Abstract
Sourdough is renowned for improving bakery products’ nutritional and quality characteristics through the enzymes produced by its microbiota. Among the enzymatic framework present in sourdough fermentation, amylase, cellulase, and peptidase are responsible for many of the properties valued in sourdough products. Furthermore, there is an increasing concern regarding the allergenic potential of gluten, which motivates the investigation of enzymatic gluten hydrolysis. This study aimed to select probiotics, isolate and identify microorganisms from sourdough, and assess their amylase, cellulase, and peptidase profiles. Additionally, a rapid screening method was developed for gluten and wheat flour hydrolysis, and gluten zymography and enzymography were performed. As a result, 18 microorganisms were isolated from sourdough and identified. The probiotic Bacillus licheniformis LMG-S 28935, and three microorganisms isolated from sourdough, the Limosilactobacillus fermentum, Pediococcus pentosaceus, and Saccharomyces cerevisiae, completed the profile of analyzed hydrolases and presented the capacity to hydrolyze gluten. These findings contribute to a better understanding of sourdough microorganisms’ hydrolase activities in the bakery science and technology field. In addition, an efficient, fast, and economical method for screening extracellular glutenase, produced by microorganisms, was applied. To our knowledge, it was the first time that amylase, cellulase, and peptidase activities were assessed from sourdough microorganisms.
1. Introduction
Natural sourdough fermentation occurs due to microbiota activity in the fermented flour, mainly due to Lactobacillus and yeasts. These microorganisms are related to baked products’ sensory and nutritional quality, and one important function is enzyme production. There are three enzymes sources in sourdough: the endogenous enzymes in flour used, enzymes from microorganisms during the fermentation process, and exogenous enzymes, which can be added to the dough [1]. This study focused on extracellular hydrolases belonging to peptidases, amylases, and cellulase classes secreted by microorganisms used as starters in sourdough.
Peptidases act in proteins present in the dough. Wheat gluten proteins account for 80% of the total wheat protein and are major determinants of its baking quality. Depending on several factors, gluten protein is associated with wheat-related disorders, and its adverse effects can cause health problems, from allergic reactions to celiac disease [2].
Gluten comprises proteins found in grains such as wheat, barley, and rye. The two main proteins of gluten are gliadin and glutenin. Gliadins are monomeric proteins and comprise the types α/β (28,000–35,000 MW), γ-gliadins (31,000–35,000 MW), ω1,2 (39,000–44,000 MW), and ω5 (49,000–55,000 MW). It is an alcohol-soluble protein responsible for most of the adverse effects of gluten on people with gluten sensitivity or intolerance. Glutenin is water- and ethanol-insoluble but soluble in dilute acids. It is a polymeric protein linked by interchain disulfide (SS) bonds with a wide MW distribution ranging from 105 to several million. Glutenin, for example, presents a varying size ranging from about 500,000 to more than 10 million [3]. Glutenin subunits exist in two types: high-molecular-weight (HMW–GS) (70,000–90,000 MW) and LMW–GS (20,000–45,000 MW) [3,4,5]. HMW-GS is responsible for wheat flour dough’s elasticity. Gliadins and LMW-GS are responsible for the viscous properties [6]. The proportion of gluten proteins varies considerably depending on genetic and environmental factors [7,8].
Gluten degradation by peptidases during sourdough fermentation is a crucial phenomenon affecting sourdough bread quality. Besides this, immunogenicity reduction is linked to decreasing gluten, explicitly weakening the polymerization ability of glutenin and hydrolyzing glutenin peptides [9]. The amino acids and peptides produced by proteolysis affect the taste of fermented foods and, in particular, are essential precursors for volatile flavor compounds [10]. In addition, it has been described that a limited amount of proteolysis during sourdough fermentation beneficially improves the bread flavor without adversely affecting texture and volume [6]. Other wheat proteins are the water/salt-soluble albumins and globulins known to cause typical IgE-mediated wheat allergy [11]. In bread wheat, a type of globulin, the α-amylase/trypsin inhibitors (ATI) represent up to 4% of total wheat proteins and consist of at least 14 types of subunit protein. Recently, ATI has been identified as an inducer of an innate immune response via toll-like receptor 4 in celiac disease and non-celiac wheat sensitivity. This protein is resistant primarily to gastrointestinal proteases and heat, and their inflammatory activity affects the intestine and peripheral organs. A recent study demonstrated that sourdough fermentation degraded ATI structure and bioactivity [12]. The study of peptidases involved in sourdough fermentation leads to a better understanding of these mechanisms and points to new possibilities for reducing the allergenicity of wheat proteins while maintaining a higher organoleptic, nutritional quality of wheat dough. Investigations revealed that a low pH environment, microbial proteolytic system, and wheat enzymes involved in sourdough enabled the allergenicity and toxicity reduction in wheat proteins during fermentation [9,13].
Another hydrolase present in sourdough is amylase. Amylase can come from the flour and the microorganisms in the sourdough, and specifically targets and hydrolyses alpha-1,4-glycosidic bonds within starch molecules, converting them into maltose and other smaller sugar units, providing a nutrient source for the yeast and lactic acid bacteria [14]. Yeast consumes the sugars from amylase, converting them into carbon dioxide and alcohol through fermentation. This carbon dioxide creates air pockets within the dough, leading to its rise during proofing and baking [15]. Lactic acid bacteria consume the sugars and produce lactic acid as a byproduct. The lactic acid contributes to the sour flavor. It helps create an acidic environment that inhibits the growth of harmful bacteria [16]. The enzyme improver helps to increase the loaf volume, lowers the crumb firmness, and keeps bread fresh for longer [14].
Cellulases are not typically studied in the microorganisms present in sourdough. However, the addition of the enzyme in sourdoughs has been studied. In Chinese steamed bread dough enriched in wheat bran, cellulase addition significantly increased the development time, stability, departure time, mixing tolerance index (MTI), extensibility, and stickiness of the regular dough. It decreased both softening and resistance to extension [17]. A study by Liu et al. [18] demonstrated that by combining α-amylase, xylanase, and cellulase, a better rheological effect is obtained with the synergism.
Enzymes in sourdough fermentation is a broad field of research, and still needs many studies, considering the multiplicity of factors involved and the complexity of the microbiota. It is interesting to work with co-cultures aiming to improve health properties and quality of bread; for instance, a co-culture of Pediococcus acidilactici and yeast improved the digestibility of wheat protein compared with single-strain fermentation [11].
The present study points out that the microbial enzymes from native sourdough microorganisms could be a strategy to improve product development for better bread with excellent properties, decreasing wheat sensitivity in the susceptible population.
2. Materials and Methods
2.1. Materials
The Brain Heart Infusion media (BHI) was obtained from Neogen. De Man, Rogosa e Sharp medium (MRS), and yeast extract were provided by Kasvi (São José dos Pinhais, PR, Brazil), and peptone was obtained from Himedia (Kennett Square, PA, USA). Soluble starch was purchased from Reagen (Colombo, PR, Brazil), gelatin from Vetec (Duque de Caxias, RJ, Brazil), and lugol solution from Laborclin (Pinhais, PR, Brazil). Other reagents were carboxymethylcellulose, Coomassie brilliant blue R-250, and dithiothreitol (DTT) (Sigma-Aldrich, Burlington, MA, USA). SDS-PAGE standards broad range was provided by Bio-Rad (Hercules, CA, USA). The flours used were white wheat flour Type 1 from Brazil, containing 12% carbohydrates, 7% proteins, 1% total fat, 5% dietary fiber, and whole wheat flour from Brazil, containing 12% carbohydrates, 8% proteins, 1% total fat, 24% dietary fiber, 1% Ca, 34% Fe. Gluten by MV Química (Barueri, SP, Brazil) containing 7.2% moisture, 0.57% ashes, 75.4% proteins, 161.00% water absorption, and 99.8% granulometry-mesh 80.
2.2. Microorganisms
2.2.1. Probiotics Strains
The Bacillus licheniformis LMG 12363 and Saccharomyces boulardii MUCL 43341 were obtained from Belgian Coordinated Collections of Microorganisms. The Bacillus subtilis LFB-FIOCRUZ 1267 obtained from the Culture Collection of Bacillus at the Fundação Oswaldo Cruz (Rio de Janeiro, RJ, Brazil) was used as a positive standard for detecting amylases, cellulases, and peptidases. The strains were cultivated in a Yeast Extract medium (5 g/L yeast extract, 5 g/L peptone, 20 g/L sucrose, 20 g/L KCl). The bacteria were cultivated at 37 °C for 24 h and yeast at 28 °C for 48 h. The strains were maintained in a medium culture agar slant tube at 4 °C for daily use and cryopreserved with glycerol 20% in a freezer at −18 °C. The Bacillus subtilis LFB-FIOCRUZ 1267 was used as a positive standard for detecting amylases, cellulases, and peptidases.
2.2.2. Isolation of Bacteria and Yeasts from Sourdough
Sourdoughs were produced based on the traditional protocol, according to Coda et al. [19] with modification. Two sourdoughs were produced: sourdough 1 (white wheat flour, WF) and sourdough 2 (whole wheat flour, WOF). In both types, 50 mL of sterile Milli-Q water was added to each result in a final dough yield (DY) (dough weight × 100/flour weight) of 200. The sourdoughs were propagated through daily back-slopping for ten days, mixing 50 g of the previous dough with 25 g of respective flours and 25 g of sterile Mili-Q water, maintaining a DY of 200. The doughs were incubated at room temperature (28 °C) for 4 days, then refrigerated at 4 °C until the 10th day. Samples were collected on the 4th and 10th day. Sourdough samples (1 g) were diluted in peptone water (9 mL) using a ten-fold dilution series. The diluted samples of lactic acid bacteria were spread-plated on an MRS medium with 0.01 g/L of nystatin and incubated anaerobically at 30 °C for 48 h, using shaker-incubator LAC−INA−800 (LACTEA, Cambuci, SP, Brazil). For total mesophilic aerobic microorganisms, the diluted samples were spread-plated on BHI, Yeast Extract medium, and Plate Count Agar (5.0 g/L enzymatic digest of casein/tryptone, 2.5 g/L yeast extract, 1.0 g/L glucose, and 20.0 g/L agar), and incubated at 30 °C for 48 h. For yeasts, the diluted samples were spread-plated on a Yeast Extract medium (according to Section 2.2.1) and incubated at 28 °C for 48–72 h. Colonies were picked randomly from plates containing 100 to 300 colonies, subcultured in the corresponding medium, and re-streaked onto the same medium with 2% agar to isolate colonies. The bacteria and yeasts were stored in agar slant tubes at 4 °C for further identification.
2.3. Microorganisms Identification
2.3.1. Matrix-Assisted Laser Desorption-Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF)
A single colony forming unit (CFU), isolated from a fresh culture, was spotted on Bruker’s ground steel target plate (Bruker S.A.S., Wissembourg, France). Then, 1 μL of 70% formic acid was overlaid on each spot. After air drying at room temperature, 1 μL of the α-cyano-4 hydroxy-cinnamic acid matrix solution was added and air dried. Finally, the MALDI-TOF MS profiles were obtained using MALDI Microflex LT (Bruker Daltonics, Bremen, Germany) [20,21].
Identification results based on MALDI-TOF were accepted at the genus or species level, following Bruker’s instructions. Only identifications with score > 2.00 were considered. Scores below 2.00 were selected for 16S rDNA analysis.
2.3.2. 16S rDNA Identification
The genomic DNA (gDNA) extraction was taken from the overgrown colonies using a commercial genomic DNA extraction kit. The integrity of the gDNA was verified through 1% agarose gel electrophoresis. The purity and concentration of the DNA samples were determined using the NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). The 16S rDNA identification was performed by the company AgregaBiotec (Porto Alegre, RS, Brazil). For the Polymerase Chain Reaction (PCR), 20 ng of gDNA, Taq DNA polymerase, 10X concentrated PCR buffer, MgCl2, dNTPs, and primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) were used. The amplicon sequencing, based on the method of Sanger, was performed in both directions for the 16S rDNA amplicon obtained by PCR. To mark the template DNA, 2.5 pmol of primer (785F 5′-GGA TTA GAT ACCCTG GTA-3′ or 907R 5′-CCG TCA ATT CMT TTR AGT TT-3′) and 0.5 μL of bigDye reagent Terminator v3.1 Cycle Sequencing Kit Applied Biosystems (Thermo Fisher Scientific, Waltham, MA, USA) in a final volume of 10 µL were used. Sequencing data were collected using the Data Collection 3 program Applied Biosystems (Thermo Fisher Scientific, Waltham, MA, USA). The resulting Data Collection files (ab1; electropherograms) were converted into FASTA files (seq; text) by Sequence Analysis Software v. 6 Applied Biosystems (Thermo Fisher Scientific, Waltham, MA, USA) under standard parameters. Data analysis for each amplicon was conducted, and the final sequence was compared to 16S rDNA sequences of the genus of interest obtained from a database [22]. An outgroup was used for phylogenetic inference by Maximum Likelihood (ML). The outgroup was selected because it shared the same family as the analyzed genus, according to data from the Genome Taxonomy Database (GTDB, gtdb.ecogenomic.org/tree, accessed on 19 April 2023). Sequences were aligned with SINA 1.2.11 [23], and positions containing gaps were removed. Finally, a phylogenetic tree was calculated to build cladograms and identify microorganisms.
2.4. Hydrolases Detection
2.4.1. Detection of Amylase, Cellulase, and Peptidase Activity on Agar Plates
The selected microorganisms were subjected to the detection of extracellular hydrolases (amylase, cellulase, and peptidase). The tests were conducted on agar plates with the enzymatic extracts obtained from each microorganism after growth in its respective culture medium. For the screening of amylase, cellulase, and peptidase detection on agar plates, the microorganisms were cultured in their specific media for 24 h. Subsequently, the cultures were centrifuged at 5000 rpm for 20 min at 4 °C using centrifuge Sorvall ST 16R (Thermo Scientific, Waltham, MA, USA). The resulting supernatants were collected as enzymatic extracts for further analysis, following the method described by Junior et al. [24]. For amylase detection, the medium consisted of 2 g/L starch, 1 g/L yeast extract, and 8.5 g/L NaCl. For cellulase detection, the medium contained carboxymethylcellulose (CMC) at a concentration of 2 g/L, 1 g/L yeast extract, and 8.5 g/L NaCl. A gelatin medium was prepared with 10 g/L gelatin, 1 g/L yeast extract, and 8.5 g/L NaCl for peptidase detection. All media used for enzymatic screening contained 2.0 g of agar. For the hydrolases assay, 70 µL of the enzymatic extract was added to a 5 mm well made in the agar plate. The plates were then incubated at 30 °C for 48 h to allow the enzymes to act. To visualize the hydrolysis halos caused by amylase and cellulase, a solution of Lugol 2% (containing 0.7% KI and 0.3% I2) was used. A solution of Coomassie blue (0.25% w/v) in a mixture of methanol and acetic acid (5:1:4 v/v/v) was employed for peptidase activity. The plates were subsequently destained using a solution of 50% methanol and 10% acetic acid (v/v). All hydrolase activities were scored as grades: “grade -” when no visible halo was present, “grade +” when the visible halo was less than 2.0 cm, and “grade ++” when the halo zone was equal to or greater than 2.0 cm [25]. The experiments were performed in triplicate.
2.4.2. Quantitative Assay for Amylase, Cellulase, and Peptidase
Amylase activity was estimated using the DNS method [26] with minor modifications. Briefly, 500 µL enzymatic extract with 500 µL of DNS was boiled at 100 °C for 5 min. The quantity of enzyme was measured at 540 nm using UV–Vis Spectrophotometer SpectraMax i3x (Molecular Devices, San Jose, CA, USA), with a blank sample as a reference (enzymatic extract inactivated by boiled at 70 °C for 30 min). One unit (U) of enzyme activity was defined as the amount of amylase to produce 1 µmol of glucose per minute under the assay conditions. Amylase activity was performed in triplicate.
The cellulase activity was measured by reducing sugars liberated from CMC (2%) solubilized in 50 nM citrate buffer, pH 5.0 using pH meter AT–355 (Alfakit, Florianópolis, SC, Brazil). This mixture was incubated for 30 min at 50 °C. The reactions were stopped by adding DNS reagent and boiling for 5 min. The absorbance was read at 540 nm. The reducing sugars formed were quantified using glucose as standard. One unit (U) was defined as the amount of enzyme that releases 1 µmol of reducing sugar per minute. The experiments were performed in triplicate [24].
The quantification of peptidase activity was performed for gelatinase. The gelatinase assay was described by Mazotto et al. [27]. The supernatants of all assays were measured by absorbance at 660 nm to determine the protein concentration through the Lowry method using BSA (bovine serum albumin) as a standard [28]. The amount of enzyme necessary to increase the protein concentration in 1 µg was defined as one unit of activity (U) [29]. The experiments were performed in triplicate.
2.4.3. Analysis of Gluten and Wheat Flour Protein Hydrolysis by Peptidases
This study used a novel screening methodology to evaluate the capability of microorganisms to hydrolyze gluten and wheat flour proteins based on Vermelho et al. [25]. In each experiment, 70 µL of crude enzymatic extracts from microorganisms with peptidase activity, as described in Section 2.4.1, were applied to the wells of Petri dishes with medium (10 g/L gluten, 20 g/L agar) and medium (10 g/L white wheat flour, 20 g/L agar), respectively. The Petri dishes were then incubated at 30 °C for 48 h. Hydrolysis zones were detected using Coomassie blue staining and washed with a destaining solution (50% v/v Methanol, 10% v/v acetic acid). The gluten hydrolysis (glutenase) and the activity of peptidases in wheat flour protein substrate were categorized based on a grading system: “grade −” indicated the absence of a visible halo, “grade +” indicated visible proteolysis limited to smaller than 2.0 cm diameter halo, and “grade ++” indicated a proteolysis zone extending 2.0 cm diameter or more. The experiments were performed in triplicate.
2.5. Analysis of Leavening Effect of Yeasts Using White Wheat Flour
Yeast samples were selected based on the better enzymatic profile as described in Section 2.4.1. Five doughs were prepared using containers with the same shape and size. Each dough consisted of 15 g of white wheat flour, 18 g of filtered water, and 0.3 g of each sample with 109 CFU/mL, and no inoculated dough (control) was used as a reference. The doughs were incubated at 35 °C for 3 h. At the end of the period, the heights of each sample were measured [30,31]. The sample with the best performance after fermentation was selected for analyses as described in Section 2.4.2, Section 2.4.3 and Section 2.6.
2.6. Electrophoretic Analyses
These experiments were carried out with the microorganisms that showed the best hydrolysis screening results and leavening ability.
Gluten Zymography and Enzymography
Proteolytic activities were characterized using 10% SDS-PAGE copolymerized with 10% of the supernatant after gluten extraction with the buffer Tris-HCl pH 8.8 1.5 M (w/v). The gel was loaded with 40 µL of enzyme extract concentrated on a 10,000 kDa membrane Amicon (Sigma-Aldrich, Burlington, MA, USA) per slot and subjected to electrophoresis at a constant voltage of 170 V, 400 mA at 4 °C for 2 h, using Mini PROTEAN® Tetra Cell (Bio-Rad, Hercules, CA, USA). After that, the gel was washed twice with Triton X100 for 15 min each. Finally, the gel was stained with Coomassie blue R250 solution for 24 h and destained with methanol and acetic acid solutions [27]. Enzymography was applied to detect gluten hydrolysis. The crude enzyme extract supernatants of microorganisms were mixed with 60 µg of gluten powder diluted in phosphate buffer (0.06 M Na2HPO4·7H2O/0.04 M KH2PO4, pH 7.2). The reaction mixtures were incubated for 10 min, 20 min, 30 min, 1 h, 2 h, and 4 h at 37 °C. The reactions were stopped by adding sample protein buffer (250 mM Tris-HCL, 5% v/v 2-mercaptoethanol, 10% w/v SDS, 30% w/v glycerol, 0.02% w/v bromophenol blue, pH 6.8), and boiled at 100 °C for 5 min. A volume of 40 µL of the samples was added per slot, and following electrophoresis, 10% SDS-PAGE gel at a constant voltage of 170 V, 400 mA at room temperature for 2 h, staining with Coomassie brilliant blue for 12 h, and then destaining in washing buffer (50% v/v Methanol, 10% v/v acetic acid) for 1 h [27].
2.7. Statistical Analysis
All statistical analyses were performed using R (v4.2) [32]. A Shapiro–Wilk test was performed to check data normality, after that Kruskal–Wallis and Dunn’s tests were used to evaluate the statistical significance of the data. Post hoc analyses were performed using the R package postHoc [33]. Statistical significance was considered when p < 0.05.
3. Results and Discussion
3.1. Microorganisms Identification
Initially, microorganisms were isolated from different wheat flour samples to screen strains producing enzymes relevant to the baking industry. The identification of the isolated microorganisms was performed through MALDI-TOF and 16S rDNA, where the selection of the strains of interest was performed through qualitative enzymatic methods. Table 1 shows the microorganisms identified in the flours (MALDI-TOF) and species similarity (16S rDNA). In total, 14 strains were identified by MALDI-TOF and 4 by 16S rDNA.

Table 1.
Microorganisms’ identification by MALDI TOF and 16S rDNA.
The isolation of genera belonging to the phylum Pseudomonadota (K. cowanii (S), and P. agglomerans (S)) in fresh sourdough aligns with previous studies that obtained similar results in sourdough cultures refreshed up to four days [34]. This phylum is commonly associated with wheat flour. In mature sourdough, the dominant group is the lactic acid bacteria (LAB) independent of using a LAB starter [35].
The decrease in pH during sourdough fermentation, with levels below 5, can inhibit the growth of Enterobacteriaceae [36]. However, a pH of 4 is a safe mark to determine mature sourdough. Nonetheless, genera such as Pantoea, Pseudomonas, and Kosakonia may persist if the fermentation temperature reaches 30 ± 1 °C, as observed in a study conducted in Brazil [37].
Related LAB species, namely, L. plantarum (S), P. pentosaceus (S), and P. acidilactici (S), were isolated after ten days of consecutive back-slopping. Among these, L. plantarum is the most frequently found hetero-fermentative species in the sourdough environment, as mentioned in 142 out of 312 research articles [38]. Although Lactobacillus is generally more abundant in sourdough, Pediococcus is found in smaller quantities or rarely occurs [39,40].
Regarding wild yeasts, the ratio of LAB to yeast in sourdough typically ranges from 100:1 to 10:1 [41]. K. unispora (S) was isolated from WOF, and R. mucilaginosa (S) from WF, both are reported as rarely in sourdough by De Vuyst et al. [42]. S. cerevisiae is the most predominant yeast in sourdough microbiota [37], and two strains, S. cerevisiae (S1) and S. cerevisiae (S2), were isolated from WF.
Bacillus presence in wheat flour is usual [43]. A study showed the importance of analyzing the role of the Bacillus genus in sourdough fermentation. They were found throughout the entire sourdough fermentation process for 14 and 39 days in the Portuguese broa sourdough production [44]. Moreover, Bacillus brevis, Bacillus cereus, Bacillus circulans, Bacillus laterosporus, Bacillus licheniformis, Bacillus macerans, Bacillus megaterium, Bacillus mycoides, Bacillus polymyxa, Bacillus pumilus, Bacillus stearothermophilus, and Bacillus subtilis were isolated from Portuguese sourdough for bread production with maize and rye [45]. Bacillus species isolated from Adhirasam, a rice-fermented doughnut from South India, were used as starter cultures and produced Adhirasam with superior quality [46]. Bacillus species are found in several fermented foods. They are recognized for producing hydrolases, extracellular polysaccharides, and lipopeptides with antimicrobial activity that could be advantageous for their use as starters in fermented foods [47]. Paraburkholderia is not related to wheat sourdough, but Brazil is known as a diversity center of this microorganism [48]. It is also associated with a fermented cereal non-alcoholic beverage from Nigeria [49].
3.2. Qualitative and Quantitative Hydrolases Analysis
After identifying the microorganisms isolated from sourdough, these microorganisms and the probiotic strains were used in qualitative enzyme assays to evaluate the production of amylase, cellulase, peptidase, wheat flour peptidase, and gluten peptidase on agar plates. Table 2 summarizes all results for the enzymes obtained with the diffusion method on agar plates. As a result, four strains stood out in the simultaneous production of the hydrolases for all evaluated substrates: L. fermentum (S), B. licheniformis LMG 12363 (P), P. Pentosaceus (S), and S. cerevisiae (S2). The B. subtilis from the FIOCRUZ collection was used as the positive control.

Table 2.
Hydrolysis screening results for isolated microorganisms (S) and acquired probiotics (P).
The enzymatic profile was heterogeneous, but it is interesting to note that some bacteria can degrade gluten, such as the probiotics B. liqueniformis LMG 12363 (P), L. fermentum (S), and the S. boulardii MUCL 43341 (P), as well the E. faecium (S2), L. plantarum (S), P. acidilactici (S), P. pentosaceus (S), K. unispora (S), and two S. cerevisiae isolates from sourdough. L. fermentum (S) and S. cerevisiae (S2) showed similarity in the enzymatic profile, with halos of hydrolysis for amylase, peptidase, and peptidase for wheat flour and gluten.
For bread technology, the amount of amylase in the dough contributes to liberating fermentable sugars for fermentation, which can interfere with the velocity of fermentation. Additionally, the enzyme acts in starch retrogradation properties, which can interfere with bread characteristics and modify the color of crumb and crust due to the Maillard reactions [50]. The strains B. licheniformis LMG 12363 (P), L. fermentum (S), P. pentosaceus (S), and S. cerevisiae (S2), showed activity for all analyzed enzymes and then were selected for quantitative enzymatic analysis and gluten electrophoresis. Although amylase activity in fermentation is essential, the balance of the amount is crucial to support the final starch structure to maintain the bread quality [13,51]. In the screening for amylase, all yeasts presented hydrolysis halo; S. cerevisiae (S2) showed the most expressive halo (++). The bacteria B. licheniformis LMG 12363 (P), L. fermentum (S), and P. pentosaceus (S) presented a hydrolysis halo, with emphasis on L. fermentum (S) (++). For amylolytic activity, B. licheniformis LMG 12363 (P) (0.413 µmol/mL ± 0.016) presented the lowest activity in the quantitative method. S. cerevisiae (S2) (3.98 µmol/mL ± 0.186) showed the highest activity, followed by P. pentosaceus (S) (3.698 µmol/mL ± 0.074) and L. fermentum (S) (2.21 µmol/mL ± 0.017) (Figure 1).
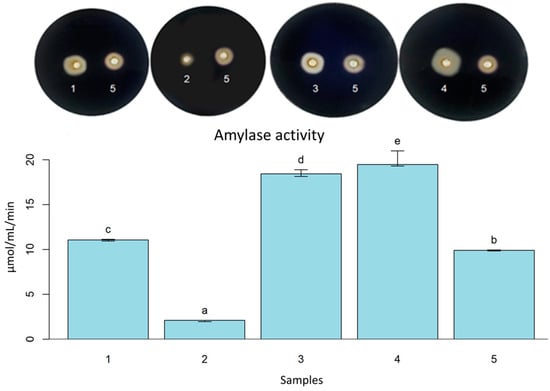
Figure 1.
Amylase activity with Petri dishes screening of L. fermentum (S) (1); B. licheniformis LMG 12363 (P) (2); P. pentosaceus (S) (3); S. cerevisiae (S2) (4); B. subtilis LFB-FIOCRUZ 1267 (5). Different letters indicate statistically significant (p < 0.05) differences between the samples.
Studies reported the amylolytic activity of B. licheniformis. The strain B. subtilis WB 600 was genetically modified to overexpress the maltogenic amylase gene from B. licheniformis. As a result, the strain promoted better bread volume and elasticity, collaborating to increase the product’s shelf life [52]. Another study reported B. licheniformis YB-1234 isolated from fermented soybeans capable of producing thermostable α-amylase [53]. In this way, our result for B. licheniformis LMG 12363 demonstrates the potential of the strain. However, further in-depth studies are needed for its use in the production of bakery products.
The strains L. plantarum and L. fermentum present amylolytic activity below pH 4.0 [54,55]; this feature may provide the availability of fermentable sugar during the entire fermentation process, which can be seen as a fermentation advantage since the amylase from wheat is deactivated in low pH [13]. Our result for the amylase activity of L. fermentum (S) was similar to the strain L. fermentum EN17-2, which exhibited α-amylase activity of 2.00 U/mL in the pH range of 3.5 to 5.5 [55].
S. cerevisiae is not recognized as a great amylase producer, except for S. cerevisiae var. diastaticus, which produces glucoamylase [56]. However, S. cerevisiae produces α-glucosidase that catalyzes the liberation of α-glucose from nonreducing ends of α-glucosides or from complex polymers with α-(1-4) bonds, such as malto-oligosaccharides, soluble starch, amylose, and glycogen [57]. A study showed that 2% potato peel increased the amylase activity of S. cerevisiae, and 4% potato peel improved cellulase activity, and the bread properties, such as retard staling and sensory quality [58]. Olasupo et al. [59] isolated and characterized an amylolytic strain of S. cerevisiae from yam tuber for the brewing industry in Nigeria. Additionally, S. cerevisiae NJJUM 13 isolated from Mangrove Environ showed amylase hydrolysis halo (2.2 cm) [60], similar to our result for S. cerevisiae (S2) with 2.1 cm. P. pentosaceus PKL-17 strain isolated from traditional pickles showed amylase, cellulase, and peptidase activity [61]. This result agrees with our work demonstrating high amylase activity by P. pentosaceus (S). However, this activity is not ubiquitous, as an α-amylase inhibitory capacity was described for a strain of P. pentosaceus. The authors suggest that this can be an interesting property. It could be used as an anti-diabetic probiotic [62].
The cellulase role in bread technology has been described to promote iron bioaccessibility [63]. This enzyme can act positively in gluten network promoting the anti-staling process, elasticity, volume, and softness of bread [64].
Enterococcus faecium (S1) and P. acidilactici (S) showed a significant hydrolysis halo in the cellulase screening. Only S. cerevisiae (S2) among yeasts exhibited cellulose hydrolysis (Table 2). In the cellulase activity, P. pentosaceus (S) showed the highest value (0.493 U/mL ± 0.058), and the B. lichehiformis LMG 12363 (P) (0.307 U/mL ± 0.020) was the smallest value close the L. fermentum (S) (0.363 U/mL ± 0.060) and S. cerevisiae (S2) (0.375 U/mL ± 0.060) (Figure 2).
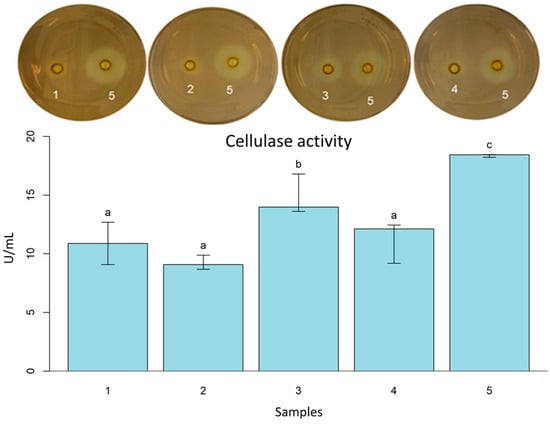
Figure 2.
Cellulase activity with Petri dishes screening of L. fermentum (S) (1); B. licheniformis LMG 12363 (P) (2); P. pentosaceus (S) (3); S. cerevisiae (S2) (4); B. subtilis LFB-FIOCRUZ 1267(5). Different letters indicate statistically significant (p < 0.05) differences between the samples.
Cellulase has been associated with improvements in bread, particularly those with a high dietary fiber content [64,65]. It has been described that the presence of peptidases and cellulases of B. licheniformis isolated from camel feces can reduce allergenic compounds and improve the nutrition properties of soybean meal, which is a source of protein for animal feeds [66]. B. licheniformis MVS1 and Bacillus sp. MVS3 isolated from hot springs showed variation in cellulase activity with changes in nitrogen sources, carbon substrates, pH, temperature, and incubation time. According to the authors, B. licheniformis MVS1 exhibited the highest cellulase activity (0.120 ± 0.012 IU/mL) when wheat straw was the carbon source. However, when cultivated in a culture medium with yeast extract and carboxymethylcellulose, the cellulase activity of B. licheniformis MVS1 was reduced to 0.041 ± 0.003 IU/mL [67].
All analyzed strains have a performance that suggests the potential for future applications in fiber-rich bakery products, such as products made with whole wheat flour or incorporating grains into the dough. However, more investigations are needed to explore using active cellulase inoculants in sourdough preparations.
The peptidase activity in bread fermentation has significant importance since it can liberate peptides with antimicrobial properties [68], flavor’s precursors, bioactive peptides, and anti-hypertensive tripeptides [13,69], as well as improve volume, texture, and reduce staling ratio when it is an appropriate amount [70]. In addition, it was observed that peptidases from Lactobacillus acidophilus 5e2 and Aspergillus ninger could reduce gliadins and coeliac-toxic peptides during the bread-making process [71]. P. acidilactici XZ31 with S. cerevisiae JM4 degraded gluten peptides and reduced gluten immunogenicity [9]. L. fermentum 3872 produced bacteriolysin BLF3872 with a lysozyme-like domain and peptidase M23 domain [72].
As shown in Table 2, some strains presented positive results for peptidases, such as B. licheniformis LMG 12363 (P), L. fermentum (S), E. faecium (S2), L. plantarum (S), P. acidilactici (S), P. pentosaceus (S), and all analyzed yeasts. Additionally, those strains presented a capacity for wheat flour and gluten hydrolysis. These findings are highly significant due to the interest in understanding the impact of peptidases on baking technology. L. fermentum (S) exhibited the highest gelatinase activity (205.64 U/mL ± 5.888). B. licheniformis LMG 12363 (P) (20.89 U/mL ± 6.841) was the second followed by S. cerevisiae (S2) (12.051 ± 4.778), and P. pentosaceus (S) (6.02 U/mL ± 5.605) (Figure 3).
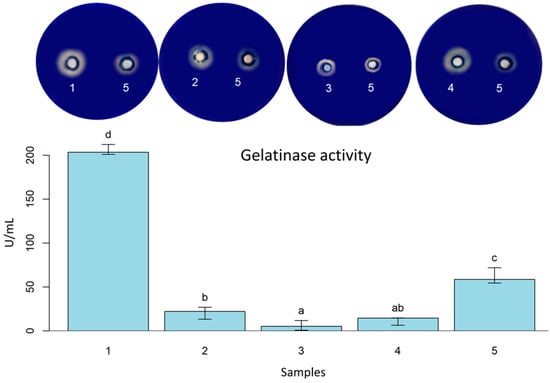
Figure 3.
Gelatinase activity with Petri dishes screening of L. fermentum (S) (1); B. licheniformis LMG 12363 (P) (2); P. pentosaceus (S) (3); S. cerevisiae (S2) (4); B. subtilis LFB-FIOCRUZ 1267 (5). Different letters indicate statistically significant (p < 0.05) differences between the samples.
Bacillus and Aspergillus are recognized as primary peptidase producers [73]. A thermostable serine protease from B. licheniformis (LMG7561) reduced staling, promoted softness, and the effect was additive to known anti-staling agents (such as amylases) [74]. In another study, a serine protease with properties to be used in the food industry was purified and characterized from B. licheniformis KB111 [75]. The Bacillus genus is involved in bread spoilage [76]. However, some Bacillus species are recognized as Generally Recognized as Safe (GRAS), and probiotics can improve nutritionally and technologically gluten products with their cells or enzymes [47]. In our study, for instance, the strain B. licheniformis LMG 12363 (P) had the highest gluten hydrolysis halo, followed by P. pentosaceus (S) and S. cerevisiae (S2). In terms of wheat flour hydrolysis, L. fermentum (S) stood out for its hydrolysis halo performance, followed by S. cerevisiae (S2) (Figure 4).
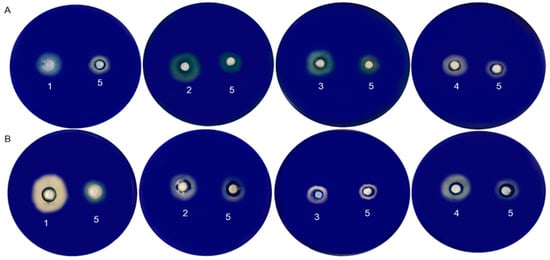
Figure 4.
Gluten protein hydrolysis screening (A) and wheat flour protein hydrolysis screening (B): L. fermentum (S) (1); Bacillus licheniformis LMG 12363 (P) (2); P. pentosaceus (S) (3); S. cerevisiae (S2) (4); B. subtilis LFB-FIOCRUZ 1267 (5).
Peptidase can break down both soluble and insoluble proteins in wheat [11,77,78]. When comparing our results of wheat flour and gluten hydrolysis, we observed that L. fermentum (S), B. licheniformis LMG 12363, P. pentosaceus (S), and S. cerevisiae (S2) presented hydrolysis halo for both types. The combination of S. cerevisiae (S2) and selected probiotic bacteria could enhance the fermentation and enzymatic activity of bakery products, resulting in novel characteristics. Further research is required to evaluate the optimal conditions and benefits of using these strains.
3.3. Leavening Effect of Yeasts
The objective of leavening dough is to generate and retain carbon dioxide (CO2) within the dough structure. CO2 production is linked to the yeast’s ability to transform available fermentable carbohydrates in the dough through ethanolic fermentation. Certain bacteria, such as obligatory heterofermentative LAB (OHLAB), also contribute to CO2 production. Combining S. cerevisiae with OHLAB from sourdough significantly improved the leavening capacity of dough [30].
The primary source of carbohydrates in wheat is starch, which accounts for 70–75% of wheat flour. Starch undergoes hydrolysis by amylases, resulting in the production of fermentable sugars. Additionally, wheat flour contains slight amounts of readily fermentable sugars such as glucose, sucrose, maltose, fructose, maltotriose, and raffinose [79]. The hydrolysis of maltose by S. cerevisiae is responsible for the speed of fermentation, and this capability may be attributed to its adaptation during its long history in bread-making [80].
In this study, more than one strain of S. cerevisiae produced hydrolases (amylases and cellulases) which can liberate carbohydrates and, subsequently, through ethanolic fermentation, produce CO2. A method for evaluating fermentative capacity was used as a tie-breaker test. In this case, the strains were inoculated in wheat flour and then placed for fermentation under controlled conditions. Figure 5A shows the fermentative performance of the evaluated strains. The height data were tested for normality using the Shapiro–Wilk test. Non-parametric tests (Kruskal–Wallis and Dunn’s test) were used to compare the height of the samples (α = 0.05). Sample 3 was the only sample that presented height with a statistical difference from the control (Figure 5B). Thus, it was possible to choose strain S. cerevisiae (S2) due to its superior fermentation volume.
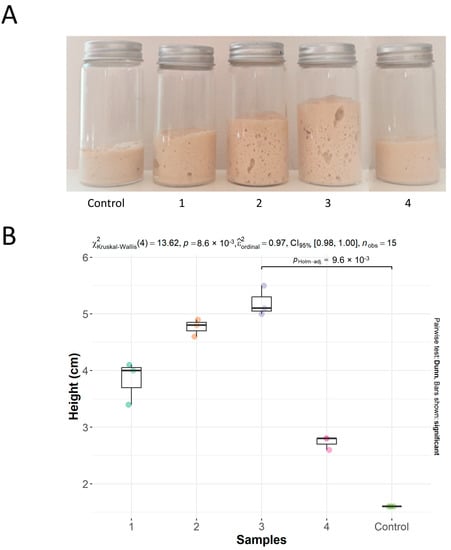
Figure 5.
(A) Dough without inoculum (Control); dough with S. boulardii MUCL 43341 (P) (1); dough with S. cerevisiae (S1) (2); dough with S. cerevisiae (S2) (3); dough with K. unispora (S) (4). (B) Boxplot of height (cm) of levain fermentation in tubes. The line represents the only sample (3) that presented height with a statistically significant difference compared to the control (p < 0.05).
K. unispora has been studied for its consistent presence in sourdough and fermented foods, making it a potentially safe option for the food industry. A recent study showed an adequate leavening capacity for this yeast [81]. However, when compared to commercial S. cerevisiae, its performance was inferior. Our findings supported this finding, as the strain K. unispora (S) did not surpass the leavening ability of the S. cerevisiae samples. The probiotic sample S. boulardii MUCL 43341 (P) presented low performance. A study carried out with S. boulardii application to sourdough found that the bread had a distinct flavor and texture with good acceptance [82]. However, studies of S. boulardii performance in dough fermentation must be more assessed. Regarding the result for the two samples of S. cerevisiae, the highlight of the S. cerevisiae (S2) strain suggests the influence of its superior enzymatic profile for amylase and cellulase activity previously analyzed (Table 2).
3.4. Enzymography and Zymography
Enzymography and zymography are techniques for the separation and visualization of enzymes that can act in the degradation of a substrate, so they can be applied to the study of gluten and its degradation [83,84].
This study investigated the enzymatic activity of the selected microorganisms’ crude extracts concerning gluten biodegradation. The enzymography technique was adopted to detect the enzymatic action on gluten at different incubation times (Figure 6). Gluten proteins are wheat storage proteins with hundreds of components that differ in solubility, structure, molecular weight, and amino acid composition. The enzymatic extracts of all microorganisms showed activity. Figure 6A shows the beginning of the experiment at time 0, where it is possible to observe no hydrolysis and consequent bands because the action of the microorganism’s peptidases did not start effectively. Gluten is a macromolecule that must be hydrolyzed to enter the acrylamide gel. The biodegradation begins slowly, and in 10 min, it is possible to observe subtle bands starting to show (Figure 6B), at 30 min (Figure 6C) and at 2 h (Figure 6D). All extracts showed activity with bands around 30,000 MW, 45,000 MW, and 50,000 MW.
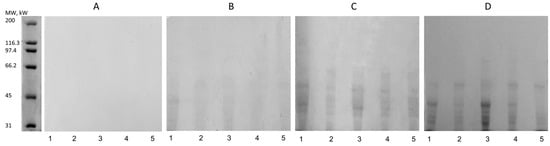
Figure 6.
Enzymography of gluten hydrolysis over time (0–2 h) by extracellular enzymes of 1: L. fermentum (S); 2: B. licheniformis LMG 12363 (P); 3: P. pentosaceus (S); 4: S. cerevisiae (S2); and 5: B. subtilis LFB-FIOCRUZ 1267 used as control. The reaction mixtures containing culture supernatant and gluten solution were incubated for 0 min (A), 10 min (B), 30 min (C), and 2 h (D).
The diversity of MW ranges shows that the byproducts of gluten biodegradation can present very diverse fragments. Figure 6D shows the result of degradation after 2 h of incubation. It can be seen that Lane 3 showed bands and possibly up to ~66.2 kDa, while the other lanes reached a maximum of ~45 kDa. del Amo-Maestro et al. [85] used SDS-PAGE analysis to investigate the digestion of gliadin by neprosin. They observed that neprosin efficiently degraded gliadin at concentrations below ~5 μM, generating fragments below the control bands. Thus, the fragments shown in Figure 7 suggest gluten degradation. However, making any statement about the specificity of enzyme action is impossible.
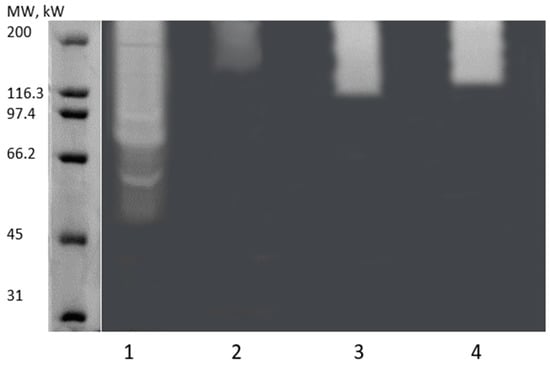
Figure 7.
Zymogram analysis of extracellular enzymes from 1: L. fermentum (S); 2: B. licheniformis LMG 12363 (P); 3: P. pentosaceus (S); and 4: S. cerevisiae (S2). The molecular standard used is shown on the left.
After verifying the enzymatic activity of gluten by enzymography, we employed the zymography technique to evaluate the MW distribution of enzymes with activity on the fraction of gluten solubilized in the buffer Tris-HCl pH 8.8 1.5 M (w/v). Thus, it is possible to visualize the active enzymes in an electrophoresis gel directly. Figure 7 shows the zymography of the four extracts evaluated where it is possible to observe the presence of different patterns of clear bands in the gels, indicating a variety of enzymes with activity on gluten.
These bands demonstrate enzymatic activity in the degradation of gluten which is a complex protein. It is possible to notice that Lane 1 stands out in the diversity of enzymes with action on gluten, then Lanes 3–4, and lastly, Lane 2, which showed a smaller band. Thus, a more pronounced enzymatic activity in the extract L. fermentum (S) is suggested. In contrast, others showed smaller or less evident bands. Thus, the enzyme extracts show enzymes in the ~45–200 kDa range.
Some peptidases can degrade gluten and have a MW range ranging from 20 to 35 kDa [86]. In the work by Ciurko et al. [87], the authors used zymography to determine the molecular weight of the proteolytic enzymes in different microbial extracts. As a result, they observed the presence of peptidases with high molecular weights, specifically with relative molecular masses of 100, 70, and 55 kDa. Another work demonstrated that Bacillus polymyxa could produce extracellular peptidase with molecular masses of 20, 35, 50, and 210 kDa [88]. Liu et al. [89] used a similar technique where they employed gliadin zymogram to analyze secreted peptidases from Burkholderia gladioli, Burkholderia cepacia, Dyella japonica, Dyella yeojuensis, Pseudomonas aeruginosa, and Serratia marcescens. The authors’ zymogram shows the occurrence of peptidases in the range of 48–180 kDa, where they isolated a serine peptidase with a molecular mass of ~51.4 kDa.
This work and other gluten-related studies have reported protein clustering in the 180–200 kDa region. Wei et al. [90] identified a proteolytic enzyme (elastase) with a molecular weight of 53 kDa that was located in the ~200 kDa band. The authors claimed that protein complex formation cannot be excluded in native PAGE. Lu et al. [91] used gliadin zymography to characterize the molecular weight of gliadin-degrading enzymes from B. cereus strains. As a result, they observed active enzyme bands in the high molecular weight region (>170 kDa) and the ~55–72 kDa region. The authors claimed that the band exists in the high region due to dimeric forms of the low molecular weight enzymes [91].
The analysis of enzyme activity through the techniques of zymography and enzymography is of great importance since they allow a direct, visual analysis with information about MW and enzyme activity on gluten. This information is fundamental for understanding the enzymatic diversity of microorganisms involved in fermentation, and helps in the selection of promising strains for the baking industry. However, it is important to highlight that these results do not precisely characterize the involved enzymes.
Future studies should be conducted to purify and identify these enzymes, and investigate their biochemical properties and potential applications in the baking industry. These findings provide a promising basis to explore the enzymes produced by isolated microorganisms as tools to improve the quality of gluten and, consequently, baked goods.
4. Conclusions
Combining and balancing microorganisms, with enzymatic versatility, can offer advantages in reducing allergenic molecules in wheat flour while preserving the properties of bakery products. Hence, enrichment of the enzyme profile of fermented dough can improve the nutritional value and quality of the final product. Screening for gluten and wheat flour hydrolysis is the first step in selecting promising strains to reduce wheat-allergenic molecules. The method we used proved to be simple and efficient in screening glutenase-producing microorganisms. This study identified several microorganisms from sourdough, including Bacillus strains. Characterizing these strains could mean more starters for use in the food industry. The addition of probiotic bacteria could represent an innovation in sourdough, improving and evolving its beneficial properties. The results of this work contributed to identify activities of hydrolases from sourdough microorganisms, which may help in the development of new applications in this type of fermentation. Furthermore, to the best of our knowledge, it was the first time that amylase, cellulase, and peptidase activities were evaluated from sourdough microorganisms.
Author Contributions
Conceptualization, experiments, and writing, I.T.A.; writing, formal analysis, and original draft preparation, F.R.P.M.; performing enzyme experiments, discussing, and integrating the data, V.S.C.; performing substrate characterization, and analyzing and discussing data; E.P.d.S.D.; conceptualization, writing, review, editing, project administration, and funding acquisition, A.B.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by the postgraduate program of the Paulo de Góes Institute of Microbiology, Federal University of Rio de Janeiro (UFRJ), through the Coordenação de Aperfeiçoamento Pessoal de Nível Superior (CAPES) [grant number 001], Conselho Nacional de Desenvolvimento Científico e Tecnológico (MCTI-CNPq) grant code [309461/2019-7], and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), “Cientista do Nosso Estado” grant code [E-26200.428/2023, 282650}.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank PNGWing.com (accessed on 5 April 2023), TogoTV (©2016 DBCLS TogoTV) and Servier Medical Art (smart.servier.com, accessed on 5 April 2023) for providing visual resources for the figures.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Di Cagno, R.; De Angelis, M.; Corsetti, A.; Lavermicocca, P.; Arnault, P.; Tossut, P.; Gallo, G.; Gobbetti, M. Interactions between Sourdough Lactic Acid Bacteria and Exogenous Enzymes: Effects on the Microbial Kinetics of Acidification and Dough Textural Properties. Food Microbiol. 2003, 20, 67–75. [Google Scholar] [CrossRef]
- Rashmi, B.S.; Gayathri, D.; Vasudha, M.; Prashantkumar, C.S.; Swamy, C.T.; Sunil, K.S.; Somaraja, P.K.; Prakash, P. Gluten Hydrolyzing Activity of Bacillus spp. Isolated from Sourdough. Microb. Cell Factories 2020, 19, 130. [Google Scholar] [CrossRef]
- Wieser, H. Chemistry of Gluten Proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef]
- Abedi, E.; Pourmohammadi, K. The Effect of Redox Agents on Conformation and Structure Characterization of Gluten Protein: An Extensive Review. Food Sci. Nutr. 2020, 8, 6301–6319. [Google Scholar] [CrossRef]
- D’Ovidio, R.; Masci, S. The Low-Molecular-Weight Glutenin Subunits of Wheat Gluten. J. Cereal Sci. 2004, 39, 321–339. [Google Scholar] [CrossRef]
- Thiele, C.; Gänzle, M.G.; Vogel, R.F. Contribution of Sourdough Lactobacilli, Yeast, and Cereal Enzymes to the Generation of Amino Acids in Dough Relevant for Bread Flavor. Cereal Chem. J. 2002, 79, 45–51. [Google Scholar] [CrossRef]
- Bonilla, J.C.; Erturk, M.Y.; Kokini, J.L. Understanding the Role of Gluten Subunits (LMW, HMW Glutenins and Gliadin) in the Networking Behavior of a Weak Soft Wheat Dough and a Strong Semolina Wheat Flour Dough and the Relationship with Linear and Non-Linear Rheology. Food Hydrocoll. 2020, 108, 106002. [Google Scholar] [CrossRef]
- Wieser, H.; Koehler, P.; Scherf, K.A. Chemistry of Wheat Gluten Proteins: Quantitative Composition. Cereal Chem. 2023, 100, 36–55. [Google Scholar] [CrossRef]
- Fu, W.; Jia, X.; Liu, C.; Meng, X.; Zhang, K.; Tao, S.; Xue, W. Sourdough Yeast-Bacteria Interactions Results in Reduced Immunogenicity by Increasing Depolymerization and Hydrolysis of Gluten. Innov. Food Sci. Emerg. Technol. 2023, 84, 103281. [Google Scholar] [CrossRef]
- Gänzle, M.G.; Loponen, J.; Gobbetti, M. Proteolysis in Sourdough Fermentations: Mechanisms and Potential for Improved Bread Quality. Trends Food Sci. Technol. 2008, 19, 513–521. [Google Scholar] [CrossRef]
- Fu, W.; Chen, C.; Liu, C.; Tao, S.; Xue, W. Changes in Wheat Protein Digestibility and Allergenicity: Role of Pediococcus acidilactici XZ31 and Yeast during Dough Fermentation. Food Sci. Hum. Wellness 2023, 12, 2381–2389. [Google Scholar] [CrossRef]
- Huang, X.; Schuppan, D.; Rojas Tovar, L.E.; Zevallos, V.F.; Loponen, J.; Gänzle, M. Sourdough Fermentation Degrades Wheat Alpha-Amylase/Trypsin Inhibitor (ATI) and Reduces Pro-Inflammatory Activity. Foods 2020, 9, 943. [Google Scholar] [CrossRef]
- Gänzle, M.G. Enzymatic and Bacterial Conversions during Sourdough Fermentation. Food Microbiol. 2014, 37, 2–10. [Google Scholar] [CrossRef]
- Chen, Y.; Eder, S.; Schubert, S.; Gorgerat, S.; Boschet, E.; Baltensperger, L.; Boschet, E.; Städeli, C.; Kuster, S.; Fischer, P.; et al. Influence of Amylase Addition on Bread Quality and Bread Staling. ACS Food Sci. Technol. 2021, 1, 1143–1150. [Google Scholar] [CrossRef]
- Maicas, S. The Role of Yeasts in Fermentation Processes. Microorganisms 2020, 8, 1142. [Google Scholar] [CrossRef]
- Ayivi, R.D.; Gyawali, R.; Krastanov, A.; Aljaloud, S.O.; Worku, M.; Tahergorabi, R.; Silva, R.C.D.; Ibrahim, S.A. Lactic Acid Bacteria: Food Safety and Human Health Applications. Dairy 2020, 1, 202–232. [Google Scholar] [CrossRef]
- Liu, W.; Brennan, M.A.; Serventi, L.; Brennan, C.S. Effect of Cellulase, Xylanase and α-Amylase Combinations on the Rheological Properties of Chinese Steamed Bread Dough Enriched in Wheat Bran. Food Chem. 2017, 234, 93–102. [Google Scholar] [CrossRef]
- Liu, W.; Brennan, M.; Tu, D.; Brennan, C. Influence of α-Amylase, Xylanase and Cellulase on the Rheological Properties of Bread Dough Enriched with Oat Bran. Sci. Rep. 2023, 13, 4534. [Google Scholar] [CrossRef]
- Coda, R.; Kianjam, M.; Pontonio, E.; Verni, M.; Di Cagno, R.; Katina, K.; Rizzello, C.G.; Gobbetti, M. Sourdough-Type Propagation of Faba Bean Flour: Dynamics of Microbial Consortia and Biochemical Implications. Int. J. Food Microbiol. 2017, 248, 10–21. [Google Scholar] [CrossRef]
- Hsieh, S.-Y.; Tseng, C.-L.; Lee, Y.-S.; Kuo, A.-J.; Sun, C.-F.; Lin, Y.-H.; Chen, J.-K. Highly Efficient Classification and Identification of Human Pathogenic Bacteria by MALDI-TOF MS. Mol. Cell. Proteomics MCP 2008, 7, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Mougin, J.; Flahaut, C.; Roquigny, R.; Bonnin-Jusserand, M.; Grard, T.; Le Bris, C. Rapid Identification of Vibrio Species of the Harveyi Clade Using MALDI-TOF MS Profiling with Main Spectral Profile Database Implemented with an In-House Database: Luvibase. Front. Microbiol. 2020, 11, 586536. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate High-Throughput Multiple Sequence Alignment of Ribosomal RNA Genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Neves Junior, A.; da Silva Cardoso, V.; Mansoldo, F.R.P.; Cedrola, S.M.L.; Reis Mansur, M.C.P.P.; Godoy, M.G.; Vermelho, A.B. A Microplate Assay for Extracellular Hydrolase Detection. J. Microbiol. Methods 2020, 175, 105948. [Google Scholar] [CrossRef] [PubMed]
- Vermelho, A.B.; Meirelles, M.N.L.; Lopes, A.; Petinate, S.D.G.; Chaia, A.A.; Branquinha, M.H. Detection of Extracellular Proteases from Microorganisms on Agar Plates. Mem. Inst. Oswaldo Cruz 1996, 91, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Mazotto, A.M.; de Melo, A.C.N.; Macrae, A.; Rosado, A.S.; Peixoto, R.; Cedrola, S.M.L.; Couri, S.; Zingali, R.B.; Villa, A.L.V.; Rabinovitch, L.; et al. Biodegradation of Feather Waste by Extracellular Keratinases and Gelatinases from Bacillus spp. World J. Microbiol. Biotechnol. 2011, 27, 1355–1365. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Dalmaso, G.Z.L.; Lage, C.A.S.; Mazotto, A.M.; Dias, E.P.d.S.; Caldas, L.A.; Ferreira, D.; Vermelho, A.B. Extracellular Peptidases from Deinococcus radiodurans. Extremophiles 2015, 19, 989–999. [Google Scholar] [CrossRef]
- El Fechtali, T.; Mimoune Reffai, Y.; Idbella, M. The Leavening Ability of Many Lactic Acid Bacteria Isolated from Spontaneous Sourdough. Emir. J. Food Agric. 2023, 35, 23–31. [Google Scholar] [CrossRef]
- Semumu, T.; Gamero, A.; Boekhout, T.; Zhou, N. Evolutionary Engineering to Improve Wickerhamomyces subpelliculosus and Kazachstania gamospora for Baking. World J. Microbiol. Biotechnol. 2022, 38, 48. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 10 March 2023).
- Labouriau, R. postHoc: Tools for Post-Hoc Analysis. R Package Version 0.1.3. 2020. Available online: https://CRAN.R-project.org/package=postHoc (accessed on 10 March 2023).
- Ercolini, D.; Pontonio, E.; De Filippis, F.; Minervini, F.; Storia, A.L.; Gobbetti, M.; Di Cagno, R. Microbial Ecology Dynamics during Rye and Wheat Sourdough Preparation. Appl. Environ. Microbiol. 2013, 79, 7827–7836. [Google Scholar] [CrossRef]
- Celano, G.; De Angelis, M.; Minervini, F.; Gobbetti, M. Different Flour Microbial Communities Drive to Sourdoughs Characterized by Diverse Bacterial Strains and Free Amino Acid Profiles. Front. Microbiol. 2016, 7, 1770. [Google Scholar] [CrossRef]
- Dinardo, F.R.; Minervini, F.; De Angelis, M.; Gobbetti, M.; Gänzle, M.G. Dynamics of Enterobacteriaceae and Lactobacilli in Model Sourdoughs Are Driven by pH and Concentrations of Sucrose and Ferulic Acid. LWT 2019, 114, 108394. [Google Scholar] [CrossRef]
- Menezes, L.A.A.; Sardaro, M.L.S.; Duarte, R.T.D.; Mazzon, R.R.; Neviani, E.; Gatti, M.; De Dea Lindner, J. Sourdough Bacterial Dynamics Revealed by Metagenomic Analysis in Brazil. Food Microbiol. 2020, 85, 103302. [Google Scholar] [CrossRef]
- Arora, K.; Ameur, H.; Polo, A.; Di Cagno, R.; Rizzello, C.G.; Gobbetti, M. Thirty Years of Knowledge on Sourdough Fermentation: A Systematic Review. Trends Food Sci. Technol. 2021, 108, 71–83. [Google Scholar] [CrossRef]
- De Vuyst, L.; Neysens, P. The Sourdough Microflora: Biodiversity and Metabolic Interactions. Trends Food Sci. Technol. 2005, 16, 43–56. [Google Scholar] [CrossRef]
- Fraberger, V.; Unger, C.; Kummer, C.; Domig, K.J. Insights into Microbial Diversity of Traditional Austrian Sourdough. LWT 2020, 127, 109358. [Google Scholar] [CrossRef]
- Minervini, F.; Di Cagno, R.; Lattanzi, A.; De Angelis, M.; Antonielli, L.; Cardinali, G.; Cappelle, S.; Gobbetti, M. Lactic Acid Bacterium and Yeast Microbiotas of 19 Sourdoughs Used for Traditional/Typical Italian Breads: Interactions between Ingredients and Microbial Species Diversity. Appl. Environ. Microbiol. 2012, 78, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Harth, H.; Van Kerrebroeck, S.; Leroy, F. Yeast Diversity of Sourdoughs and Associated Metabolic Properties and Functionalities. Int. J. Food Microbiol. 2016, 239, 26–34. [Google Scholar] [CrossRef]
- Van Kerrebroeck, S.; Bastos, F.C.C.; Harth, H.; De Vuyst, L. A Low pH Does Not Determine the Community Dynamics of Spontaneously Developed Backslopped Liquid Wheat Sourdoughs but Does Influence Their Metabolite Kinetics. Int. J. Food Microbiol. 2016, 239, 54–64. [Google Scholar] [CrossRef]
- Rocha, J.M.; Malcata, F.X. Microbial Ecology Dynamics in Portuguese Broa Sourdough. J. Food Qual. 2016, 39, 634–648. [Google Scholar] [CrossRef]
- Rocha, J.M.; Malcata, F.X. On the Microbiological Profile of Traditional Portuguese Sourdough. J. Food Prot. 1999, 62, 1416–1429. [Google Scholar] [CrossRef] [PubMed]
- Anisha, A.H.N.; Anandham, R.; Kwon, S.W.; Gandhi, P.I.; Gopal, N.O. Evaluation of Bacillus spp. as Dough Starters for Adhirasam-A Traditional Rice Based Fermented Food of Southern India. Braz. J. Microbiol. 2015, 46, 1183–1191. [Google Scholar] [CrossRef]
- Li, Z.; Zheng, M.; Zheng, J.; Gänzle, M.G. Bacillus Species in Food Fermentations: An Underappreciated Group of Organisms for Safe Use in Food Fermentations. Curr. Opin. Food Sci. 2023, 50, 101007. [Google Scholar] [CrossRef]
- Paulitsch, F.; Dos Reis, F.B.; Hungria, M. Twenty Years of Paradigm-Breaking Studies of Taxonomy and Symbiotic Nitrogen Fixation by Beta-Rhizobia, and Indication of Brazil as a Hotspot of Paraburkholderia Diversity. Arch. Microbiol. 2021, 203, 4785–4803. [Google Scholar] [CrossRef]
- Ndukwe, J.K.; Aduba, C.C.; Ughamba, K.T.; Chukwu, K.O.; Eze, C.N.; Nwaiwu, O.; Onyeaka, H. Diet Diversification and Priming with Kunu: An Indigenous Probiotic Cereal-Based Non-Alcoholic Beverage in Nigeria. Beverages 2023, 9, 14. [Google Scholar] [CrossRef]
- Atudorei, D.; Mironeasa, S.; Codină, G.G. Dough Rheological Behavior and Bread Quality as Affected by Addition of Soybean Flour in a Germinated Form. Foods 2023, 12, 1316. [Google Scholar] [CrossRef] [PubMed]
- Rakita, S.; Torbica, A.; Dokic, L.; Tomic, J.; Pojic, M.; Hadnadjev, M.; Hadnadjev-Dapcevic, T. Alpha-Amylase Activity in Wheat Flour and Breadmaking Properties in Relation to Different Climatic Conditions. Food Feed Res. 2015, 42, 91–99. [Google Scholar] [CrossRef]
- Ruan, Y.; Xu, Y.; Zhang, W.; Zhang, R. A New Maltogenic Amylase from Bacillus licheniformis R-53 Significantly Improves Bread Quality and Extends Shelf Life. Food Chem. 2021, 344, 128599. [Google Scholar] [CrossRef]
- Song, S.H. Analysis of Microflora Profile in Korean Traditional Nuruk. J. Microbiol. Biotechnol. 2013, 23, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Oguntoyinbo, F.A.; Narbad, A. Molecular Characterization of Lactic Acid Bacteria and in Situ Amylase Expression during Traditional Fermentation of Cereal Foods. Food Microbiol. 2012, 31, 254–262. [Google Scholar] [CrossRef]
- Khusniati, T.; Gresi Hatmaya, P.; Amir, M.; Rachmach, J.; Safriana, V.; Sulistiani. Characterization of α-Amylase and Protease from Indigenous Lactobacillus fermentum EN17-2 and Its Use in Tuber Paste Flour. IOP Conf. Ser. Earth Environ. Sci. 2020, 439, 012059. [Google Scholar] [CrossRef]
- Knox, A.M.; Du Preez, J.C.; Kilian, S.G. Starch Fermentation Characteristics of Saccharomyces cerevisiae Strains Transformed with Amylase Genes from Lipomyces kononenkoae and Saccharomycopsis fibuligera. Enzyme Microb. Technol. 2004, 34, 453–460. [Google Scholar] [CrossRef]
- del Moral, S.; Barradas-Dermitz, D.M.; Aguilar-Uscanga, M.G. Production and Biochemical Characterization of α-Glucosidase from Aspergillus niger ITV-01 Isolated from Sugar Cane Bagasse. 3 Biotech 2017, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Najmalddin, H.; Yurdugül, S.; Hamzah, H. Screening of Enzyme Activities for Improvement of Bread Quality by Potato Peel Addition to the Yeast Growth Medium. Food Biosci. 2023, 51, 102239. [Google Scholar] [CrossRef]
- Olasupo, N.A.; Teniola, O.D.; Okosun, R.; Omowaye, A.; Olatope, S.O.; Scott-Emuakpor, M.B. Studies on an Amylolytic Strain of Saccharomyces cerevisiae Isolated from Yam Tuber. J. Basic Microbiol. 1996, 36, 283–288. [Google Scholar] [CrossRef]
- Jayalakshmi, N.; Umamaheswari, G. Production and Optimization of Amylase Enzyme from Saccharomyces cerevisiae by Mangrove Environ. Int. J. Sci. Res. 2017, 6, 2524–2526. [Google Scholar]
- Monika; Savitri; Kumar, V.; Kumari, A.; Angmo, K.; Bhalla, T.C. Isolation and Characterization of Lactic Acid Bacteria from Traditional Pickles of Himachal Pradesh, India. J. Food Sci. Technol. 2017, 54, 1945–1952. [Google Scholar] [CrossRef]
- Kim, S.; Hong, S.; Lim, S.-D. Physiological Characteristics and Anti-Diabetic Effect of Pediococcus pentosaceus KI62. Food Sci. Anim. Resour. 2021, 41, 274–287. [Google Scholar] [CrossRef]
- Baye, K.; Guyot, J.-P.; Icard-Vernière, C.; Rochette, I.; Mouquet-Rivier, C. Enzymatic Degradation of Phytate, Polyphenols and Dietary Fibers in Ethiopian Injera Flours: Effect on Iron Bioaccessibility. Food Chem. 2015, 174, 60–67. [Google Scholar] [CrossRef]
- Pourmohammadi, K.; Abedi, E. Hydrolytic Enzymes and Their Directly and Indirectly Effects on Gluten and Dough Properties: An Extensive Review. Food Sci. Nutr. 2021, 9, 3988–4006. [Google Scholar] [CrossRef]
- Messia, M.C.; Reale, A.; Maiuro, L.; Candigliota, T.; Sorrentino, E.; Marconi, E. Effects of Pre-Fermented Wheat Bran on Dough and Bread Characteristics. J. Cereal Sci. 2016, 69, 138–144. [Google Scholar] [CrossRef]
- Qi, N.; Zhan, X.; Milmine, J.; Sahar, M.; Chang, K.-H.; Li, J. Isolation and Characterization of a Novel Hydrolase-Producing Probiotic Bacillus licheniformis and Its Application in the Fermentation of Soybean Meal. Front. Nutr. 2023, 10, 1123422. [Google Scholar] [CrossRef] [PubMed]
- Acharya, S.; Chaudhary, A. Optimization of Fermentation Conditions for Cellulases Production by Bacillus licheniformis MVS1 and Bacillus sp. MVS3 Isolated from Indian Hot Spring. Braz. Arch. Biol. Technol. 2012, 55, 497–503. [Google Scholar] [CrossRef]
- Demirbaş, F.; İspirli, H.; Kurnaz, A.A.; Yilmaz, M.T.; Dertli, E. Antimicrobial and Functional Properties of Lactic Acid Bacteria Isolated from Sourdoughs. LWT Food Sci. Technol. 2017, 79, 361–366. [Google Scholar] [CrossRef]
- Galli, V.; Mazzoli, L.; Luti, S.; Venturi, M.; Guerrini, S.; Paoli, P.; Vincenzini, M.; Granchi, L.; Pazzagli, L. Effect of Selected Strains of Lactobacilli on the Antioxidant and Anti-Inflammatory Properties of Sourdough. Int. J. Food Microbiol. 2018, 286, 55–65. [Google Scholar] [CrossRef]
- Gu, M.; Hong, T.; Ma, Y.; Xi, J.; Zhao, Q.; Xu, D.; Jin, Y.; Wu, F.; Xu, X. Effects of a Commercial Peptidase on Rheology, Microstructure, Gluten Properties of Wheat Dough and Bread Quality. LWT 2022, 160, 113266. [Google Scholar] [CrossRef]
- Brzozowski, B.; Stasiewicz, K.; Ostolski, M.; Adamczak, M. Reducing Immunoreactivity of Gliadins and Coeliac-Toxic Peptides Using Peptidases from L. acidophilus 5e2 and A. niger. Catalysts 2020, 10, 923. [Google Scholar] [CrossRef]
- Abramov, V.M.; Kosarev, I.V.; Machulin, A.V.; Priputnevich, T.V.; Deryusheva, E.I.; Nemashkalova, E.L.; Chikileva, I.O.; Abashina, T.N.; Panin, A.N.; Melnikov, V.G.; et al. Limosilactobacillus fermentum 3872 That Produces Class III Bacteriocin Forms Co-Aggregates with the Antibiotic-Resistant Staphylococcus aureus Strains and Induces Their Lethal Damage. Antibiotics 2023, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Souza, T.S.P.D.; De Andrade, C.J.; Koblitz, M.G.B.; Fai, A.E.C. Microbial Peptidase in Food Processing: Current State of the Art and Future Trends. Catal. Lett. 2023, 153, 114–137. [Google Scholar] [CrossRef]
- Arnaut, F.; Verte, F.; Vekemans, N. Method and Composition for the Prevention or Retarding of Staling of Bakery Products. U.S. Patent US9456616B2, 4 October 2016. [Google Scholar]
- Foophow, T.; Sittipol, D.; Rukying, N.; Phoohinkong, W.; Jongruja, N. Purification and Characterization of a Novel Extracellular Haloprotease Vpr from Bacillus licheniformis Strain KB111. Food Technol. Biotechnol. 2022, 60, 225–236. [Google Scholar] [CrossRef]
- Pacher, N.; Burtscher, J.; Johler, S.; Etter, D.; Bender, D.; Fieseler, L.; Domig, K.J. Ropiness in Bread—A Re-Emerging Spoilage Phenomenon. Foods 2022, 11, 3021. [Google Scholar] [CrossRef] [PubMed]
- Hailegiorgis, D.; Mekonnen, F.; Hailu, F.; Lee, C.A.; Yun, S.J. Composition and Molecular Weight Distribution of Albumin and Globulin Protein Isolates from Durum Wheat Genotypes. Am. J. Plant Sci. 2020, 11, 137–147. [Google Scholar] [CrossRef]
- Goesaert, H.; Brijs, K.; Veraverbeke, W.S.; Courtin, C.M.; Gebruers, K.; Delcour, J.A. Wheat Flour Constituents: How They Impact Bread Quality, and How to Impact Their Functionality. Trends Food Sci. Technol. 2005, 16, 12–30. [Google Scholar] [CrossRef]
- Mietton, L.; Samson, M.-F.; Marlin, T.; Godet, T.; Nolleau, V.; Guezenec, S.; Segond, D.; Nidelet, T.; Desclaux, D.; Sicard, D. Impact of Leavening Agent and Wheat Variety on Bread Organoleptic and Nutritional Quality. Microorganisms 2022, 10, 1416. [Google Scholar] [CrossRef] [PubMed]
- Carbonetto, B.; Ramsayer, J.; Nidelet, T.; Legrand, J.; Sicard, D. Bakery Yeasts, a New Model for Studies in Ecology and Evolution. Yeast 2018, 35, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Korcari, D.; Ricci, G.; Capusoni, C.; Fortina, M.G. Physiological Performance of Kazachstania unispora in Sourdough Environments. World J. Microbiol. Biotechnol. 2021, 37, 88. [Google Scholar] [CrossRef]
- Marolia, K.Z.; Khan, B.K.; Raval, N.; Sharma, Y. Production of Bio-Flavored Sourdough Bread. Afr. J. Biol. Sci. 2022, 4, 127. [Google Scholar] [CrossRef]
- Vandooren, J.; Geurts, N.; Martens, E.; Van Den Steen, P.E.; Opdenakker, G. Zymography Methods for Visualizing Hydrolytic Enzymes. Nat. Methods 2013, 10, 211–220. [Google Scholar] [CrossRef]
- Vermelho, A.B.; Mazotto, A.M.; De Melo, A.C.N.; Vieira, F.H.C.; Duarte, T.R.; Macrae, A.; Nishikawa, M.M.; Da Silva Bon, E.P. Identification of a Candida parapsilosis Strain Producing Extracellular Serine Peptidase with Keratinolytic Activity. Mycopathologia 2010, 169, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Del Amo-Maestro, L.; Mendes, S.R.; Rodríguez-Banqueri, A.; Garzon-Flores, L.; Girbal, M.; Rodríguez-Lagunas, M.J.; Guevara, T.; Franch, À.; Pérez-Cano, F.J.; Eckhard, U.; et al. Molecular and in Vivo Studies of a Glutamate-Class Prolyl-Endopeptidase for Coeliac Disease Therapy. Nat. Commun. 2022, 13, 4446. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, S.; Pérez-Andrés, J.; Martínez-Blanco, H.; Ferrero, M.A.; Vaquero, L.; Vivas, S.; Casqueiro, J.; Rodríguez-Aparicio, L.B. The Human Digestive Tract Has Proteases Capable of Gluten Hydrolysis. Mol. Metab. 2017, 6, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Ciurko, D.; Łaba, W.; Żarowska, B.; Janek, T. Enzymatic Hydrolysis Using Bacterial Cultures as a Novel Method for Obtaining Antioxidant Peptides from Brewers’ Spent Grain. RSC Adv. 2021, 11, 4688–4700. [Google Scholar] [CrossRef]
- Lal, S.; Tabacchioni, S. Ecology and Biotechnological Potential of Paenibacillus polymyxa: A Minireview. Indian J. Microbiol. 2009, 49, 2–10. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Lee, C.-C.; Hsu, J.-H.; Leu, W.-M.; Meng, M. Efficient Hydrolysis of Gluten-Derived Celiac Disease-Triggering Immunogenic Peptides by a Bacterial Serine Protease from Burkholderia gladioli. Biomolecules 2021, 11, 451. [Google Scholar] [CrossRef]
- Wei, G.; Tian, N.; Valery, A.C.; Zhong, Y.; Schuppan, D.; Helmerhorst, E.J. Identification of Pseudolysin (lasB) as an Aciduric Gluten-Degrading Enzyme with High Therapeutic Potential for Celiac Disease. Am. J. Gastroenterol. 2015, 110, 899–908. [Google Scholar] [CrossRef]
- Lu, J.; Wu, Y.; Yuan, J.; Yuan, J.; Wang, Z.; Gao, J.; Chen, H. Characterization of Bacillus cereus AFA01 Capable of Degrading Gluten and Celiac-Immunotoxic Peptides. Foods 2021, 10, 1725. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).