Assembly and Source of the Lithobiontic Microbial Community in Limestone
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
2.2. Sample Processing and Sequencing
2.3. Quantification of the Community Assembly Process
2.4. Microbial Source Tracking
2.5. Statistical Analyses
3. Results
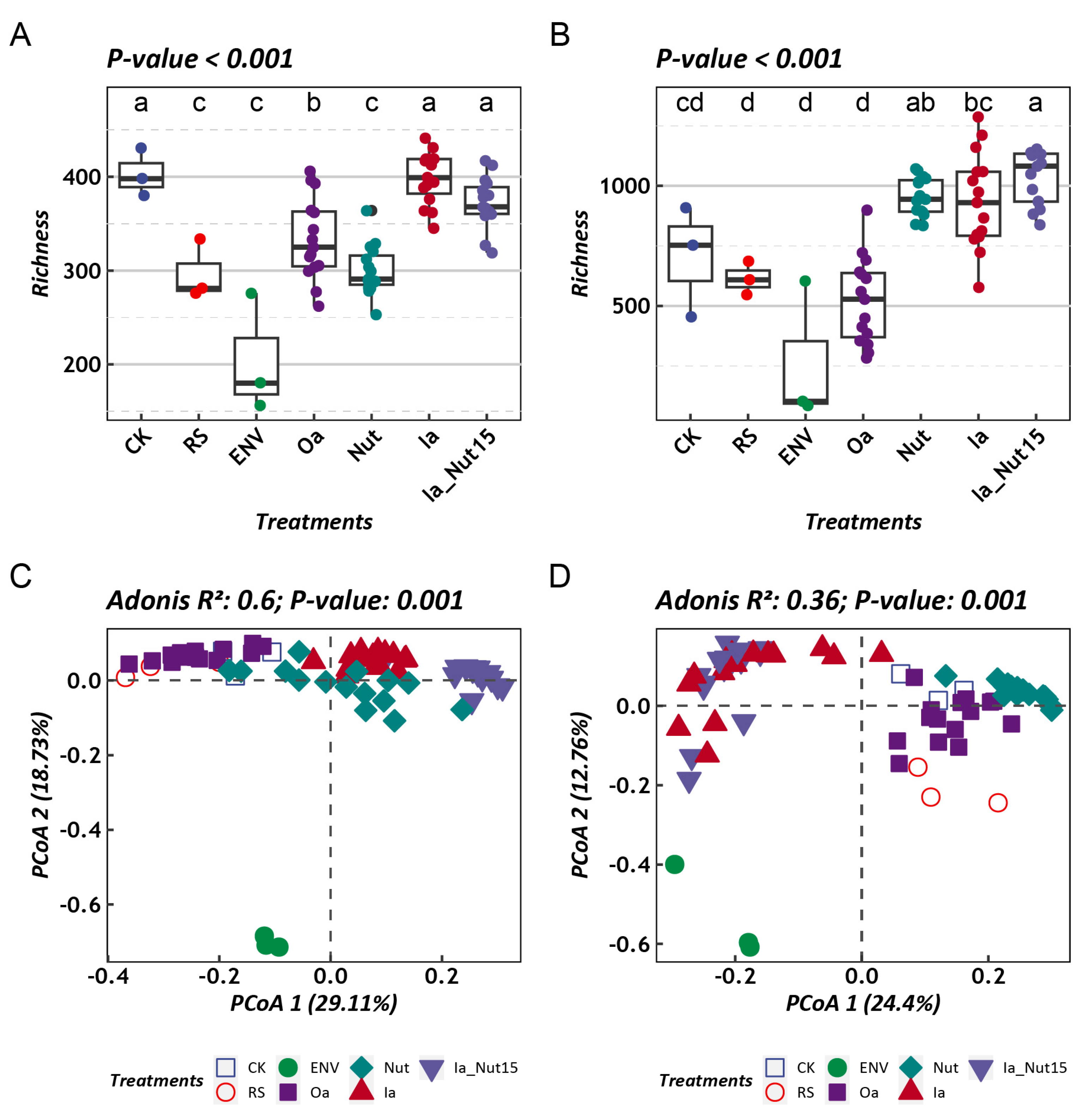
3.1. Microbial Community Composition and Diversity in Different Habitats
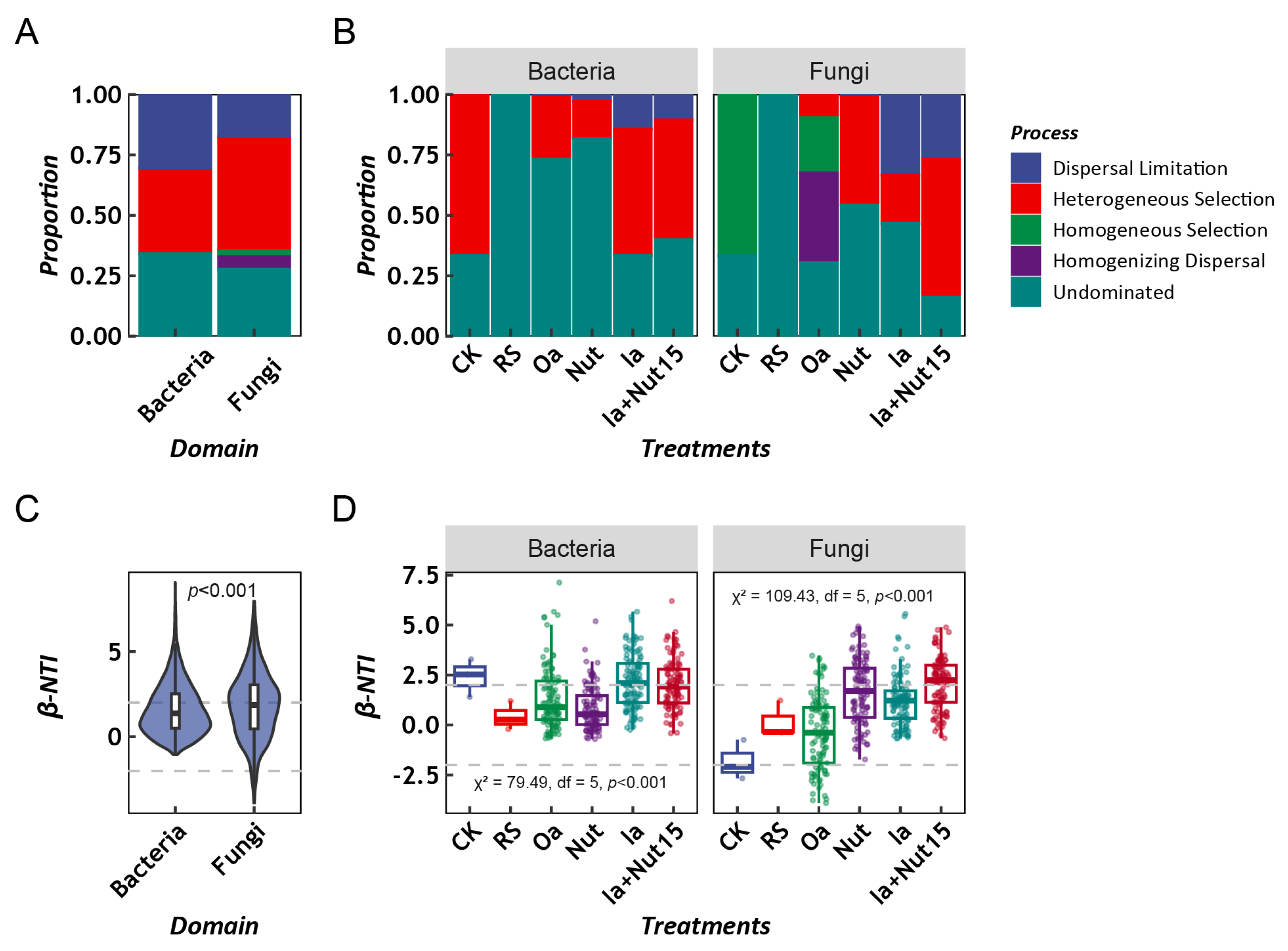
3.2. Assembly Process of LLMCs under Different Treatment Conditions
3.3. Factors Influencing the Assembly Process of LLMCs under Different Treatment Conditions
3.4. Source Tracking of Fungal and Bacterial Communities
3.5. Contribution of Different Treatments to the Sources of Bacterial and Fungal Community Composition
4. Discussion
4.1. Diversity of Microorganisms Inhabiting Limestone
4.2. The Surrounding Environment Can Account for a Certain Proportion of the Source of LLMCs
4.3. Inorganic Acids and Nutrient Treatments Promote Fungal Colonization of Limestone in the Surrounding Environment While Inhibiting Bacterial Colonization
4.4. The Community Construction Process of Limestone LMs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, J.; Ning, D. Stochastic community assembly: Does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 2017, 81, 1–32. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Konopka, A.E. Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 2015, 6, 370. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, W.; Deng, Y.; Jiang, Y.H.; Xue, K.; He, Z.; Van Nostrand, J.D.; Wu, L.; Yang, Y.; Wang, A. Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. mBio 2013, 4, e00584-12. [Google Scholar] [CrossRef] [PubMed]
- Golubic, S.; Friedmann, E.I.; Schneider, J. The lithobiontic ecological niche, with special reference to microorganisms. J. Sediment. Res. 1981, 51, 475–478. [Google Scholar] [CrossRef]
- Zhang, G.; Gong, C.; Gu, J.; Katayama, Y.; Someya, T.; Gu, J.-D. Biochemical reactions and mechanisms involved in the biodeterioration of stone world cultural heritage under the tropical climate conditions. Int. Biodeterior. Biodegrad. 2019, 143, 104723. [Google Scholar] [CrossRef]
- Sand, W.; Bock, E. Biodeterioration of mineral materials by microorganisms—Biogenic sulfuric and nitric acid corrosion of concrete and natural stone. Geomicrobiol. J. 1991, 9, 129–138. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, Q.; Li, F.; Zhao, X.; Wang, Y.; Zhang, L.; Liu, J.; Yan, L.; Yu, L. Nutrient availability and acid erosion determine the early colonization of limestone by lithobiontic microorganisms. Front. Microbiol. 2023, 14, 1194871. [Google Scholar] [CrossRef]
- Chen, J.; Li, F.; Zhao, X.; Wang, Y.; Zhang, L.; Yan, L.; Yu, L. Change in composition and potential functional genes of microbial communities on carbonatite rinds with different weathering times. Front. Microbiol. 2022, 13, 1024672. [Google Scholar] [CrossRef]
- Medici, G.; Lorenzi, V.; Sbarbati, C.; Manetta, M.; Petitta, M. Structural classification, discharge statistics, and recession analysis from the springs of the Gran Sasso (Italy) carbonate aquifer; comparison with selected analogues worldwide. Sustainability 2023, 15, 10125. [Google Scholar] [CrossRef]
- Goldscheider, N.; Chen, Z.; Auler, A.S.; Bakalowicz, M.; Broda, S.; Drew, D.; Hartmann, J.; Jiang, G.; Moosdorf, N.; Stevanovic, Z. Global distribution of carbonate rocks and karst water resources. Hydrogeol. J. 2020, 28, 1661–1677. [Google Scholar] [CrossRef]
- Pinheiro, A.C.; Mesquita, N.; Trovão, J.; Soares, F.; Tiago, I.; Coelho, C.; de Carvalho, H.P.; Gil, F.; Catarino, L.; Piñar, G.; et al. Limestone biodeterioration: A review on the Portuguese cultural heritage scenario. J. Cult. Herit. 2019, 36, 275–285. [Google Scholar] [CrossRef]
- Liu, X.; Koestler, R.J.; Warscheid, T.; Katayama, Y.; Gu, J.-D. Microbial deterioration and sustainable conservation of stone monuments and buildings. Nat. Sustain. 2020, 3, 991–1004. [Google Scholar] [CrossRef]
- Emmanuel, S.; Levenson, Y. Limestone weathering rates accelerated by micron-scale grain detachment. Geology 2014, 42, 751–754. [Google Scholar] [CrossRef]
- Noiriel, C.; Luquot, L.; Madé, B.; Raimbault, L.; Gouze, P.; van der Lee, J. Changes in reactive surface area during limestone dissolution: An experimental and modelling study. Chem. Geol. 2009, 265, 160–170. [Google Scholar] [CrossRef]
- Jones, R.J. Aspects of the biological weathering of Limestone pavement. Proc. Geol. Assoc. 1965, 76, 421-IN428. [Google Scholar] [CrossRef]
- Walker, J.J.; Pace, N.R. Endolithic microbial ecosystems. Annu. Rev. Microbiol. 2007, 61, 331–347. [Google Scholar] [CrossRef] [PubMed]
- Teske, A.P.; Edgcomb, V.P. Editorial: Insights in extreme microbiology: 2021. Front. Microbiol. 2022, 13, 1119051. [Google Scholar] [CrossRef]
- Miller, A.Z.; Sanmartín, P.; Pereira-Pardo, L.; Dionísio, A.; Saiz-Jimenez, C.; Macedo, M.F.; Prieto, B. Bioreceptivity of building stones: A review. Sci. Total Environ. 2012, 426, 1–12. [Google Scholar] [CrossRef]
- Cappitelli, F.; Principi, P.; Pedrazzani, R.; Toniolo, L.; Sorlini, C. Bacterial and fungal deterioration of the Milan Cathedral marble treated with protective synthetic resins. Sci. Total Environ. 2007, 385, 172–181. [Google Scholar] [CrossRef]
- Gambino, M.; Lepri, G.; Štovícek, A.; Ghazayarn, L.; Villa, F.; Gillor, O.; Cappitelli, F. The tombstones at the Monumental Cemetery of Milano select for a specialized microbial community. Int. Biodeterior. Biodegrad. 2021, 164, 105298. [Google Scholar] [CrossRef]
- Choe, Y.-H.; Kim, M.; Lee, Y.K. Distinct microbial communities in adjacent rock and soil substrates on a high arctic polar desert. Front. Microbiol. 2021, 11, 607396. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Stewart, P.S.; Klapper, I.; Jacob, J.M.; Cappitelli, F.J.B. Subaerial biofilms on outdoor stone monuments: Changing the perspective toward an ecological framework. Bioscience 2016, 66, 285–294. [Google Scholar] [CrossRef]
- Li, T.; Hu, Y.; Zhang, B.; Yang, X. Role of Fungi in the Formation of Patinas on Feilaifeng Limestone, China. Microb. Ecol. 2018, 76, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Wieler, N.; Ginat, H.; Gillor, O.; Angel, R. The origin and role of biological rock crusts in rocky desert weathering. Biogeosciences 2019, 16, 1133–1145. [Google Scholar] [CrossRef]
- Wang, S.J.; Liu, Q.M.; Zhang, D.F. Karst rocky desertification in southwestern China: Geomorphology, landuse, impact and rehabilitation. Land Degrad. Dev. 2004, 15, 115–121. [Google Scholar] [CrossRef]
- He, G.; Zhang, Z.; Zhang, J.; Huang, X. Soil organic carbon dynamics and driving factors in typical cultivated Land on the Karst Plateau. Int. J. Environ. Res. Public Health 2020, 17, 5697. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Hervé, M. RVAideMemoire: Testing and Plotting Procedures for Biostatistics. 2022, 1–149. Available online: https://CRAN.R-project.org/package=RVAideMemoire (accessed on 29 June 2023).
- Conover, W.J. Practical Nonparametric Statistics, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1999; Volume 350. [Google Scholar]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Shenhav, L.; Thompson, M.; Joseph, T.A.; Briscoe, L.; Furman, O.; Bogumil, D.; Mizrahi, I.; Pe’er, I.; Halperin, E. FEAST: Fast expectation-maximization for microbial source tracking. Nat. Methods 2019, 16, 627–632. [Google Scholar] [CrossRef]
- Kane, M.; Emerson, J.W.; Weston, S. Scalable Strategies for Computing with Massive Data. J. Stat. Softw. 2013, 55, 1–19. [Google Scholar] [CrossRef]
- Ning, D.; Yuan, M.; Wu, L.; Zhang, Y.; Guo, X.; Zhou, X.; Yang, Y.; Arkin, A.P.; Firestone, M.K.; Zhou, J. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 2020, 11, 4717. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, G.N.; Rogers, C.E. Symbolic description of factorial models for analysis of variance. J. R. Stat. Soc. Ser. C Appl. Stat. 1973, 22, 392–399. [Google Scholar] [CrossRef]
- Cuezva, S.; Sanchez-Moral, S.; Saiz-Jimenez, C.; Cañaveras, J. Microbial Communities and Associated Mineral Fabrics in Altamira Cave, Spain. Int. J. Speleol. 2009, 38, 9. [Google Scholar] [CrossRef]
- Abdel Ghany, T.M.; Omar, A.M.; Elwkeel, F.M.; Al Abboud, M.A.; Alawlaqi, M.M. Fungal deterioration of limestone false-door monument. Heliyon 2019, 5, e02673. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef]
- de Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef]
- Dini-Andreote, F.; Stegen, J.C.; van Elsas, J.D.; Salles, J.F. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. USA 2015, 112, E1326–E1332. [Google Scholar] [CrossRef]
- Martiny, A.C.; Jørgensen, T.M.; Albrechtsen, H.J.; Arvin, E.; Molin, S. Long-term succession of structure and diversity of a biofilm formed in a model drinking water distribution system. Appl. Environ. Microbiol. 2003, 69, 6899–6907. [Google Scholar] [CrossRef]
- Badri, D.V.; Chaparro, J.M.; Zhang, R.; Shen, Q.; Vivanco, J.M. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 2013, 288, 4502–4512. [Google Scholar] [CrossRef]
- Inceoğlu, Ö.; Al-Soud, W.A.; Salles, J.F.; Semenov, A.V.; van Elsas, J.D. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS ONE 2011, 6, e23321. [Google Scholar] [CrossRef] [PubMed]
- Chase, J.M. Drought mediates the importance of stochastic community assembly. Proc. Natl. Acad. Sci. USA 2007, 104, 17430–17434. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Li, S.; Avera, B.N.; Strahm, B.D.; Badgley, B.D. Soil bacterial and fungal communities show distinct recovery patterns during forest ecosystem restoration. Appl. Environ. Microbiol. 2017, 83, e00966-17. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.K.; Nemergut, D.R.; Darcy, J.L.; Lynch, R. Do bacterial and fungal communities assemble differently during primary succession? Mol. Ecol. 2014, 23, 254–258. [Google Scholar] [CrossRef]
- Ren, J.; Liu, X.; Yang, W.; Yang, X.; Li, W.; Xia, Q.; Li, J.; Gao, Z.; Yang, Z. Rhizosphere soil properties, microbial community, and enzyme activities: Short-term responses to partial substitution of chemical fertilizer with organic manure. J. Environ. Manag. 2021, 299, 113650. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Li, F.; Zhao, X.; Wang, Y.; Zhang, L.; Liu, F.; Yang, D.; Yan, L.; Yu, L. Assembly and Source of the Lithobiontic Microbial Community in Limestone. Fermentation 2023, 9, 672. https://doi.org/10.3390/fermentation9070672
Chen J, Li F, Zhao X, Wang Y, Zhang L, Liu F, Yang D, Yan L, Yu L. Assembly and Source of the Lithobiontic Microbial Community in Limestone. Fermentation. 2023; 9(7):672. https://doi.org/10.3390/fermentation9070672
Chicago/Turabian StyleChen, Jin, Fangbing Li, Xiangwei Zhao, Yang Wang, Limin Zhang, Feng Liu, Dan Yang, Lingbin Yan, and Lifei Yu. 2023. "Assembly and Source of the Lithobiontic Microbial Community in Limestone" Fermentation 9, no. 7: 672. https://doi.org/10.3390/fermentation9070672
APA StyleChen, J., Li, F., Zhao, X., Wang, Y., Zhang, L., Liu, F., Yang, D., Yan, L., & Yu, L. (2023). Assembly and Source of the Lithobiontic Microbial Community in Limestone. Fermentation, 9(7), 672. https://doi.org/10.3390/fermentation9070672





