cleyet-Marel, J.C. Early modifications of Brassica napus root system architecture induced by a plant growth-promoting Phyllobacterium strain. New Phytol. 2003, 160, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Flores-Félix, J.D.; Velázquez, E.; García-Fraile, P.; González-Andrés, F.; Silva, L.R.; Rivas, R. Rhizobium and Phyllobacterium bacterial inoculants increase bioactive compounds and quality of strawberries cultivated in field conditions. Food Res. Int. 2018, 111, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Bresson, J.; Varoquaux, F.; Bontpart, T.; Touraine, B.; Vile, D. The PGPR strain Phyllobacterium brassicacearum STM 196 induces a reproductive delay and physiological changes that result in improved drought tolerance in Arabidopsis. New Phytol. 2013, 200, 558–569. [Google Scholar] [CrossRef]
- Kechid, M.; Desbrosses, G.; Gamet, L.; Castaings, L.; Varoquaux, F.; Djekoun, A.; Touraine, B. Arabidopsis growth-promotion and root architecture responses to the beneficial rhizobacterium Phyllobacterium brassicacearum strain STM196 are independent of the nitrate assimilatory pathway. Plants 2022, 11, 128. [Google Scholar] [CrossRef] [PubMed]
- Valverde, A.; Velázquez, E.; Fernández-Santos, F.; Vizcaino, N.; Rivas, R.; Mateos, P.F.; Martínez-Molina, E.; Igual, J.M.; Willems, A. Phyllobacterium trifolii sp. nov.; nodulating Trifolium and Lupinus in Spanish soils. Int. J. Syst. Evol. Microbiol. 2005, 55, 1985–1989. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Yadav, S.; Sharma, R.; Dalela, M.; Celin, S.M.; Sharma, A.; Sharma, S. Augmentation of stimulated Pelomonas aquatica dispersible granules enhances remediation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) contaminated soil. Environ. Technol. Innov. 2020, 17, 100594. [Google Scholar] [CrossRef]
- Somal, M.K.; Karnwal, A. Effect of stress tolerance endophytic bacteria on the growth of Rheum emodi under abiotic stress. Plant Biol. J. 2022. [Google Scholar] [CrossRef]
- Doudoroff, M. The oxidative assimilation of sugars and related substances by Pseudomonas saccharophila with a contribution to the problem of the direct respiration of di-and polysaccharides. Enzymologia 1940, 9, 59–72. [Google Scholar]
- Nagar, S.; Shaw, A.K.; Anand, S.; Celin, S.M.; Rai, P.K. Biodegradation of octogen and hexogen by Pelomonas aquatica strain WS2-R2A-65 under aerobic condition. Environ. Technol. 2020, 43, 1003–1012. [Google Scholar] [CrossRef]
- Gomila, M.; Bowien, B.; Falsen, E.; Moore, E.R.; Lalucat, J. Description of Pelomonas aquatica sp. nov. and Pelomonas puraquae sp. nov., isolated from industrial and haemodialysis water. Int. J. Syst. Evol. Microbiol. 2007, 57, 2629–2635. [Google Scholar] [CrossRef] [PubMed]
- Abraham, W.R.; Estrela, A.B.; Rohde, M.; Smit, J.; Vancanneyt, M. Prosthecate sphingomonads: Proposal of Sphingomonas canadensis sp. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 3214–3219. [Google Scholar] [CrossRef]
- Willison, J.C. Isolation and characterization of a novel sphingomonad capable of growth with chrysene as sole carbon and energy source. FEMS Microbiol. Lett. 2004, 241, 143–150. [Google Scholar] [CrossRef]
- Wittich, R.M.; Wilkes, H.; Sinnwell, V.; Francke, W.; Fortnagel, P. Metabolism of dibenzo-p-dioxin by Sphingomonas spp. strain RW1. Appl. Environ. Microbiol. 1992, 58, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Ashtaputre, A.A.; Shah, A.K. Studies on a viscous, gel-forming exopolysaccharide from Sphingomonas paucimobilis GS1. Appl. Environ. Microbiol. 1995, 61, 1159–1162. [Google Scholar] [CrossRef] [PubMed]
- Tangaromsuk, J.; Pokethitiyook, P.; Kruatrachue, M.; Upatham, E.S. Cadmium biosorption by Sphingomonas paucimobilis biomass. Bioresour. Technol. 2002, 85, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Feng, L.; Liu, K.; Cheng, Y.; Hou, N.; Li, C. Optimization of cold-active CMCase production by psychrotrophic Sphingomonas sp. FLX-7 from the cold region of China. Cellulose 2016, 23, 1335–1347. [Google Scholar] [CrossRef]
- Halo, B.A.; Khan, A.L.; Waqas, M.; Al-Harrasi, A.; Hussain, J.; Ali, L.; Adnan, M.; Lee, I.J. Endophytic bacteria (Sphingomonas sp. LK11) and gibberellin can improve Solanum lycopersicum growth and oxidative stress under salinity. J. Plant. Interact. 2015, 10, 117–125. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, X.; Cao, Z.; Zhao, K.; Wang, S.; Chen, M.; Hu, X. Growth-promoting Sphingomonas paucimobilis ZJSH 1 associated with Dendrobium officinale through phytohormone production and nitrogen fixation. Microb. Biotechnol. 2014, 7, 611–620. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, F.; Huang, Y.; Zhou, M.; Gao, J.; Yan, T.; Sheng, H.; An, L. Sphingomonas sp. Cra20 increases plant growth rate and alters rhizosphere microbial community structure of Arabidopsis thaliana under drought stress. Front. Microbiol. 2019, 10, 1221. [Google Scholar] [CrossRef] [PubMed]
- Videira, S.S.; De Araujo, J.L.S.; da Silva Rodrigues, L.; Baldani, V.L.D.; Baldani, J.I. Occurrence and diversity of nitrogen-fixing Sphingomonas bacteria associated with rice plants grown in Brazil. FEMS Microbiol. Lett. 2009, 293, 11–19. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Egorova, M.A.; Leontieva, M.R.; Malakho, S.G.; Kolomeitseva, G.L.; Netrusov, A.I. Dendrobium nobile Lindl. seed germination in co-cultures with diverse associated bacteria. Plant Growth Regul. 2016, 80, 79–91. [Google Scholar] [CrossRef]
- Lowman, S.; Kim-Dura, S.; Mei, C.; Nowak, J. Strategies for enhancement of switchgrass (Panicum virgatum L.) performance under limited nitrogen supply based on utilization of N-fixing bacterial endophytes. Plant Soil 2016, 405, 47–63. [Google Scholar] [CrossRef]
- Karni, M.; Zidon, D.; Polak, P.; Zalevsky, Z.; Shefi, O. Thermal degradation of DNA. DNA Cell Biol. 2013, 32, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Bauer, T.; Weller, P.; Hammes, W.P.; Hertel, C. The effect of processing parameters on DNA degradation in food. Eur. Food Res. Technol. 2003, 217, 338–343. [Google Scholar] [CrossRef]

| Heat treatment | T121: Steam 121 °C with 2 bar pressure |
| (sterilization/pasteurization) | T100: Hot air 100 °C for 1 h |
| T85: Hot air 85 °C for 1 h | |
| T75: Hot air 75 °C for 1 h | |
| Shiitake inoculation | Inoculated vs. non-inoculated |
| Incubation | At 20 °C; in Respicond respirator |
| Measurement/analysis | CO2 release ratio (mg/h); CO2 release accumulated (mg); Profile of microbial community structure and taxonomy by PLFA analysis; and ITS 18S and 16S rRNA gene sequencing. |
| Inoculated | Non-Inoculated | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Classs | Order | Family | Genus | Species | T121 | T100 | T85 | T75 | T85 | T75 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus | B. circulans | 0.20 | 0.35 | 0.31 | 0.11 | 0.49 | 18.60 |
| B. subterraneus | <0.01 | <0.01 | 0.01 | <0.01 | |||||||
| B. spp. | 0.18 | 0.32 | 0.25 | 0.09 | 19.91 | 0.88 | |||||
| Cohnella | C. xylanilytica | 0.30 | 0.44 | 0.48 | 0.15 | 49.95 | 0.42 | ||||
| Paenibacillaceae | Paenibacillus | P. alginolyticus | 0.01 | 0.01 | <0.01 | <0.01 | 0.01 | 0.20 | |||
| P. favisporus | 0.12 | 0.21 | 0.17 | 0.09 | 20.96 | 3.28 | |||||
| P. lautus | 0.01 | 0.02 | 0.01 | <0.01 | 0.03 | 0.63 | |||||
| P. odorifer | 0.11 | 0.13 | 0.36 | 0.06 | 0.23 | 12.02 | |||||
| P. spp. i76 | 0.01 | 0.01 | 0.01 | <0.01 | 0.01 | 1.48 | |||||
| P. spp. | 0.37 | 0.78 | 0.31 | 0.31 | 2.36 | 53.49 | |||||
| Proteobacteria | α-proteobacteria | Rhizobiales | Phyllobacteriaceae | Phyllobacterium | Ph. Spp. | 47.57 | 30.94 | 14.32 | 22.63 | 2.50 | 4.32 |
| Sphingomonadales | Sphingomonadaceae | Sphingomonas | S. leidyi | 36.40 | 38.8 | 7.73 | 41.36 | 1.74 | 2.76 | ||
| β-proteobacteria | Burkholdderiales | Alcaligenaceae | Achromobacter | A. xylosoxidans | <0.01 | 0.09 | 9.18 | 0.03 | 0.04 | 0.04 | |
| Comamonadaceae | Delftia | D. spp. | 0.16 | 0.39 | 26.11 | 0.17 | 0.06 | 0.04 | |||
| Pelomonas | P. spp. | 14.24 | 27.21 | 17.46 | 34.33 | 1.19 | 1.54 | ||||
| γ-proteobacteria | Enterobacteriales | Enterobacteriaceae | Serratia | S. spp. | 0.10 | 0.15 | 17.24 | 0.33 | 0.13 | 0.05 | |
| Pseudomonas | P. spp. | 0.09 | 0.07 | 6.02 | 0.15 | 0.03 | 0.03 | ||||
| Ratio of the identified ones to all bacterial sequences of the top 10 genera % | 96.01 | 84.26 | 90.36 | 82.94 | 92.82 | 91.84 | |||||
| Bacterium | Habitat | Symbiosis | Major Finding/Characteristic | Reference |
|---|---|---|---|---|
| Phyllobacterium spp. | ||||
| Ph. spp. CLE16 | Root of Zostera marina | PGPB | Nitrogen fixing | [27] |
| Ph. sophorae spp. CCBAU 3422 | Root nodule of Sophora flavescens | PGPR | Nodule formation and nitrogen fixation | [34] |
| Ph. spp. (29-15) | Root of Brassica napus | PGPB | Enhanced root length | [35] |
| Ph. (PEPV15) | Legume nodules of Phaseolus vulgaris | PGPB | Increased in citric acid. vitamin C. epicatechin and other bioactive compounds | [36] |
| Ph. rubiacearum | Black, white, and red mangroves | PGPB | Co-cultivation increased nitrogen fixation and phosphate solubilization. | [30] |
| Ph. Brassicacearum STM196 | Root of Brassica napus | PGPR | Increased in biomass. drought resistance and water-use efficiency | [37] |
| Ph. brassicacearum STM196 | Roots of canola plants | PGPR | Stimulating growth | [38] |
| Ph. strain PETP02 | Nodules of Trifolium pratense | PGPR | Induced nodules in roots | [39] |
| Pelomonas spp. | ||||
| P. aquatica strain 12868 | Soil | − | Remediation of hexahydro-1.3.5-trinitro-1.3.5-triazine (RDX) | [40] |
| P. spp. MRB1 | Roots of aquatic plants | PGPB | Increased in growth. biomass and chlorophyll content | [31] |
| P. spp. MRB3 | Roots of aquatic plants | PGPB | Enhanced in biomass. chlorophyll and high growth-promoting effect | [31] |
| P. aquatica AIS1S | Abutilon indicum | PGPR | Seed germination under salt and heavy metal stress conditions | [41] |
| P. saccharophila | Mud | − | Hydrogen-oxidizing bacterium | [42] |
| P. aquatica strain WS2-R2A-65 | Nitramine explosive-contaminated effluent | − | Biodegradation of octogen and hexogen | [43] |
| P. strains CCUG 52769T | Haemodialysis water | − | Presence of nifH (nitrogenase) and hoxG (hydrogenase) genes | [44] |
| Sphingomonas spp. | ||||
| S. canadensis strain FWC47T | Pulp mill sludge pond | − | Positive towards α-chymotrypsin. β-galactosidase activities and hydrolysed aesculin | [45] |
| S. spp. strain CHY-1 | Soil (heavily contaminated with PAHs from a coal gasification site) | − | Capable of degrading PAHs such as naphthalene. phenanthrene and chrysene | [46] |
| S. wittichii RW1 | Water from the river Elbe, Germany | − | Biodegradation of biaryl ethers dibenzo-p-dioxin and dibenzofuran | [47] |
| S. paucimobilis GS1 | Soil | − | Produce EPS with high mechanical and heat resistance | [48] |
| S. paucimobilis strain BKK1 | Municipal wastewater treatment plant | − | Highly efficient for cadmium removal from solution | [49] |
| S. spp. FLX-7 | Soil | − | Degrades cellulose at low temperatures | [50] |
| S. spp. LK11 | Leaves of Tephrosia apollinea | PGPR | Improve salinity tolerance to tomato crops, increase biomass and growth. | [51] |
| S. paucimobilis ZJSH1 | Root of Dendrobium officinale | PGPB | Phytohormone production and nitrogen fixation | [52] |
| S. spp. Cra20 | Root of Leontopodium leontopodioides | PGPR | Promotes growth rate under water-deficit | [53] |
| S. spp. strains BR12245. BR12249. BR12253. BR12195. and BR12200 | Washed root of rice plant | PGPR | Nitrogen fixing | [54] |
| S. spp. (JQ660212.1) | Aerial roots of Dendrobium moschatum | PGPR | Promotion of seed germination | [55] |
| S. spp. strain NSL | Seeds of Panicum virgatum cv. Alamo | PGPB | Nitrogen fixation and promote growth under low nitrogen | [56] |
| S. leidyi | Stachylidium bicolor mycelium | Endo-bacterial symbiosis | Secondary metabolites in Ascomycota host | [33] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eilertsen, L.; Hultberg, M.; Lee, N.; Saroj, P.; Swaine, M.; Chen, F.; Xiong, S. The Microbial Community in a Substrate of Solid-State Fermentation by Lentinula edodes: A Preliminary Study. Fermentation 2023, 9, 736. https://doi.org/10.3390/fermentation9080736
Eilertsen L, Hultberg M, Lee N, Saroj P, Swaine M, Chen F, Xiong S. The Microbial Community in a Substrate of Solid-State Fermentation by Lentinula edodes: A Preliminary Study. Fermentation. 2023; 9(8):736. https://doi.org/10.3390/fermentation9080736
Chicago/Turabian StyleEilertsen, Lill, Malin Hultberg, Natuschka Lee, Paramjeet Saroj, Mark Swaine, Feng Chen, and Shaojun Xiong. 2023. "The Microbial Community in a Substrate of Solid-State Fermentation by Lentinula edodes: A Preliminary Study" Fermentation 9, no. 8: 736. https://doi.org/10.3390/fermentation9080736
APA StyleEilertsen, L., Hultberg, M., Lee, N., Saroj, P., Swaine, M., Chen, F., & Xiong, S. (2023). The Microbial Community in a Substrate of Solid-State Fermentation by Lentinula edodes: A Preliminary Study. Fermentation, 9(8), 736. https://doi.org/10.3390/fermentation9080736







