Abstract
Metabolic engineering is a promising strategy to realize green synthesis of valued chemicals derived from petroleum. According to the literature, cell factories for producing L-aspartate and its derivatives (β-alanine, ectoine, 3-hydroxypropionate, D-pantothenic acid and L-homoserine) have been developed. In this review, we firstly introduced the functions, applications and markets of L-aspartate and its derivatives. Then, the current research progress on microbial production of them was elaborated in detail. Finally, we have discussed the limiting factors and given some suggestions for realizing applications of engineered bacteria in the industry, including metabolic engineering of the bacteria to increase the titer, yield and productivity of the target products, fermentation condition optimization and downstream purification. With the development of novel technologies and increased investments in synthetic biology, it is promising to realize sustainable production of L-aspartate and its derivatives at the industrial scale in the future.
1. Introduction
Metabolic engineering is a field of biotechnology that focuses on the manipulation and modification of metabolic pathways in cells to enhance the production of desirable products. This involves the use of genetic engineering techniques to alter the DNA of microorganisms, plants and animals in order to optimize their biochemical processes and improve the production of specific compounds. The main goal of metabolic engineering is to design or modify metabolic pathways to produce desired products in large quantities with high efficiency. This can involve introducing genes from other organisms or even synthesizing new genes to produce needed enzymes that enable the desired metabolic reactions [1]. Examples of products that can be produced using metabolic engineering include biofuels [2], bioplastics [3], pharmaceuticals [4], flavors [5], fragrances [6] and food additives [7]. Metabolic engineering is also used in the production of enzymes [8] and in the development of medical treatments for certain diseases [9]. The key advantages of metabolic engineering include the ability to produce large quantities of products in an environmentally sustainable manner with reduced dependence on non-renewable resources [10]. It also enables the creation of new and novel compounds with unique properties that can have a wide range of applications in different industries.
With the help of metabolic engineering, the microbial cell factories for the production of L-aspartate and its derivatives are realized. In this paper, we summarize the research progress on microbial production of L-aspartate and its derivatives: β-alanine, ectoine, 3-hydroxypropionate (3-HP), D-pantothenic acid and homoserine.
2. Metabolic Engineering of Microbials to Produce L-Aspartate and Its Derivatives
2.1. Developing Cell Factories to Produce L-Aspartate
L-aspartate is an amino acid that is naturally found in many fruits and vegetables, as well as in animal proteins. It has various functions, including its role as a precursor for the synthesis of other amino acids and for the production of energy in the body [11]. L-aspartate has various applications in the food and pharmaceutical industries. In the food industry, it is commonly used as a flavor enhancer and a sweetener in many diet and low-calorie products [8,12]. In the pharmaceutical industry, L-aspartate is used to treat symptoms of liver disease and to help improve brain function [13]. The market for L-aspartate is expected to grow in the coming years, as the demand for low-calorie and diet products continues to rise. The global market for L-aspartate was 93.15 million dollars in 2021 and is projected to grow at a CAGR of 6.20% from 2022 to 2029. The increasing demand for sports and energy drinks is also expected to drive the growth of the L-aspartate market. The Asia-Pacific region is anticipated to be the fastest-growing market for L-aspartate, due to the growing demand for low-calorie products and the increasing health awareness among consumers (https://www.databridgemarketresearch.com/reports/global-aspartic-acid-market, accessed on 3 May 2023).
There are several ways to produce L-aspartate at an industrial scale, including extraction from natural sources, chemical synthesis, enzymatic processes and fermentation. The choice of the production method depends on several factors, such as cost, efficiency and sustainability. Currently, the most widely used method is enzymatic conversion of the precursors, fumarate and ammonia, catalyzed by aspartase. For example, E. coli JCL1258/pBAW2/pASP400 produced over 77 g/L of L-aspartate from fumarate with a conversion yield of 83% [14]. Since fumarate is derived from petrochemicals, the enzymatic process is not an environmentally friendly synthetic technology [15]. For L-aspartate biosynthesis by fermentation, several types of bacteria have been engineered, including Escherichia coli (E. coli), Corynebacterium glutamicum (C. glutamicum) and Brevibacterium flavum (B. flavum) (Table 1). For L-aspartate cell factory construction, researchers focus on pathway modification [15,16] and overcoming the rate-limiting steps of L-aspartate biosynthesis (Figure 1) [13,17].
To date, the best cell factory for L-aspartate biosynthesis with glucose as a carbon source is the E. coli developed by Piao et al., producing 33.1 g/L of L-aspartate (Table 1). However, the yield was only 0.39 g/g, which was about 27% of the theoretical value when oxaloacetate/fumarate, the direct substrate for L-aspartate biosynthesis, was supplied by the reductive branch of the TCA cycle from glucose [15]. The researchers have focused on pathway modification to increase phosphoenolpyruvate (PEP), oxaloacetate (OAA), L-glutamate and CO2 supply and optimizing fermentation conditions to improve L-aspartate biosynthesis [15]. For engineering C. glutamicum ATCC13032 to produce L-aspartate, several genes were inactivated since they consume pyruvate (ldhA and avtA) or fumarate (sdhCAB), and aspB, encoding L-aspartate aminotransferase, was overexpressed [18]. It could produce 5.72 g/L of L-aspartate with a yield of 0.75 g/g. As for B. flavum 70, it was developed after several rounds of mutations and could produce 22.6 g/L of L-aspartate (Table 1). When maleate is used as the substrate, it is firstly converted to fumarate by maleate cis-trans isomerase (MaiA) and then to L-aspartate by the engineered E. coli. In this process, a titer of L-aspartate of 419.8 g/L with a conversion ratio of 0.72 was achieved [13]. Above all, with glucose as the substrate, the yield of L-aspartate is much lower than the theoretical value (1.48 g/g). Low yield will waste the substrate and increase the production cost. For realizing cost-effective industrial production of L-aspartate by fermentation using low-cost substrates (glucose), there is still much work to do, such as improving cell growth by pathway modification and fermentation medium optimization.

Table 1.
Summary of microbial production of L-aspartate.
Table 1.
Summary of microbial production of L-aspartate.
| Organism | Metabolic Engineering Strategies | Substrate | Titer (g/L) | Yield a (g/g) | Fermentation Strategy | Reference |
|---|---|---|---|---|---|---|
| Engineered cell factories | ||||||
| E. coli XAR31 | Introducing and overexpressing CgaspC, Cgppc, Mspck, glk, bt-ca, acs, Cgasp, BsrocG and CR, deleting genes involved in byproduct biosynthesis, developing a cofactor self-sufficient system, optimizing the fermentation conditions | glucose | 33.1 | 0.39 | fed-batch | [15] |
| C. glutamicum SLV. pEKEx3-aspB | Deleting genes involved in byproduct biosynthesis (sdhCAB, ldhA and avtA) | glucose | ~5.72 a | 0.75 | flask | [18] |
| B. flavum 70 | Developing several mutations: a citrate synthase-defective glutamate auxotroph, S-(2-aminoethyl)-L-cysteine-resistant mutant, a methionine-insensitive revertant and hosphoenolpyruvate carboxylase, a supplement of biotin | glucose | 22.6 | 0.22 | flask | [16] |
| Enzyme catalysis | ||||||
| E. coli pMA-RBS4-G27A/G171A | Co-overexpressing maleate cis-trans isomerase (MaiA) mutant and aspartase (AspA) on the plasmid and optimizing their activity ratio by ribosome binding site (RBS) regulation | maleate | 419.8 | 0.72 | 5-L fermenter | [13] |
| E. coli JCL1258/pBAW2/pASP400 | Overexpressing aspC and tyrB on plasmid pASP400 and overexpressing parB and aspA on plasmid pBAW2 | fumarate | 77.60 a | 0.83 | [14] | |
a represents that the data were derived by calculating according to the literature.
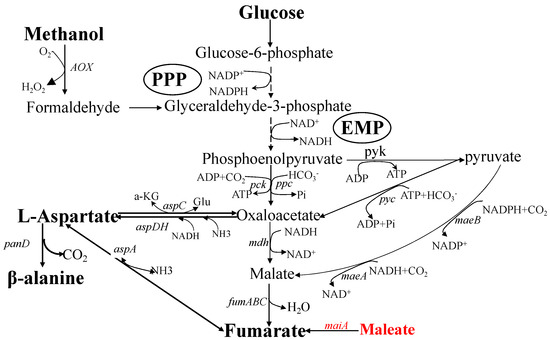
Figure 1.
Biosynthetic pathways of L-aspartate and β-alanine. Here shows the metabolic pathways for L-aspartate and β-alanine biosynthesis with glucose as the substrate. Phosphoenolpyruvate is produced from glucose through EMP, then it can be converted to oxaloacetate or fumarate, which are the direct substrates for L-aspartate. Pyruvate derived from phosphoenolpyruvate can be metabolized to OAA or malate. Malate is catalyzed by fumarase to produce fumarate. β-alanine is derived from L-aspartate with aspartate decarboxylase [19]. ppc, phosphoenolpyruvate carboxylase; pck, phosphoenolpyruvate carboxykinase; pyk, pyruvate kinase; pyc, pyruvate carboxylase; mdh, malate dehydrogenase; maeA, malate dehydrogenase; maeB, malate dehydrogenase; fumABC, fumarase; aspA, aspartate ammonia-lyase; aspDH, NADH-dependent aspartate dehydrogenase; aspC, aspartate transaminase; AOX, alcohol oxidase; maiA, maleate cis-trans isomerase.
2.2. Developing Cell Factories to Produce β-Alanine
β-alanine is a non-essential amino acid that is naturally synthesized by the liver. It is used to synthesize the dipeptide carnosine, a powerful antioxidant [20]. β-alanine has various applications in the food and fitness industries. As a food additive, β-alanine is used as a flavor enhancer and acidity regulator. It is particularly useful in meat products, as it offers a pleasant taste and acts as a natural preservative [21]. In fitness industries, it is commonly used as a sports nutrition supplement to help increase endurance, delay fatigue and improve exercise performance [22]. The global market for β-alanine is expected to grow in the coming years, driven by the increasing demand for sports nutrition supplements and functional foods. The global market revenue of β-alanine was 75 million USD in 2019 and will reach 99 million USD in 2031, with a CAGR of 4.65% during 2023–2031 (https://www.marketwatch.com/press-release/beta-alanine-market-global-industry-share-trends-size-growth-opportunity-and-forecast-2023-2031-2023-04-14, accessed on 3 May 2023).
Currently, β-alanine is mainly produced via chemical synthesis, which involves toxic precursors and operates under harsh conditions [23]. For enzymatic processes, β-alanine can be derived from L-aspartate and fumarate. When L-aspartate, the precursor of β-alanine, was fed to E. coli expressing L-aspartate-α-decarboxylase, over 271 g/L of β-alanine was produced at a conversion rate of over 92% (Table 2). Although the highest conversion efficiency could be 97.2% when L-aspartate was used as the substrate, the cost for β-alanine biosynthesis is too high since the market price of L-aspartate is around $5000/ton, while the price for β-alanine is $6000/ton (Table 2). When fumarate is used as the substrate, it needs two enzymes, L-aspartate ammonia-lyase and L-aspartate-α-decarboxylase, to finish the β-alanine biosynthesis, and the highest titer can reach 200.3 g/L with a conversion efficiency of over 90% (Table 2). There have been a few studies on the production of β-alanine with bacteria through metabolic engineering. The bacteria used for constructing a β-alanine-producing cell factory include E. coli, C. glutamicum, B. megaterium and Pichia pastoris (Table 2). Glucose and methanol are used as carbon sources for β-alanine biosynthesis. Glucose is metabolized through the pentose phosphate pathway or EMP to produce PEP, which is converted to OAA, the substrate for L-aspartate biosynthesis. L-aspartate is converted to β-alanine by L-aspartate decarboxylase (Figure 1). With glucose as the substrate, the best strain for β-alanine biosynthesis is from C. glutamicum, and the highest reported titer of β-alanine was 166.6 g/L with a productivity of 1.74 g/(L.h) [24] (Table 2). However, the yield was only 0.28 g/g glucose (the maximum theoretical yield is 0.99 g/g glucose) due to the use of the pentose phosphate pathway and aerobic fermentation instead of anaerobic conditions for producing β-alanine. The metabolic engineering strategies they adapted were introducing L-aspartate 1-decarboxylases (encoded by panD) from B. subtilis, improving the supply of OAA and L-aspartate and speeding up the secretion of β-alanine. With glucose as the substrate, the highest yield was 0.75 g/g with engineered E. coli [15] (Table 2). When methanol was added to a culture of methylotrophic Pichia pastoris 2ADC-Spe, it was first converted to glyceraldehyde-3-phosphate with formaldehyde as an intermediate and then to β-alanine (Figure 1). However, the titer of β-alanine was only 5.6 g/L (Table 2). Except substrate optimization, some researchers tried to develop new methods to improve β-alanine production as well. For example, Dr. Alper’s group has developed biosensor-assisted directed evolution and found ribonuclease E (encoded by rne) had a negative influence on β-alanine biosynthesis. The final strain, E. coli eBA32, could produce 34.8 g/L of β-alanine with fed-batch fermentation in 37 h [22]. Above all, this shows that a highly efficient β-alanine-producing cell factory can be realized in the future.

Table 2.
Summary of microbial production of β-alanine.
2.3. Developing Cell Factories to Produce Ectoine
Ectoine is a naturally occurring organic molecule. It functions as a protective agent, preventing damage to biological structures from harsh environmental conditions such as osmotic and thermal stress [34]. Ectoine also has water retention properties, allowing it to maintain hydration levels in cells, which is essential for the survival of organisms [35]. One potential application of ectoine is in the cosmetics industry, for its water-binding properties, which are key qualities for hydrating skin and hair. Besides this, ectoine has pharmaceutical applications for the treatment of skin disorders, eye diseases, and respiratory diseases as it has anti-inflammatory and antioxidant properties [36]. According to a report by businessresearchinsights, the market for ectoine was 20 million USD in 2021 and is expected to reach 31 million USD by 2028, growing at a CAGR of 6.6% from 2023 to 2028. This growth is driven by the increasing demand for natural-based cosmetics and personal-care products, as well as the growing awareness of the benefits of ectoine in healthcare (https://www.businessresearchinsights.com/market-reports/ectoine-market-100579, accessed on 3 May 2023).
Ectoine is currently produced by chemical synthesis, biocatalytic approach and fermentation. Ectoine can be chemically synthesized using chemical building blocks, such as glycine or sarcosine [37]. However, this method is not commonly used due to its low yield, high cost and low efficiency compared to biocatalytic and fermentation methods [37]. Ectoine can also be biosynthesized from its precursor, L-2,4-diaminobutyric acid (DABA), using an enzyme called ectoine synthase [38]. DABA is produced by certain bacteria and plants and can be chemically synthesized [39]. For fermentation, ectoine biosynthesis is realized in kinds of bacteria. Since ectoine is a compatible solute, it is produced by halophilic bacteria in response to high salt concentrations in their environments. Several halophilic bacteria are natural producers of ectoine in response to salt stress (Table 3). Among the natural producers, the best performer is H. elongate 1A01717, and this bacterium could produce 15.9 g/L of ectoine with glucose as the substrate. Some natural producers can convert glutamate to ectoine with L-aspartate as an intermediate (Figure 2). The key strategy was optimizing the fermentation conditions, such as the culture medium and NaCl concentration. With glutamate as the feedstock, the best performer is H.salina DSM 5928T, which could produce 14.86 g/L ectoine with a yield of 0.14 g/g at a productivity of about 0.32 g/(L.h) (Table 3). Polypeptone and yeast extract can also be the carbon sources for ectoine biosynthesis (Table 3). When glycerol was added to ectoine biosynthetic medium, it served as the source of acetyl-CoA (AcCoA) in the step converting L-2,4-diaminobutyrate to N-acetyl-2,4-diaminobutyrate (Figure 2). In the industry, halophiles are used to produce ectoine with fermentation on a large scale. However, high concentrations of salt could corrode the equipment [40]. It is urgent to realize ectoine biosynthesis under low-salt conditions. Luckily, with the development of metabolic engineering and new technologies, that is not a dream anymore.

Table 3.
Summary of microbial production of ectoine.
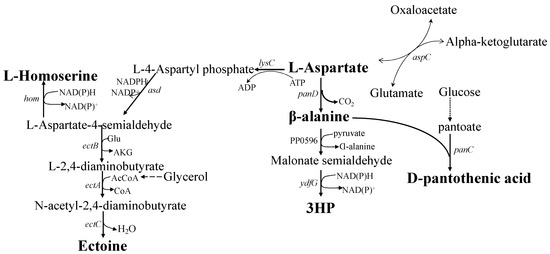
Figure 2.
Pathways for producing L-aspartate derivatives with L-aspartate as the substrate. lysC, aspartokinase; asd, aspartate-semialdehyde dehydrogenase; hom, L-homoserine dehydrogenase; ectB, L-2,4-diaminobutyrate transaminase; ectA, 2,4-diaminobutyrate acetyltransferase; ectC, ectoine synthase; panD, aspartate decarboxylase; PP0596, β-alanine-pyruvate transaminase; ydfG, 3-hydroxyacid dehydrogenase; panC, pantothenate synthetase; aspC, aspartate transaminase.
For metabolic engineering of bacteria to produce ectoine, two strategies are used. One approach is to introduce the genes that encode the enzymes involved in the ectoine biosynthetic pathway into a bacterial host that has a relatively clear genome background and well-developed gene operation method. For example, the ectABC genes, encoding the three enzymes required for ectoine biosynthesis, are cloned from Halomonas elongata and introduced into a bacterial host such as E. coli [44]. This is combined with additional manipulation to increase precursor L-aspartate production. According to the literature, the best engineered strain for ectoine biosynthesis is C. glutamicum ectABCopt, carrying the ectoine pathway from Pseudomonas stutzeri that was expressed from synthetic promoters. After fermentation condition optimization, C. glutamicum ectABCopt produced about 65 g/L of ectoine with a productivity of 2.3 g/(L.h) at the beginning of the feed phase [47]. An engineered E. coli strain ET11 produced 53.2 g/L of ectoine with a yield of 0.33 g ectoine/g glucose during fed-batch fermentation [41] (Table 3). The metabolic engineering strategies they used were introducing the ectABC gene cluster from Halomonas venusta ZH, regulating the copy numbers of ectA, ectB and ectC and eliminating byproduct metabolic pathways. For improving the ectoine production further, they optimized the fermentation medium as well. In summary, metabolic engineering strategies have yielded promising results for realizing ectoine biosynthesis for industrial use.
2.4. Developing Cell Factories to Produce 3-Hydroxypropionate
3-hydroxypropionate (3-HP) is a naturally occurring organic acid and a precursor chemical to produce various value-added chemicals such as acrylates, acrylic acid, malic acid and 1,3-propanediol [69]. One potential application of 3-HP is as a building block chemical for biodegradable polymers, potentially replacing petroleum-based plastics in environmentally conscious products [70]. The global market for 3-HP and its related compounds is expected to grow significantly in the coming years. According to a report by marketwatch, the market for 3-HP was 117.14 million USD in 2022 and is expected to reach 153.81 million USD by 2028 with a CAGR of 4.64% (https://www.marketwatch.com/press-release/3-hydroxypropionic-acid-market-research-2023-2030-2023-06-15, accessed on 3 May 2023). This growth is driven by the increasing demand for sustainable and eco-friendly materials, as well as the increasing investment in renewable chemicals and biofuels.
Currently, 3-HP is produced via chemical synthesis and cell factory. 3-HP can be produced through chemical synthesis using acrolein, formaldehyde and hydrogen cyanide [71]. This method is not economically feasible due to the high cost of raw materials and environmental concerns. For realizing 3-HP biosynthesis with bacteria, many researchers have focused on optimizing the 3-HP biosynthetic pathway and tried different feedstocks to increase the titer and yield of 3-HP during fermentation (Table 4). The feedstocks can be a single sugar (such as glucose, xylose, glycerol, malonate, acrylic acid, 1,3-propanediol, ethanol and sorbitol), a sugar combination (glycerol and acetate, glucose and cellobiose) or a complex mixture (such as fatty acids (FAs), mechanically refined corn stover hydrolysate) (Table 4). After β-alanine is produced from L-aspartate, it is converted to 3-HP via malonate semialdehyde, an important intermediate for 3-HP biosynthesis. Except 1,3-propanediol and acrylic acid, all of the substrates mentioned above can join 3-HP biosynthesis via malonate semialdehyde (Figure 3). Glycerol is the most promising substrate for 3-HP biosynthesis since it is a byproduct of biodiesel and just needs two steps to complete the biosynthetic process. K. pneumoniae is the natural producer of 3-HP. After overexpressing of PuuC, the best engineered performer of K. pneumoniae could produce 102.61 g/L of 3-HP with glycerol as the carbon source [72]. E. coli is the most popular strain used for engineering, and it is modified to produce 3-HP from kinds of sugars as well (Table 4). To date, the best one is introducing dhaB1234, gdrAB and ydcW from K. pneumoniae into E. coli to realize 3-HP biosynthesis with glycerol as substrate, and the titer has reached 76.2 g/L at a productivity of 1.89 g/(L.h) [73]. That is promising for industrial use. Yeast has also been engineered to produce 3-HP via the malonyl-CoA pathway, and the titer has reached 71.09 g/L, which is the highest value with glucose as the substrate [74]. The interesting thing is that 3-HP biosynthesis was finished in the mitochondria. Except overexpressing malonyl-CoA reductase (MCR), they also optimized the expression of POS5 and IDP1 to improve NADPH supply. In addition, they found an ACC1 mutant could improve 3-HP production as well. When 1,3-propanediol is used as the substrate, there are only two steps needed to realize 3-HP biosynthesis, and the best performer is engineered Halomonas bluephagenesis TD27, which could produce 154 g/L of 3-HP with a yield of 0.93 g/g 1,3-propanediol (Table 4). The metabolic engineering strategies were deleting the 3-HP degradation pathway and overexpressing alcohol dehydrogenases (AdhP) to improve 3-HP biosynthesis [73]. Halomonas bluephagenesis is promising for industrial use since it can be cultured under an open and unsterile condition with continuous process [75]. Except the organisms mentioned above, 3-HP biosynthesis has also been realized in Schizosaccharomyces pombe, Lactobacillus reuteri, Debaryomyces hansenii, Rhodococcus erythropolis, Lentilactobacillus diolivorans and Gluconobacter oxydans (Table 4). The chassis cells and its cultivation conditions have great influence on 3-HP biosynthesis. Above all, it is promising to realize green biosynthesis of 3-HP via metabolic engineering in the industry.

Table 4.
Summary of microbial production of 3-HP.
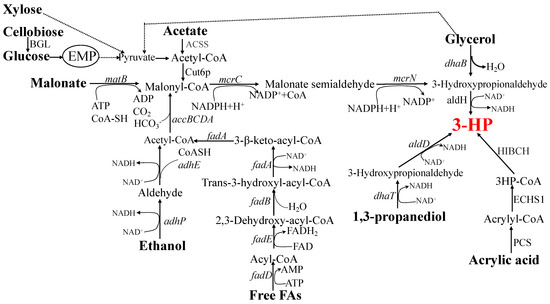
Figure 3.
Biosynthetic pathways of 3-HP. The substrate used for 3-HP biosynthesis is marked in bold. dhaB, glycerol dehydratase; aldH, aldehyde dehydrogenase; matB, malonyl-CoA synthetase; mcrC, malonyl-CoA reductase C-domain; mcrN, malonyl-CoA reductase N-domain; fadD, fatty acyl-CoA synthetase; fadE, acyl-CoA dehydrogenase; fadB, α component of the fatty acid oxidation complex; fadA, β component of the fatty acid oxidation complex; accBCDA, acetyl-CoA carboxyltransferase complex; adhP, alcohol dehydrogenase; adhE, alcohol/aldehyde dehydrogenase; ACSS, acetyl-CoA synthetase; Cut6p, acetyl-CoA/biotin carboxylase; BGL, beta-glucosidase; dhaT, alcohol dehydrogenase; aldD, aldehyde dehydrogenase; PCS, acrylyl-CoA (propionyl-CoA) synthetase; ECHS1, enoyl-CoA hydratase, HIBCH, 3-hydroxyisobutyryl-CoA hydrolase.
2.5. Current Process for Developing Cell Factories to Produce D-Pantothenic Acid
D-pantothenic acid is a water-soluble B-vitamin (Vitamin B5) that plays a crucial role in energy metabolism and the synthesis of various compounds, such as fatty acids, cholesterol and steroid hormones [110]. The global market of D-pantothenic acid was valued at about 460.3 million USD in 2020 and is expected to exhibit a CAGR of 6.19% over the forecast period (2021–2028). It is primarily driven by the application of D-pantothenic acid as an ingredient in dietary supplements and animal feed. It is also used in the production of cosmetics, pharmaceuticals and food additives. (https://www.globenewswire.com/en/news-release/2021/12/14/2351835/0/en/At-6-2-CAGR-Global-Pantothenic-Acid-Market-to-Reach-US-750-7-Million-by-2028-Says-Coherent-Market-Insights-CMI.html, accessed on 3 May 2023).
Synthetic methods for producing D-pantothenic acid include chemical synthesis, enzymatic catalysis and microbial fermentation. Currently, D-pantothenic acid is mainly produced by chemical synthesis and enzymatic catalysis [111]. Chemical synthesis involves several steps and requires some toxic chemicals, such as hydrocyanic acid and sodium cyanide, which cause wastewater pollution [112]. For enzymatic catalysis, pantothenate synthetase can catalyze pantoate and β-alanine to produce D-pantothenic acid. For example, when pantothenate synthetase was overexpressed in E. coli or in Bacillus megaterium (B. megaterium), D-pantothenic acid was biosynthesized after pantoate and β-alanine were added into the culture medium (Table 5). The titer of D-pantothenic acid in E. coli was 97.1 g/L at a productivity of 3.0 g/(L.h) [113], while that in B. megaterium was about 45.56 g/L with fed-batch fermentation [114]. However, since pantoate is much more expensive from commercial sources, enzymatic process is not a good choice for D-pantothenic acid synthesis in the industry [115]. For microbial fermentation, D-pantothenic acid biosynthesis was realized with glucose and β-alanine as feedstocks since glucose could be converted to pantoate through the valine biosynthetic pathway combined with overexpression of panB from different kinds of organisms (Figure 2). With this strategy, E. coli DPAL 8 could produce 66.39 g/L of D-pantothenic acid with a yield of 0.27 g/g glucose after optimizing the fermentation conditions (Table 5). For genome modification, several genes involved in pantoate biosynthesis were overexpressed, such as pck, maeB, ilvD, ilvBN and cycA. Pathways for byproduct biosynthesis were deleted or downregulated in E. coli DPAL 8 [116]. L-isoleucine and citric acid are used for D-pantothenic acid biosynthesis also since they can increase ATP and NADPH supply via the TCA cycle [117]. L-isoleucine is also beneficial for improving the availability of CoA. When L-isoleucine and glucose were used to feed the engineered strain, the best performer, E. coli ECPA, could produce 39.1 g/L of D-pantothenic acid with a yield of 0.175 g/g glucose at a productivity of 0.58 g/(L.h) (Table 5). Furthermore, some engineered strains can use glucose as the only substrate for D-pantothenic acid biosynthesis also, including E. coli, C. glutamicum and Saccharomyces cerevisiae (Table 5). Moreover, the highest titer of D-pantothenic acid reached 68.3 g/L with a yield of 0.36 g/g and a productivity of 0.794 g/(L.h) in E. coli DPA02/pT-ppnk. The metabolic engineering strategies were overexpressing ppnk and deleting genes involved in byproduct biosynthesis, such as aceF and mdh [118]. Overall, metabolic engineering is a powerful tool for realizing D-pantothenic acid commercialization.

Table 5.
Summary of microbial production of D-pantothenic acid.
2.6. Developing Cell Factories to Produce L-Homoserine
L-homoserine is an amino acid and functions as an intermediate in multiple metabolic pathways, including the synthesis of various essential amino acids, such as methionine and threonine, and the production of certain pharmaceuticals and specialty chemicals [125]. The market for L-homoserine is relatively small compared to other amino acids due to its inefficient production and expensive price [126].
In the industry, L-homoserine can be produced via chemical synthesis, enzymatic synthesis and microbial fermentation. Chemical synthesis is expensive and complicated, and enzymatic synthesis has limited scalability. Therefore, microbial fermentation is the most promising method for producing L-homoserine. Microbial sources of L-homoserine biosynthesis are bacteria such as E. coli and C. glutamicum, and glucose is usually used as the feedstock. For producing L-homoserine from glucose, the biosynthetic pathway is shown in Figure 2. Glucose is metabolized to L-aspartate (Figure 2) and then to L-homoserine catalyzed by aspartokinase (lysC), aspartate-semialdehyde dehydrogenase (asd) and L-homoserine dehydrogenase [127] (Figure 2). To date, the best producer of L-homoserine is E. coli W-18/pM2/pR1, and the titer could reach 110 g/L with a yield of 0.64 g/g at a productivity of 1.82 g/(L.h) (Table 6). The metabolic engineering strategies were improving precursor supply, such as OAA and L-aspartate, by overexpressing glf, ppc, aspA, glk, asd, metL and rhtA and decreasing byproduct biosynthesis, such as lactate and acetate, by deleting lysA, thrB, metA, ldhA, adhE, pflB, ptsG, iclR and arcA. They also deleted lacI and regulated key genes’ expression with the lac promoter [126]. The fermentation strategy was fed-batch and two stage bioreaction: the growth stage and the production stage [126]. For L-homoserine biosynthesis, the highest productivity was 1.96 g/(L.h), and it was realized by engineering E. coli BW25113 after redox balance regulation and competitive and degradative pathway deletion [128]. Since C. glutamicum is successfully engineered to produce kinds of amino acids, some researchers have also engineered it to produce L-homoserine with different sugars. Among them, the best performer is C. glutamicum Cg18-1, which could produce 63.5 g/L of L-homoserine with a yield of 0.25 g/g glucose (Table 6). Their work focused on improving NADPH supply by regulating specific genes’ expression, such as pntAB and ppnK, and pathway modification, such as enhancing the pentose phosphate pathway (PPP) and introducing the Entner–Doudoroff (ED) pathway [129]. Since the productivity was lower than the industry demand (≥2 g/L/h) and the yield is less than 50% of the theoretical value, there is still a distance to achieve L-homoserine biosynthesis with a cell factory in the industry.

Table 6.
Summary of microbial production of L-homoserine.
3. Perspective
Metabolic engineering is a promising method for realizing desired products biosynthesis with a cell factory. Nowadays, except the natural producers, four kinds of microbials, E. coli, B. subtilis, C. glutamicum and S. cerevisiae, are popular for producing kinds of compounds with metabolic engineering since their genome backgrounds are relatively clear and the gene editing methods are well developed. There are several factors that block the commercializing of the engineered strains, and the detailed information is described as the following:
- (1)
- metabolic engineering of the bacteria to increase the titer, yield and productivity of the target products
After the biosynthetic pathway is clear, the next step is optimizing the pathway to improve the titer and yield of the target product, usually by balancing the supply and consumption of the cofactors (NADH, NADPH, FADH2), deleting the competitive pathways, regulating the expression of genes involved in the biosynthesis and increasing the key enzymes’ activity as well as specificity. Shi et al. have developed a cell factory to produce isobutanol under anaerobic conditions with a high yield of 0.92 mol/mol glucose [136]. The strategies they applied were deleting competitive pathways, such as biosynthetic pathways of ethanol, acetate and lactate, regulating key genes’ expression (alsS, ilvC, ilvD, kivD and adhA) with strong artificial promoters and increasing the conversion speed between NADH and NADPH by activating transhydrogenase and NAD kinase together. With the development of bioinformatics, pathway optimization becomes more rational and more accurate.
Aspartate ammonia-lyase, an important enzyme for L-aspartate biosynthesis, is allosterically regulated by L-aspartate. In addition, the activity of phosphoenolpyruvate carboxylase, catalyzing phosphoenolpyruvate to oxaloacetate, is also inhibited by a high concentration of L-aspartate. This problem should be solved for developing a high-performance cell factory for L-aspartate biosynthesis. Protein engineering is relatively difficult since it is time-consuming and usually unsuccessful. Luckily, with the development of new technologies (e.g., Alphafold), it becomes easier and more predictable. Fei et al. have developed a dual-fluorescence reporter system to screen L-aspartate-α-decarboxylase variants with a high-throughput method and found one mutant with increased activity and stability [137]. This mutant was further applied for β-alanine biosynthesis in E. coli Nissle 1917 [25]. The growth of the engineered strain is another factor that the researchers need to consider since it will influence the productivity of the target products. The engineered strain for L-aspartate biosynthesis needs relatively enriched fermentation medium (yeast extract added) since the bacteria could not grow well with mineral medium only [15].
After the target product is biosynthesized in the cell, the next step is to transfer it from intracellular to extracellular in order to release its inhibition to the enzymes involved in its biosynthesis. To realize this, the secretion mechanism of the target product needs to be investigated. For L-aspartate, we only know that its uptake is realized by the C4-dicarboxylate transporter [138]. However, little information is given for its secretion. Ghiffary et al. have found a β-alanine exporter in C. glutamicum, which had a great influence on the titer of β-alanine [24].
- (2)
- fermentation condition optimization
Some bacteria are natural producers of the target product, while the fermentation strategies of those bacteria are not well developed. For example, many halophilic bacteria, such as Halomonas sp. [51] and Sinobaca sp. [57], have the ability to produce ectoine, but we have little data about its scale-up fermentation.
The engineered strain cannot enter into the industry until the fermentation cost is competitive with the up-to-date synthetic method. The fermentation cost includes the medium cost for strain growth, the substrates for target product biosynthesis and the fermentation conditions, such as sterile treatment, pH control, dissolved oxygen (DO) control and feeding strategy. Some researchers have focused on optimizing the fermentation process. For example, E. coli ET01 is an engineered producer for ectoine biosynthesis. For improving ectoine production, Dong et al. have optimized the fermentation condition, such as feeding strategies and DO levels. Finally, 47.8 g/L of ectoine has been produced with two-stage fermentation [44]. A Clostridium pasteurianum strain for 1,3-propanediol biosynthesis developed by Dr. Zeng’s group could be fermented with medium of low cost and unsterile treatment. The fermentation cost was decreased by 50% [139]. As a renewable energy source, biomass is promising to be the substrate for valuable product biosynthesis. Nowadays, many researchers try to find an economic way to break the biomass into monosaccharides with less toxic side products produced. If it is successful, the fermentation cost will be dramatically decreased.
- (3)
- downstream processing
The purification cost is another important factor needing to be taken into account. The methods for purification are determined by the characteristic of the target product, the culture medium and the side products. Ectoine was produced by H. elongata with fermentation. Chen et al. have designed a strategy with multiple steps to purify it, including microfiltration, desalination, cation exchange, decolorizing with activated carbon, refining with methanol, crystallization and centrifugation. However, the yield is only 43%, and the process is time consuming. The method is not ready for commercial use [68].
Above all, there are many scientific problems that should be solved before a biosynthesized product moves into commercialization. Luckily, with the development of novel technologies, such as synthetic biology and bioinformatics, the engineering process becomes predictable and faster. According to the literature, to improve 3-HP production in Aspergillus niger, several genes were selected and modified according to proteomic and metabolomic analysis [81]. After protein engineering of ketopantoate hydroxymethyltransferase from C. glutamicum and overexpressing panB, CgKPHMT-K25A/E189S and panC, E. coli W3110 DPA-11 could produce 41.17 g/L of D-pantothenic acid [119]. Moreover, the fermentation condition of D-pantothenic acid in Escherichia DPA21 was optimized according to comparative transcriptome and metabolomics analysis [122]. In the future, with the development of genome editing technology such as CRISPR/Cas9, some natural producers of 3-HP can be engineered. As a result, this gives us more choice to realize 3-HP biosynthesis in the industry.
The cost of fermentation and downstream purification has great influence on the product’s selling price and are of great concern to the fermentation company. We believe these problems will be solved by the cooperation of researchers from different areas.
Author Contributions
Conceptualization, A.S., Y.L., B.J. and G.Z.; methodology, A.S.; validation, G.Z.; writing—original draft preparation, A.S.; writing—review and editing, Y.L. and B.J.; supervision, G.Z. and Y.Y. All authors have read and agreed to the published version of the manuscript.
Funding
The work was financially supported by the Scientific Research Foundation of Xianghu Laboratory.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the related references.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Choi, K.R.; Jang, W.D.; Yang, D.; Cho, J.S.; Park, D.; Lee, S.Y. Systems metabolic engineering strategies: Integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019, 37, 817–837. [Google Scholar] [PubMed]
- Choi, K.Y.; Wernick, D.G.; Tat, C.A.; Liao, J.C. Consolidated conversion of protein waste into biofuels and ammonia using Bacillus subtilis. Metab. Eng. 2014, 23, 53–61. [Google Scholar] [PubMed]
- Angermayr, S.A.; Gorchs Rovira, A.; Hellingwerf, K.J. Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends Biotechnol. 2015, 33, 352–361. [Google Scholar] [PubMed]
- Antoniewicz, M.R. A guide to metabolic flux analysis in metabolic engineering: Methods, tools and applications. Metab. Eng. 2021, 63, 2–12. [Google Scholar] [PubMed]
- Huang, L.; Ho, C.T.; Wang, Y. Biosynthetic pathways and metabolic engineering of spice flavors. Crit. Rev. Food Sci. Nutr. 2021, 61, 2047–2060. [Google Scholar]
- Zhu, K.; Kong, J.; Zhao, B.; Rong, L.; Liu, S.; Lu, Z.; Zhang, C.; Xiao, D.; Pushpanathan, K.; Foo, J.L.; et al. Metabolic engineering of microbes for monoterpenoid production. Biotechnol. Adv. 2021, 53, 107837. [Google Scholar]
- Yang, D.; Park, S.Y.; Park, Y.S.; Eun, H.; Lee, S.Y. Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 2020, 38, 745–765. [Google Scholar]
- Sinha, R.; Shukla, P. Current trends in protein engineering: Updates and progress. Curr. Protein Pept. Sci. 2019, 20, 398–407. [Google Scholar]
- Yarmush, M.L.; Banta, S. Metabolic engineering: Advances in modeling and intervention in health and disease. Annu. Rev. Biomed. Eng. 2003, 5, 349–381. [Google Scholar]
- Jarboe, L.R.; Zhang, X.; Wang, X.; Moore, J.C.; Shanmugam, K.T.; Ingram, L.O. Metabolic engineering for production of biorenewable fuels and chemicals: Contributions of synthetic biology. J. Biomed. Biotechnol. 2010, 2010, 761042. [Google Scholar]
- Han, M.; Zhang, C.; Suglo, P.; Sun, S.; Wang, M.; Su, T. L-aspartate: An essential metabolite for plant growth and stress acclimation. Molecules 2021, 26, 1887. [Google Scholar]
- Czarnecka, K.; Pilarz, A.; Rogut, A.; Maj, P.; Szymańska, J.; Olejnik, Ł.; Szymański, P. Aspartame-true or false? Narrative review of safety analysis of general use in products. Nutrients 2021, 13, 1957. [Google Scholar] [PubMed]
- Liu, Z.; Yu, L.; Zhou, L.; Zhou, Z. One-pot biosynthesis of L-aspartate from maleate via an engineered strain containing a dual-enzyme system. Appl. Environ. Microbiol. 2019, 85, 19. [Google Scholar]
- Chao, Y.P.; Lo, T.E.; Luo, N.S. Selective production of L-aspartic acid and L-phenylalanine by coupling reactions of aspartase and aminotransferase in Escherichia coli. Enzyme Microb. Technol. 2000, 27, 19–25. [Google Scholar] [CrossRef]
- Piao, X.; Wang, L.; Lin, B.; Chen, H.; Liu, W.; Tao, Y. Metabolic engineering of Escherichia coli for production of L-aspartate and its derivative β-alanine with high stoichiometric yield. Metab. Eng. 2019, 54, 244–254. [Google Scholar]
- Mori, M.; Shiio, I. Production of aspartic acid and enzymatic alteration in pyruvate kinase mutants of Brevibacterium flavum. Agric. Biol. Chem. 1984, 48, 9. [Google Scholar] [CrossRef]
- Tajima, T.; Hamada, M.; Nakashimada, Y.; Kato, J. Efficient aspartic acid production by a psychrophile-based simple biocatalyst. J. Ind. Microbiol. Biotechnol. 2015, 42, 1319–1324. [Google Scholar] [PubMed]
- Ziert, C. Metabolic Engineering of Corynebacterium glutamicum for the Production of L-Aspartate and Its Derivatives β-Alanine and Ectoine; Universitätsbibliothek Bielefeld: Bielefeld, Germany, 2014. [Google Scholar]
- Padhi, S.; Dash, M.; Sahu, R.; Panda, P. Urinary tract infection due to Paenibacillus alvei in a chronic kidney disease: A rare case report. J. Lab. Physicians 2013, 5, 133–135. [Google Scholar] [PubMed]
- Hoffman, J.R.; Varanoske, A.; Stout, J.R. Effects of β-Alanine supplementation on carnosine elevation and physiological performance. Adv. Food Nutr. Res. 2018, 84, 183–206. [Google Scholar]
- Kopec, W.; Jamroz, D.; Wiliczkiewicz, A.; Biazik, E.; Pudlo, A.; Korzeniowska, M.; Hikawczuk, T.; Skiba, T. Antioxidative characteristics of chicken breast meat and blood after diet supplementation with carnosine, L-histidine, and β-alanine. Antioxidants 2020, 9, 1093. [Google Scholar]
- Yuan, S.F.; Nair, P.H.; Borbon, D.; Coleman, S.M.; Fan, P.H.; Lin, W.L.; Alper, H.S. Metabolic engineering of E. coli for β-alanine production using a multi-biosensor enabled approach. Metab. Eng. 2022, 74, 24–35. [Google Scholar] [PubMed]
- Wang, P.; Zhou, H.Y.; Li, B.; Ding, W.Q.; Liu, Z.Q.; Zheng, Y.G. Multiplex modification of Escherichia coli for enhanced β-alanine biosynthesis through metabolic engineering. Bioresour. Technol. 2021, 342, 126050. [Google Scholar] [CrossRef] [PubMed]
- Ghiffary, M.R.; Prabowo, C.P.S.; Adidjaja, J.J.; Lee, S.Y.; Kim, H.U. Systems metabolic engineering of Corynebacterium glutamicum for the efficient production of β-alanine. Metab. Eng. 2022, 74, 121–129. [Google Scholar] [PubMed]
- Hu, S.; Fei, M.; Fu, B.; Yu, M.; Yuan, P.; Tang, B.; Yang, H.; Sun, D. Development of probiotic E. coli Nissle 1917 for β-alanine production by using protein and metabolic engineering. Appl. Microbiol. Biotechnol. 2023, 107, 2277–2288. [Google Scholar] [PubMed]
- Li, B.; Zhang, B.; Wang, P.; Cai, X.; Chen, Y.Y.; Yang, Y.F.; Liu, Z.Q.; Zheng, Y.G. Rerouting fluxes of the central carbon metabolism and relieving mechanism-based inactivation of l-Aspartate-α-decarboxylase for fermentative production of β-Alanine in Escherichia coli. ACS Synth. Biol. 2022, 11, 1908–1918. [Google Scholar]
- Zou, X.; Guo, L.; Huang, L.; Li, M.; Zhang, S.; Yang, A.; Zhang, Y.; Zhu, L.; Zhang, H.; Zhang, J.; et al. Pathway construction and metabolic engineering for fermentative production of β-alanine in Escherichia coli. Appl. Microbiol. Biotechnol. 2020, 104, 2545–2559. [Google Scholar]
- Wang, J.Y.; Rao, Z.M.; Xu, J.Z.; Zhang, W.G. Enhancing β-alanine production from glucose in genetically modified Corynebacterium glutamicum by metabolic pathway engineering. Appl. Microbiol. Biotechnol. 2021, 105, 9153–9166. [Google Scholar]
- Tadi SR, R.; Nehru, G.; Sivaprakasam, S. Metabolic Engineering of Bacillus megaterium for the Production of β-alanine. Biotechnol. Bioprocess Eng. 2022, 27, 909–920. [Google Scholar]
- Miao, L.; Li, Y.; Zhu, T. Metabolic engineering of methylotrophic Pichia pastoris for the production of β-alanine. Bioresour. Bioprocess. 2021, 8, 89. [Google Scholar] [CrossRef]
- Shen, Y.; Zhao, L.; Li, Y.; Zhang, L.; Shi, G. Synthesis of β-alanine from L-aspartate using L-aspartate-α-decarboxylase from Corynebacterium glutamicum. Biotechnol. Lett. 2014, 36, 1681–1686. [Google Scholar]
- Wang, L.; Piao, X.; Cui, S.; Hu, M.; Tao, Y. Enhanced production of β-alanine through co-expressing two different subtypes of L-aspartate-α-decarboxylase. J. Ind. Microbiol. Biotechnol. 2020, 47, 465–474. [Google Scholar]
- Tadi SR, R.; Nehru, G.; Sivaprakasam, S. One-Pot biosynthesis of 3-aminopropionic acid from fumaric acid using recombinant Bacillus megaterium Containing a linear dual-enzyme cascade. Appl. Biochem. Biotechnol. 2022, 194, 1740–1754. [Google Scholar]
- Bownik, A.; Stępniewska, Z. Ectoine as a promising protective agent in humans and animals. Arh Hig. Rada Toksikol. 2016, 67, 260–265. [Google Scholar] [PubMed]
- Guzmán, H.; Van-Thuoc, D.; Martín, J.; Hatti-Kaul, R.; Quillaguamán, J. A process for the production of ectoine and poly(3-hydroxybutyrate) by Halomonas boliviensis. Appl. Microbiol. Biotechnol. 2009, 84, 1069–1077. [Google Scholar] [CrossRef]
- Sattar, O.I.A.; Abuseada, H.H.M.; Emara, M.S.; Rabee, M. Green electrochemical and chromatographic quantifications of the extremolyte ectoine in halophilic bacterial cultures and related pharmaceutical preparations. J. Pharm. Biomed. Anal. 2022, 213, 114680. [Google Scholar]
- Peters, P.; Galinski, E.A.; Trüper, H.G. The biosynthesis of ectoine. FEMS Microbiol. Lett. 1990, 71, 157–162. [Google Scholar]
- Czech, L.; Höppner, A.; Kobus, S.; Seubert, A.; Riclea, R.; Dickschat, J.S.; Heider, J.; Smits SH, J.; Bremer, E. Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis. Sci. Rep. 2019, 9, 364. [Google Scholar] [PubMed]
- Mantas MJ, Q.; Nunn, P.B.; Ke, Z.; Codd, G.A.; Barker, D. Genomic insights into the biosynthesis and physiology of the cyanobacterial neurotoxin 2,4-diaminobutanoic acid (2,4-DAB). Phytochemistry 2021, 192, 112953. [Google Scholar]
- Fallet, C.; Rohe, P.; Franco-Lara, E. Process optimization of the integrated synthesis and secretion of ectoine and hydroxyectoine under hyper/hypo-osmotic stress. Biotechnol. Bioeng. 2010, 107, 124–133. [Google Scholar] [PubMed]
- Zhang, H.; Liang, Z.; Zhao, M.; Ma, Y.; Luo, Z.; Li, S.; Xu, H. Metabolic engineering of Escherichia coli for ectoine production with a fermentation strategy of supplementing the amino donor. Front. Bioeng. Biotechnol. 2022, 10, 824859. [Google Scholar] [CrossRef]
- Ning, Y.; Wu, X.; Zhang, C.; Xu, Q.; Chen, N.; Xie, X. Pathway construction and metabolic engineering for fermentative production of ectoine in Escherichia coli. Metab. Eng. 2016, 36, 10–18. [Google Scholar] [PubMed]
- Kang, J.Y.; Lee, B.; Kim, J.A.; Kim, M.S.; Kim, C.H. Identification and characterization of an ectoine biosynthesis gene cluster from Aestuariispira ectoiniformans sp. nov., isolated from seawater. Microbiol. Res. 2022, 254, 126898. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Zhang, H.; Wang, X.; Ma, J.; Lei, P.; Xu, H.; Li, S. Enhancing ectoine production by recombinant Escherichia coli through step-wise fermentation optimization strategy based on kinetic analysis. Bioprocess Biosyst. Eng. 2021, 44, 1557–1566. [Google Scholar] [PubMed]
- He, Y.Z.; Gong, J.; Yu, H.Y.; Tao, Y.; Zhang, S.; Dong, Z.Y. High production of ectoine from aspartate and glycerol by use of whole-cell biocatalysis in recombinant Escherichia coli. Microb. Cell Fact. 2015, 14, 55. [Google Scholar]
- Chen, J.; Liu, P.; Chu, X.; Zhang, H.; Rowley, D.C.; Wang, H. Metabolic pathway construction and optimization of Escherichia coli for high-level ectoine production. Curr. Microbiol. 2020, 77, 1412–1418. [Google Scholar] [CrossRef]
- Gießelmann, G.; Dietrich, D.; Jungmann, L.; Kohlstedt, M.; Jeon, E.J.; Yim, S.S.; Sommer, F.; Zimmer, D.; Mühlhaus, T.; Schroda, M.; et al. Metabolic engineering of Corynebacterium glutamicum for high-level ectoine production: Design, combinatorial assembly, and implementation of a transcriptionally balanced heterologous ectoine pathway. Biotechnol. J. 2019, 14, e1800417. [Google Scholar] [CrossRef]
- Becker, J.; Schäfer, R.; Kohlstedt, M.; Harder, B.J.; Borchert, N.S.; Stöveken, N.; Bremer, E.; Wittmann, C. Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microb. Cell Fact. 2013, 12, 110. [Google Scholar]
- Jiang, A.; Song, Y.; You, J.; Zhang, X.; Xu, M.; Rao, Z. High-yield ectoine production in engineered Corynebacterium glutamicum by fine metabolic regulation via plug-in repressor library. Bioresour. Technol. 2022, 362, 127802. [Google Scholar]
- Zhao, Q.; Li, S.; Lv, P.; Sun, S.; Ma, C.; Xu, P.; Su, H.; Yang, C. High ectoine production by an engineered Halomonas hydrothermalis Y2 in a reduced salinity medium. Microb. Cell Fact. 2019, 18, 184. [Google Scholar]
- Ma, H.; Zhao, Y.; Huang, W.; Zhang, L.; Wu, F.; Ye, J.; Chen, G.Q. Rational flux-tuning of Halomonas bluephagenesis for co-production of bioplastic PHB and ectoine. Nat. Commun. 2020, 11, 3313. [Google Scholar]
- Salar-García, M.J.; Bernal, V.; Pastor, J.M.; Salvador, M.; Argandoña, M.; Nieto, J.J.; Vargas, C.; Cánovas, M. Understanding the interplay of carbon and nitrogen supply for ectoines production and metabolic overflow in high density cultures of Chromohalobacter salexigens. Microb. Cell Fact. 2017, 16, 23. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Wang, Y.; Oshima, A.; Zhang, L.; Miyake, H.; Sasaki, H.; Ishida, A. Efficient cyclic system to yield ectoine using Brevibacterium sp. JCM 6894 subjected to osmotic downshock. Biotechnol. Bioeng. 2008, 99, 941–948. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Onraedt, A.E.; Walcarius, B.A.; Soetaert, W.K.; Vandamme, E.J. Optimization of ectoine synthesis through fed-batch fermentation of Brevibacterium epidermis. Biotechnol. Prog. 2005, 21, 1206–1212. [Google Scholar] [CrossRef] [PubMed]
- Van-Thuoc, D.; Guzmán, H.; Thi-Hang, M.; Hatti-Kaul, R. Ectoine production by Halomonas boliviensis: Optimization using response surface methodology. Mar. Biotechnol. 2010, 12, 586–593. [Google Scholar] [CrossRef]
- Chen, W.C.; Hsu, C.C.; Lan, J.C.; Chang, Y.K.; Wang, L.F.; Wei, Y.H. Production and characterization of ectoine using a moderately halophilic strain Halomonas salina BCRC17875. J. Biosci. Bioeng. 2018, 125, 578–584. [Google Scholar] [CrossRef]
- Chen, S.Y.; Peng, T.C.; Huang, S.Z.; Chien, C.C. Isolation of an ectoine-producing Sinobaca sp. and identification of genes that are involved in ectoine biosynthesis. FEMS Microbiol. Lett. 2022, 369, fnac046. [Google Scholar] [CrossRef]
- Zhang, L.H.; Lang, Y.J.; Nagata, S. Efficient production of ectoine using ectoine-excreting strain. Extremophiles 2009, 13, 717–724. [Google Scholar] [CrossRef]
- Fatollahi, P.; Ghasemi, M.; Yazdian, F.; Sadeghi, A. Ectoine production in bioreactor by Halomonas elongata DSM2581: Using MWCNT and Fe-nanoparticle. Biotechnol. Prog. 2021, 37, e3073. [Google Scholar] [CrossRef]
- Chen, W.C.; Yuan, F.W.; Wang, L.F.; Chien, C.C.; Wei, Y.H. Ectoine production with indigenous Marinococcussp. MAR2 isolated from the marine environment. Prep. Biochem. Biotechnol. 2020, 50, 74–81. [Google Scholar] [CrossRef]
- Lang, Y.J.; Bai, L.; Ren, Y.N.; Zhang, L.H.; Nagata, S. Production of ectoine through a combined process that uses both growing and resting cells of Halomonas salina DSM 5928T. Extremophiles 2011, 15, 303–310. [Google Scholar] [CrossRef]
- Wei, Y.H.; Yuan, F.W.; Chen, W.C.; Chen, S.Y. Production and characterization of ectoine by Marinococcus sp. ECT1 isolated from a high-salinity environment. J. Biosci. Bioeng. 2011, 111, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Wang, Y.B. Accumulation of ectoine in the halotolerant Brevibacterium sp. JCM 6894. J. Biosci. Bioeng. 2001, 91, 288–293. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Gao, X.; Xing, J.; Wang, R.; Zhu, D.; Shen, G. Comparative genomic analysis of Halomonas campaniensis wild-type and ultraviolet radiation-mutated strains reveal genomic differences associated with increased ectoine production. Int. Microbiol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, Z.; Wang, J.; Mohisn, A.; Liu, H.; Zhang, Y.; Zhuang, Y.; Guo, M. Physiological metabolic topology analysis of Halomonas elongata DSM 2581. Biotechnol. Bioeng. 2022, 119, 3509–3525. [Google Scholar] [CrossRef] [PubMed]
- Van-Thuoc, D.; Guzmán, H.; Quillaguamán, J.; Hatti-Kaul, R. High productivity of ectoines by Halomonas boliviensis using a combined two-step fed-batch culture and milking process. J. Biotechnol. 2010, 147, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, H.; Frikha-Dammak, D.; Fakhfakh, J.; Chamkha, M.; Hassairi, I.; Allouche, N.; Sayadi, S.; Maalej, S. The saltern-derived Paludifilum halophilum DSM 102817T is a new high-yield ectoines producer in minimal medium and under salt stress conditions. 3 Biotech 2020, 10, 533. [Google Scholar] [CrossRef]
- Chen, R.; Zhu, L.; Lv, L.; Yao, S.; Li, B.; Qian, J. Optimization of the extraction and purification of the compatible solute ectoine from Halomonas elongate in the laboratory experiment of a commercial production project. World J. Microbiol. Biotechnol. 2017, 33, 116. [Google Scholar] [CrossRef]
- Yu, W.; Cao, X.; Gao, J.; Zhou, Y.J. Overproduction of 3-hydroxypropionate in a super yeast chassis. Bioresour. Technol. 2022, 361, 127690. [Google Scholar] [CrossRef]
- Choi, S.; Song, C.W.; Shin, J.H.; Lee, S.Y. Biorefineries for the production of top building block chemicals and their derivatives. Metab. Eng. 2015, 28, 223–239. [Google Scholar] [CrossRef]
- Della Pina, C.; Falletta, E.; Rossi, M. A green approach to chemical building blocks. The case of 3-hydroxypropanoic acid. Green Chem. 2011, 13, 1624–1632. [Google Scholar] [CrossRef]
- Zhao, P.; Ma, C.; Xu, L.; Tian, P. Exploiting tandem repetitive promoters for high-level production of 3-hydroxypropionic acid. Appl. Microbiol. Biotechnol. 2019, 103, 4017–4031. [Google Scholar] [CrossRef]
- Kim, J.W.; Ko, Y.S.; Chae, T.U.; Lee, S.Y. High-level production of 3-hydroxypropionic acid from glycerol as a sole carbon source using metabolically engineered Escherichia coli. Biotechnol. Bioeng. 2020, 117, 2139–2152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Su, M.; Chen, Y.; Wang, Z.; Nielsen, J.; Liu, Z. Engineering yeast mitochondrial metabolism for 3-hydroxypropionate production. Biotechnol. Biofuels Bioprod. 2023, 16, 64. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.R.; Yan, X.; Yu, L.P.; Liu, X.Y.; Chen, G.Q. Hyperproduction of 3-hydroxypropionate by Halomonas bluephagenesis. Nat. Commun. 2021, 12, 1513. [Google Scholar] [CrossRef]
- Liu, B.; Xiang, S.; Zhao, G.; Wang, B.; Ma, Y.; Liu, W.; Tao, Y. Efficient production of 3-hydroxypropionate from fatty acids feedstock in Escherichia coli. Metab. Eng. 2019, 51, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Xiong, T.; Qin, H.B.; Wu, H.; Liu, Z.Q.; Zheng, Y.G. 3-Hydroxypropionic acid production by recombinant Escherichia coli ZJU-3HP01 using glycerol-glucose dual-substrate fermentative strategy. Biotechnol. Appl. Biochem. 2017, 64, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, M.; Ashok, S.; Park, S. Production of 3-hydroxypropionic acid from glycerol by acid tolerant Escherichia coli. J. Ind. Microbiol. Biotechnol. 2014, 41, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Chaves, G.L.; Batista, R.S.; Cunha, J.S.; Oliveira, D.B.; da Silva, M.R.; Pisani GF, D.; Selistre-de-Araújo, H.S.; Zangirolami, T.C.; da Silva, A.J. Improving 3-hydroxypropionic acid production in E. coli by in silico prediction of new metabolic targets. New Biotechnol. 2022, 72, 80–88. [Google Scholar] [CrossRef]
- Heo, W.; Kim, J.H.; Kim, S.; Kim, K.H.; Kim, H.J.; Seo, J.H. Enhanced production of 3-hydroxypropionic acid from glucose and xylose by alleviation of metabolic congestion due to glycerol flux in engineered Escherichia coli. Bioresour. Technol. 2019, 285, 121320. [Google Scholar] [CrossRef]
- Dai, Z.; Pomraning, K.R.; Deng, S.; Kim, J.; Campbell, K.B.; Robles, A.L.; Hofstad, B.A.; Munoz, N.; Gao, Y.; Lemmon, T.; et al. Metabolic engineering to improve production of 3-hydroxypropionic acid from corn-stover hydrolysate in Aspergillus species. Biotechnol. Biofuels Bioprod. 2023, 16, 53. [Google Scholar] [CrossRef]
- Liang, B.; Zhang, X.; Meng, C.; Wang, L.; Yang, J. Directed evolution of tripartite ATP-independent periplasmic transporter for 3-Hydroxypropionate biosynthesis. Appl. Microbiol. Biotechnol. 2023, 107, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ding, Y.; Zhang, R.; Liu, H.; Xian, M.; Zhao, G. Functional balance between enzymes in malonyl-CoA pathway for 3-hydroxypropionate biosynthesis. Metab. Eng. 2016, 34, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Rathnasingh, C.; Raj, S.M.; Jo, J.E.; Park, S. Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol. Bioeng. 2009, 104, 729–739. [Google Scholar] [PubMed]
- Kwak, S.; Park, Y.C.; Seo, J.H. Biosynthesis of 3-hydroxypropionic acid from glycerol in recombinant Escherichia coli expressing Lactobacillus brevis dhaB and dhaR gene clusters and E. coli K-12 aldH. Bioresour. Technol. 2013, 135, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.S.; Kim, Y.S.; Lee, C.M.; Lee, J.H.; Jung, W.S.; Ahn, J.H.; Song, S.H.; Choi, I.S.; Cho, K.M. Metabolic engineering of 3-hydroxypropionic acid biosynthesis in Escherichia coli. Biotechnol. Bioeng. 2015, 112, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Jung, I.Y.; Lee, J.W.; Min, W.K.; Park, Y.C.; Seo, J.H. Simultaneous conversion of glucose and xylose to 3-hydroxypropionic acid in engineered Escherichia coli by modulation of sugar transport and glycerol synthesis. Bioresour. Technol. 2015, 198, 709–716. [Google Scholar] [CrossRef]
- Lee, T.Y.; Min, W.K.; Kim, H.J.; Seo, J.H. Improved production of 3-hydroxypropionic acid in engineered Escherichia coli by rebalancing heterologous and endogenous synthetic pathways. Bioresour. Technol. 2020, 299, 122600. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, Y.; Xu, M.; Fei, Q.; Gu, Y.; Luo, Y.; Wu, H. Efficient biosynthesis of 3-hydroxypropionic acid from ethanol in metabolically engineered Escherichia coli. Bioresour. Technol. 2022, 363, 127907. [Google Scholar] [CrossRef]
- Kildegaard, K.R.; Jensen, N.B.; Schneider, K.; Czarnotta, E.; Özdemir, E.; Klein, T.; Maury, J.; Ebert, B.E.; Christensen, H.B.; Chen, Y.; et al. Engineering and systems-level analysis of Saccharomyces cerevisiae for production of 3-hydroxypropionic acid via malonyl-CoA reductase-dependent pathway. Microb. Cell Fact. 2016, 15, 53. [Google Scholar] [CrossRef]
- Fina, A.; Brêda, G.C.; Pérez-Trujillo, M.; Freire DM, G.; Almeida, R.V.; Albiol, J.; Ferrer, P. Benchmarking recombinant Pichia pastoris for 3-hydroxypropionic acid production from glycerol. Microb. Biotechnol. 2021, 14, 1671–1682. [Google Scholar] [CrossRef]
- Luo, L.H.; Seo, J.W.; Heo, S.Y.; Oh, B.R.; Kim, D.H.; Kim, C.H. Identification and characterization of Klebsiella pneumoniae aldehyde dehydrogenases increasing production of 3-hydroxypropionic acid from glycerol. Bioprocess Biosyst. Eng. 2013, 36, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Li, Y.; Ge, X.; Tian, P. 3-Hydroxypropionaldehyde-specific aldehyde dehydrogenase from Bacillus subtilis catalyzes 3-hydroxypropionic acid production in Klebsiella pneumoniae. Biotechnol. Lett. 2015, 37, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Xian, M.; Liu, W.; Xu, C.; Zhang, H.; Zhao, G. Biosynthesis of poly(3-hydroxypropionate) from glycerol using engineered Klebsiella pneumoniae strain without vitamin B12. Bioengineered 2015, 6, 77–81. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, X.; Chen, L.; Wang, X.; Tian, P. Physiological investigations of the influences of byproduct pathways on 3-hydroxypropionic acid production in Klebsiella pneumoniae. J. Basic Microbiol. 2019, 59, 1195–1207. [Google Scholar] [CrossRef]
- Zhao, P.; Ren, M.; Ge, X.; Tian, P.; Tan, T. Development of orthogonal T7 expression system in Klebsiella pneumoniae. Biotechnol. Bioeng. 2020, 117, 2446–2459. [Google Scholar] [CrossRef]
- Wang, X.; Sa, N.; Wang, F.H.; Tian, P.F. Engineered constitutive pathway in Klebsiella pneumoniae for 3-hydroxypropionic acid production and implications for decoupling glycerol dissimilation pathways. Curr. Microbiol. 2013, 66, 293–299. [Google Scholar] [CrossRef]
- Luo, L.H.; Seo, J.W.; Baek, J.O.; Oh, B.R.; Heo, S.Y.; Hong, W.K.; Kim, D.H.; Kim, C.H. Identification and characterization of the propanediol utilization protein PduP of Lactobacillus reuteri for 3-hydroxypropionic acid production from glycerol. Appl. Microbiol. Biotechnol. 2011, 89, 697–703. [Google Scholar] [CrossRef]
- Ashok, S.; Sankaranarayanan, M.; Ko, Y.; Jae, K.E.; Ainala, S.K.; Kumar, V.; Park, S. Production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae ΔdhaTΔyqhD which can produce vitamin B12 naturally. Biotechnol. Bioeng. 2013, 110, 511–524. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Ge, X.; Tian, P. High Production of 3-Hydroxypropionic acid in Klebsiella pneumoniae by systematic optimization of glycerol metabolism. Sci. Rep. 2016, 6, 26932. [Google Scholar] [CrossRef]
- Suyama, A.; Higuchi, Y.; Urushihara, M.; Maeda, Y.; Takegawa, K. Production of 3-hydroxypropionic acid via the malonyl-CoA pathway using recombinant fission yeast strains. J. Biosci. Bioeng. 2017, 124, 392–399. [Google Scholar] [CrossRef]
- Takayama, S.; Ozaki, A.; Konishi, R.; Otomo, C.; Kishida, M.; Hirata, Y.; Matsumoto, T.; Tanaka, T.; Kondo, A. Enhancing 3-hydroxypropionic acid production in combination with sugar supply engineering by cell surface-display and metabolic engineering of Schizosaccharomyces pombe. Microb. Cell Fact. 2018, 17, 176. [Google Scholar] [CrossRef] [PubMed]
- Suppuram, P.; Ramakrishnan, G.G.; Subramanian, R. An integrated process for the production of 1,3-propanediol, lactate and 3-hydroxypropionic acid by an engineered Lactobacillus reuteri. Biosci. Biotechnol. Biochem. 2019, 83, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Burgé, G.; Saulou-Bérion, C.; Moussa, M.; Pollet, B.; Flourat, A.; Allais, F.; Athès, V.; Spinnler, H.E. Diversity of Lactobacillus reuteri strains in converting glycerol into 3-hydroxypropionic acid. Appl. Biochem. Biotechnol. 2015, 177, 923–939. [Google Scholar] [CrossRef]
- Li, W.; Wang, T.; Dong, Y.; Li, T. Screening, identification, and low-energy ion modified breeding of a yeast strain producing high level of 3-hydroxypropionic acid. Microbiologyopen 2020, 9, e00956. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Park, S.J.; Park, O.J.; Cho, J.; Rhee, J.W. Production of 3-hydroxypropionic acid from acrylic acid by newly isolated rhodococcus erythropolis LG12. J. Microbiol. Biotechnol. 2009, 19, 474–481. [Google Scholar] [CrossRef]
- Russmayer, H.; Ergoth, S.; Marx, H.; Sauer, M. Process engineering towards an oxidative cellular state improves 3-hydroxypropionic acid production with Lentilactobacillus diolivorans. Bioresour. Technol. 2023, 382, 129160. [Google Scholar] [CrossRef]
- Sun, L.; Yu, F.; Zheng, Y. [Biosynthesis of 3-hydroxypropionic acid from 1,3-propanediol by Gluconobacter oxydans ZJB09112]. Sheng Wu Gong Cheng Xue Bao 2012, 28, 498–507. [Google Scholar]
- Zhao, L.; Lin, J.; Wang, H.; Xie, J.; Wei, D. Development of a two-step process for production of 3-hydroxypropionic acid from glycerol using Klebsiella pneumoniae and Gluconobacter oxydans. Bioprocess Biosyst. Eng. 2015, 38, 2487–2495. [Google Scholar] [CrossRef]
- Tahiliani, A.G.; Beinlich, C.J. Pantothenic acid in health and disease. Vitam. Horm. 1991, 46, 165–228. [Google Scholar]
- Zhao, K.; Tang, H.; Zhang, B.; Zou, S.; Liu, Z.; Zheng, Y. Microbial production of vitamin B5: Current status and prospects. Crit. Rev. Biotechnol. 2022, 9, 1–21. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Z. Cloning and expression of D-lactonohydrolase cDNA from Fusarium moniliforme in Saccharomyces cerevisiae. Biotechnol. Lett. 2004, 26, 1861–1865. [Google Scholar] [CrossRef] [PubMed]
- Tigu, F.; Zhang, J.; Liu, G.; Cai, Z.; Li, Y. A highly active pantothenate synthetase from Corynebacterium glutamicum enables the production of D-pantothenic acid with high productivity. Appl. Microbiol. Biotechnol. 2018, 102, 6039–6046. [Google Scholar] [CrossRef] [PubMed]
- Tadi SR, R.; Nehru, G.; Allampalli SS, P.; Sivaprakasam, S. Engineering precursor and co-factor supply to enhance D-pantothenic acid production in Bacillus megaterium. Bioprocess Biosyst. Eng. 2022, 45, 843–854. [Google Scholar] [CrossRef]
- Tadi SR, R.; Nehru, G.; Limaye, A.M.; Sivaprakasam, S. High-level expression and optimization of pantoate-β-alanine ligase in Bacillus megaterium for the enhanced biocatalytic production of D-pantothenic acid. J. Food Sci. Technol. 2022, 59, 917–926. [Google Scholar] [CrossRef]
- Li, B.; Zhang, B.; Wang, P.; Cai, X.; Tang, Y.Q.; Jin, J.Y.; Liang, J.X.; Liu, Z.Q.; Zheng, Y.G. Targeting metabolic driving and minimization of by-products synthesis for high-yield production of D-pantothenate in Escherichia coli. Biotechnol. J. 2022, 17, e2100431. [Google Scholar] [PubMed]
- Zou, S.P.; Wang, Z.J.; Zhao, K.; Zhang, B.; Niu, K.; Liu, Z.Q.; Zheng, Y.G. High-level production of d-pantothenic acid from glucose by fed-batch cultivation of Escherichia coli. Biotechnol. Appl. Biochem. 2021, 68, 1227–1235. [Google Scholar] [CrossRef]
- Zou, S.; Zhao, K.; Tang, H.; Zhang, Z.; Zhang, B.; Liu, Z.; Zheng, Y. Improved production of D-pantothenic acid in Escherichia coli by integrated strain engineering and fermentation strategies. J. Biotechnol. 2021, 339, 65–72. [Google Scholar] [CrossRef]
- Cai, X.; Shi, X.; Liu, S.Q.; Qiang, Y.; Shen, J.D.; Zhang, B.; Liu, Z.Q.; Zheng, Y.G. Hot spot-based engineering of ketopantoate hydroxymethyltransferase for the improvement of D-pantothenic acid production in Escherichia coli. J. Biotechnol. 2023, 364, 40–49. [Google Scholar] [CrossRef]
- Zou, S.P.; Zhao, K.; Wang, Z.J.; Zhang, B.; Liu, Z.Q.; Zheng, Y.G. Overproduction of D-pantothenic acid via fermentation conditions optimization and isoleucine feeding from recombinant Escherichia coli W3110. 3 Biotech 2021, 11, 295. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, X.M.; Wang, W.; Liu, Z.Q.; Zheng, Y.G. Metabolic engineering of Escherichia coli for d-pantothenic acid production. Food Chem. 2019, 294, 267–275. [Google Scholar]
- Wang, P.; Zhou, H.Y.; Zhou, J.P.; Li, B.; Liu, Z.Q.; Zheng, Y.G. Module engineering coupled with omics strategies for enhancing D-pantothenate production in Escherichia coli. Bioresour. Technol. 2022, 352, 127024. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Wang, T.; Bo, T.; Cai, N.; Yuan, M.; Wu, C.; Jiang, H.; Peng, H.; Chen, N.; Li, Y. Enhanced production of D-pantothenic acid in Corynebacterium glutamicum using an efficient CRISPR-Cpf1 genome editing method. Microb. Cell Fact. 2023, 22, 3. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Sun, X.; Yuan, Y.; Chen, Q.; Ou, Z.; Deng, Z.; Ma, T.; Liu, T. Metabolic Engineering of Saccharomyces cerevisiae for Vitamin B5 Production. J. Agric. Food Chem. 2023, 71, 7408–7417. [Google Scholar] [CrossRef]
- Li, H.; Wang, B.S.; Li, Y.R.; Zhang, L.; Ding, Z.Y.; Gu, Z.H.; Shi, G.Y. Metabolic engineering of Escherichia coli W3110 for the production of L-methionine. J. Ind. Microbiol. Biotechnol. 2017, 44, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Vo, T.M.; Park, S. Metabolic engineering of Escherichia coli W3110 for efficient production of homoserine from glucose. Metab. Eng. 2022, 73, 104–113. [Google Scholar] [CrossRef]
- Muller, M.; Lee, C.M.; Gasiunas, G.; Davis, T.H.; Cradick, T.J.; Siksnys, V.; Bao, G.; Cathomen, T.; Mussolino, C. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol. Ther. 2016, 24, 636–644. [Google Scholar] [CrossRef]
- Mu, Q.; Zhang, S.; Mao, X.; Tao, Y.; Yu, B. Highly efficient production of L-homoserine in Escherichia coli by engineering a redox balance route. Metab. Eng. 2021, 67, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, L.; Yu, S.; Zhou, J. Dual-channel glycolysis balances cofactor supply for L-homoserine biosynthesis in Corynebacterium glutamicum. Bioresour. Technol. 2023, 369, 128473. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, M.; Zhao, G.; Zhang, W.; Li, Y.; Lin, B.; Xu, Q.; Chen, N.; Zhang, C. High-level production of L-homoserine using a non-induced, non-auxotrophic Escherichia coli chassis through metabolic engineering. Bioresour. Technol. 2021, 327, 124814. [Google Scholar] [CrossRef]
- Cai, M.; Zhao, Z.; Li, X.; Xu, Y.; Xu, M.; Rao, Z. Development of a nonauxotrophic L-homoserine hyperproducer in Escherichia coli by systems metabolic engineering. Metab. Eng. 2022, 73, 270–279. [Google Scholar] [CrossRef]
- Liu, M.; Lou, J.; Gu, J.; Lyu, X.M.; Wang, F.Q.; Wei, D.Z. Increasing L-homoserine production in Escherichia coli by engineering the central metabolic pathways. J. Biotechnol. 2020, 314–315, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.Y.; Wang, F.Q.; Zhao, J.; Tao, X.Y.; Liu, M.; Wei, D.Z. Engineering Escherichia coli for L-homoserine production. J. Basic Microbiol. 2023, 63, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, S.; Du, G.; Chen, J.; Zhou, J. Efficient production of L-homoserine in Corynebacterium glutamicum ATCC 13032 by redistribution of metabolic flux. Biochem. Eng. J. 2020, 161, 107665. [Google Scholar] [CrossRef]
- Plachý, J.; Ulbert, S.; Pelechová, J.; Krumphanzl, V. Fermentation production of L-homoserine by Corynebacterium sp. and its possible use in the preparation of threonine and lysine. Folia Microbiol. 1985, 30, 485–492. [Google Scholar] [CrossRef]
- Shi, A.; Zhu, X.; Lu, J.; Zhang, X.; Ma, Y. Activating transhydrogenase and NAD kinase in combination for improving isobutanol production. Metab. Eng. 2013, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fei, M.; Mao, X.; Chen, Y.; Lu, Y.; Wang, L.; Yang, J.; Qiu, J.; Sun, D. Development of a dual-fluorescence reporter system for high-throughput screening of L-aspartate-α-decarboxylase. Acta. Biochim. Biophys. Sin. 2020, 52, 1420–1426. [Google Scholar] [CrossRef] [PubMed]
- Strecker, A.; Schubert, C.; Zedler, S.; Steinmetz, P.; Unden, G. DcuA of aerobically grown Escherichia coli serves as a nitrogen shuttle (L-aspartate/fumarate) for nitrogen uptake. Mol. Microbiol. 2018, 109, 801–811. [Google Scholar] [CrossRef]
- Zhang, C.; Sharma, S.; Wang, W.; Zeng, A.P. A novel downstream process for highly pure 1,3-propanediol from an efficient fed-batch fermentation of raw glycerol by Clostridium pasteurianum. Eng. Life Sci. 2021, 21, 351–363. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).