Abstract
The microbial production of fuel ethanol is an attractive and sustainable biotechnological approach. This study presents a metabolic engineering strategy of Zymomonas mobilis aimed at coproducing bioethanol and fatty acids. The increased flux of fatty acids stabilizes the cell membrane and thus counteracts the progressively higher ethanol toxicity. In a glucose medium, the highest ethanol titer achieved was 146.7 g/kg of broth, surpassing the wild-type Z. mobilis CP4 and angel yeast by 30% and 45%, respectively. The recombinant strain exhibited a total fatty acid titer of 0.4 g/L from 230 g/L total sugar solution (5 L bioreactor), representing a 12-fold increase compared to the wild-type Z. mobilis CP4. Furthermore, when using a 4:2:1 mixture of glucose: xylose: mannose (w/v), an ethanol concentration of 142.8 g/kg of broth was attained, only 2.66% lower than that of the glucose-only medium. These findings highlight the enormous potential of this genetically engineered strain for the sustainable production of ethanol and fatty acids from lignocellulosic renewable carbon sources.
1. Introduction
Ethanol, the most prevalent commercial biofuel nowadays that accounts for 72% of global biofuel production [1], is more environmentally friendly than traditional fuels in the sense that upon burning, the former produces less carbon monoxide and hydrocarbons than the latter, and therefore does not contribute as much to the greenhouse effect [2]. Furthermore, ethanol has a higher octane number, a property that makes ethanol a reliable fuel additive that enhances the antiknock quality of fuel [3]. Traditional food crop raw materials (cereals, corn kernels, and molasses, etc.) usually lead to potential threats to food security, therefore limiting their application for bioethanol production [4]. Lignocellulosic biomass produced by solar energy through plant photosynthesis is the most prevalent biowaste on Earth with an output of 1012 tons/year and is expected to become a substitute for food crop raw materials [5]. Developing bioethanol pathways via lignocellulose can not only significantly alleviate energy and food security issues but also sustain the ecological environment.
Due to its antimicrobial properties, bioethanol is commonly produced in a diluted form, which leads to an insufficient fuel-grade concentration (>99.2 wt%) upon fermentation [6]. Therefore, to address this issue, the production of bioethanol requires a subsequent recovery and dehydration process [5]. Ethanol recycling is energy-intensive, meaning maximizing the ethanol concentration during fermentation is crucial for minimizing the energy demand in the overall production process [7]. To achieve high ethanol concentrations, the pretreatment process of lignocellulosic biomass is always carried out at high solids loadings: first, by destroying its rigid structure through physical or chemical processes and subsequently by maximizing its hydrolysis to fermentable sugars using highly efficient engineered enzymes [8,9,10]. Recent advancements in corn stover substrate treatment have enabled the extraction of 230 g/L monosaccharides via deacetylation, mechanical refining, and high solids enzymatic hydrolysis, without any concentration or purification steps [11]. Although various techniques, such as activated carbon adsorption and biodetoxification using microorganisms, have been developed to mitigate the inhibitory effects of acetic, furanic, and phenolic compounds derived from lignocellulosic pretreatment on the fermentation process [12], the intricate composition of its hydrolysate still presents a significant challenge when designing fermentation strains. This challenge necessitates the use of strains capable of enduring multiple stressors, including low nutrient conditions in the hydrolysate, anaerobic environments, high temperatures, acidic stress, ethanol toxicity, and osmotic pressure caused by high sugar-specific gravity [13].
Zymomonas mobilis, a naturally occurring anaerobic fermentative microorganism found in high-sugar liquids, has attracted a lot of attention in the science community in recent years for its exceptional ethanol tolerance. It not only exhibits tolerance to HG environmental stress, thanks to its self-flocculation mechanism, but also facilitates cell separation by low-cost sedimentation rather than centrifugation [14], making Z. mobilis an optimal candidate for HG fermentation. It utilizes the Entner–Doudoroff (ED, blue arrows in Figure 1) pathway and possesses high expression levels of pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH) genes, which contribute to its exceptional ethanol fermentation capability. Compared to the Embden–Meyerhof–Parnas (EMP, solid black arrows in Figure 1) pathway in Saccharomyces cerevisiae, the ED pathway reduces adenosine triphosphate (ATP) output by 50% [15] due to the uncoupling pathway that metabolizes the key intermediate (2-keto-3-deoxy-6 phosphogluconate, KDPG), directly to pyruvate without ATP output (Figure 1), thereby decoupling ethanol production from cell growth. Consequently, Z. mobilis accumulates less biomass and generates less heat during ethanol fermentation. In other words, Z. mobilis uses more sugar for ethanol production while reducing the investment associated with the cooling process. Additionally, its smaller size (1–2 × 2–6 μm) confers a higher specific surface area and sugar uptake efficiency compared to S. cerevisiae (2–10 × 4–20 μm) [16].
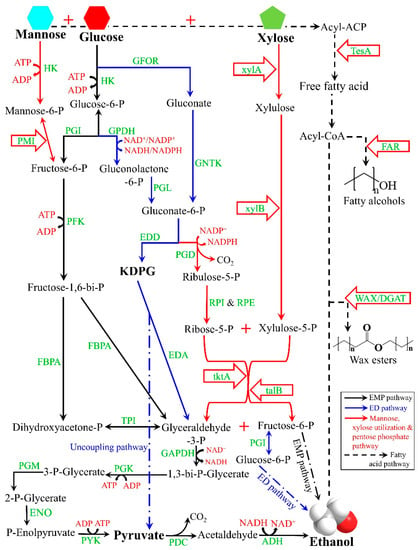
Figure 1.
Glycolytic pathways of Saccharomyces cerevisiae (EMP pathway) and Zymomonas mobilis (ED pathway) and metabolic engineering strategies of Z. mobilis to convert lignocellulose biomass under high-gravity stress. Abbreviations: EDA, 2-keto-3-deoxy-gluconate aldolase; HK, hexokinase; xylA, xylose isomerase; xylB, xylulokinase; GFOR, glucose-fructose oxidoreductase; PGI, phosphoglucoisomerase; GPDH, glucose-6-phosphate dehydrogenase; PGL, phos-phogluconolactonase; GNTK, gluconate kinase; EDD, 6-phosphogluconate dehydratase; PGD, 6-phosphogluconate dehydrogenase; RPI, ribulose-5-phosphate isomerase; RPE, ribulose-5-phosphate epimerase; PFK, phosphofructokinase; FBPA, fructose-1,6-bisphosphate aldolase; FAR, fatty acyl-CoA reductase; TPI, triose phosphate isomerase; tktA, transaldolase; talB, transketolase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; PGM, phosphoglyceromutase; ENO, enolase; PYK, pyruvate kinase; WAX/DGAT, wax ester synthase/acyl-CoA-diacylglycerol acyltransferase.
However, wild-type (WT) Z. mobilis can only metabolize glucose, fructose, and sucrose. Additionally, its ability to produce ethanol from sucrose is significantly impaired by fructan formation, rendering it unsuitable for ethanol production from raw materials such as sugarcane juice or molasses. Furthermore, although glucose is the main sugar of starch-based grains, which are used in the ethanol industry, there are also maltose and maltotriose in the hydrolyzate of starch-based grains. Therefore, starch-based grains cannot be used in Z. mobilis but can be used for S. cerevisiae fermentation [16]. Such concerns, therefore, seem to have deprived the eligibility of Z. mobilis as an intermediate for bioethanol production from food crops.
The lignocellulosic ethanol industry creates opportunities to explore the strengths of Z. mobilis. Lignocellulosic biomass mainly consists of cellulose, hemicellulose, and lignin [17]. Glucose is the only sugar released during cellulose hydrolysis, while the important components of hemicellulose hydrolyzate are xylose and mannose. Previous efforts to develop Z. mobilis or yeast chassis for lignocellulosic ethanol production have focused on addressing their narrow substrate spectrum. Specifically, three pathways for xylose utilization have been described, namely the Dahms pathway, the X-1-P pathway, and the Xylose–Xylulokinase isomerase pathway (red arrows in Figure 1) [18]. However, for the harsh environment of lignocellulosic hydrolysis products, the current work is still focused on upstream pretreatment processes, and little work has been reported on the development of Z. mobilis chassis to improve their resistance.
Acetic acid produced by the deacetylation of hemicellulose during lignocellulosic pretreatment [12] is the main inhibitor of the fermentation process. When the pH of the fermentation drops below 5.0, 36% of acetic acid becomes nonionized and nondissociated and is therefore able to penetrate the membrane into the cytoplasm, leading to uncoupling, anion accumulation, and cytoplasmic acidification [19]. In addition, the accumulation of ethanol on the cell membrane hinders ATP synthesis and decreases proton dynamics, thus interfering with the ability of the membrane to act as a barrier and disrupting important procedures such as transport and energy transfer [20]. To mitigate these effects, cells typically increase the flux of fatty acid pathways to stabilize the membrane structure, as lipids are key membrane components.
Jojoba (Simmondsia chinensis) is a woody shrub native to the arid regions of North America. It produces seeds that contain a liquid wax known as jojoba oil. Its main components are long-chain fatty alcohols synthesized by fatty acyl-CoA reductase (FAR, black dashed arrows in Figure 1), which are thought to play a key role in the survival and adaptation of jojoba trees in their natural environment, such as water retention, nutrient storage, and as a protective barrier [21]. Acinetobacter baylyi is a Gram-negative bacterium capable of the intracellular accumulation of high levels of wax esters [22]. Wax esters are neutral lipids formed by the linkage of wax ester synthase/acyl-CoA-diacylglycerol acyltransferase (WAX/DGAT) in A. baylyi esterifying an acyl-CoA (from a fatty acid) and a fatty alcohol, and play a key role in the formation and stabilization of A. baylyi’s membrane [23]. Researchers have expressed the FAR gene of jojoba and the WAX/DGAT gene of A. baylyi in yeast and other microorganisms to produce fatty alcohols and wax esters for various industrial uses while advancing the understanding of lipid biosynthetic pathways and enzymatic mechanisms involved in microbial lipid metabolism. Notably, the hybrid neutral lipids-containing yeast or E. coli cells were more resistant to heat, organic solvents, and freezing stresses than the WT strains [24]. Furthermore, systems biology and transcriptome analyses showed that dynamic changes in lipid composition likewise represent an evolutionary adaptation of Z. mobilis to survive under ethanol stress [25,26].
The main work of this study was to build on previous studies by integrating the FAR gene of Simmondsia chinensis and the WAX/DGAT gene of A. baylyi into the genomic DNA of Z. mobilis CP4 through Tn5 transposon-mediated translocation to address its tolerance to various stresses. The ethanol fermentability and fatty acid composition of the engineered Z. mobilis strains were then detected under high sugar osmosis, and the effect of coupling the ED pathway and the pentose phosphate pathway (also known as the hexose phosphate shunt) on ethanol production was evaluated.
2. Materials and Methods
2.1. Preparation of Strains
The strains utilized in this study are listed in Table 1. The only exception is Saccharomyces cerevisiae S288C (angel yeast) provided by China Oil and Foodstuffs Corporation (COFCO, Beijing, China). All other strains were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The target strains were inoculated into 10 mL of medium containing 100 g/L glucose, 10 g/L yeast extract, 2 g/L KH2PO4, and 100 mg/L ampicillin (pH 6.5). Mg2+, commonly used in fermentation, was not added to the inoculum medium because Mg2+ increases bacterial survival in the presence of antibiotics by modulating ribosomes [27], thereby increasing the likelihood of bacterial contamination.

Table 1.
The strains, genes, and primers used in this work. Restriction cleavage sites in primer sequences are indicated by underscores and protective bases are indicated by italics.
The culture was incubated anaerobically at 33 °C for approximately 12 h until the cells reached the late exponential or early stationary phase (as indicated by visible cell settling, measured the optical density with a spectrophotometer at λ = 600 nm). Then, centrifuged the above culture at 2000× g for 2 min at room temperature. Next, the cell precipitate was resuspended in 10 mL of sterile distilled water and washed 3 times. Once washed, the resulting cells were resuspended in 100 g of the medium used for fermentation studies. The fixed composition of the fermentation medium consisted of 1.0 g/L urea, 0.5 g/L MgSO4·7H2O, and 1.0 g/L KH2PO4. Then, added 80–370 g/L total sugar (glucose, xylose, and mannose) and 0–10 g/L yeast extract depending on the specific experimental conditions, adjusted the initial pH with KOH to 4.5–9.0, fermented at 100 rpm and 25–43 °C (water bath). Shake flasks equipped with corked S-shaped plastic valves were used, and the insides of the plastic valves were sealed with paraffin oil, which maintained an anaerobic environment and allowed CO2 produced during fermentation to escape through bubbling. The comparative scale-up was performed in a 5 L bioreactor (100 mL culture to 5 L medium containing 230 g/L glucose and 10 g/L yeast extract, pH 7.0) and cultured the mixture anaerobically at 100 rpm, 33 °C for 60 h.
2.2. Construction of Tn5 Transposon Vector
Plasmids and genomic DNA were isolated using HiSpeed Plasmid Midi Kit (Qiagen, Hilden, Germany) and Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA), respectively. PCR, restriction enzyme digestion, and cloning experiments were performed using standard molecular biology methods. The PvuI and PshAI restriction sites of the commercial EZ-Tn5 pMOD plasmid were replaced with SfiI sites, and this modification generated a pUC19-based plasmid featured by the presence of multiple cloning sites between 2 19 bp mosaic ends (MEs, specially identified by Tn5 transposase). A mutated tetracycline resistance gene without SalI site (a 1.4 kb clip from p34S-TC, GenBank: AF062082) was subsequently inserted into its BamHI site, termed pTN-TC (Scheme 1). Overexpression was achieved by fusing two powerful constitutive promoters of Z. mobilis CP4, which are the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter and the enolase (ENO) promoter [28]. The GAPDH promoter was amplified using the primer pair 5-CCGCTCGAGGTTCGATCAACAACCCGAATCCTA and 5-CGGGATCCCTAACTTATTAAGTAGCTATTATATTC. The ENO promoter was amplified using the primer pair 5-GAAGATCTCTCCAGTTATCTCAATACGTAAC and 5-GGAATTCAACCTTTCTTAAAAATCTTTTAGACG. The digested GAPDH DNA with BamHI and ENO DNA with BglII were mixed and incubated with T4 DNA ligase for 12 h at 18 °C to generate a fused Gap/Eno promoter.
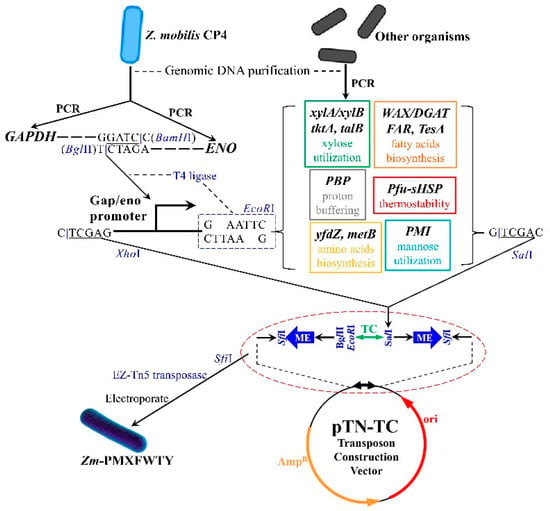
Scheme 1.
Multivariate modular construction strategy for recombinant Zm-PMXFWTY.
PCR amplification was performed using the templates and primers listed in Table 1 (the primers and some of the genes were synthesized by Tsingke Biotechnology Co., Ltd., Wuhan, China), and altered the codon by mutation to delete the SalI site in the xylA and tktA gene sequences, followed by digestion of a PCR product and the above Gap/Eno fusion promoter using EcoRI. The complementarity of the EcoRI restriction sites resulted in the joining of the two DNA fragments, forming an operon. Two different operons were digested with SalI and XhoI, respectively. Upon mixing the two DNA fragments, compatible ends of the XhoI and SalI restriction sites joined to create a “scar” site of four base pairs (5-TCGA). Since this scar site is a hybrid of the XhoI and SalI sites, neither enzyme recognizes it. The beginning and ending sequences remained stable throughout digestion and ligation, allowing for the use of more operons in subsequent assembly steps.
After digesting the fusion operon with SalI and XhoI, it was recovered from agarose gel and the DNA concentration was determined. Then, inserted the purified DNA sequence into the SalI site of pTN-TC to create a Tn5 transposon vector. Next, digested the Tn5 transposon vector with SfiI to obtain transposons, recovered them from agarose gel, and then mixed them with EZ-Tn5 transposase. The mixture was incubated at room temperature for 30 min to facilitate stable association of transposase to Tn5 transposon DNA. Finally, stored the mixture at −20 °C.
Z. mobilis CP4 receptor cells were obtained by centrifuging 400 mL of fresh CP4 cells (OD600 = 0.4) at 2000× g, washing the cells twice with an equal volume of ice-cold 10% glycerol, and then resuspending them with 2 mL of 10% glycerol. Mixed 100 µL aliquots of the receptor cells with 1 µL of the above DNA–Tn5 transposase mixture in 0.2 mm gap electroporation cuvettes. Electroporated the cells using a Bio-Rad Gene Pulser Xcell with parameters set to 6.0 ms and 1500 V. Immediately after electroporation, diluted the cells into 1 mL of SOC medium and plated them on agar plates containing 50 μg/mL tetracycline, 100 μg/mL ampicillin, and 25 g/L mannose.
2.3. Isolation and Identification of Recombinant Z. mobilis
After 2 days of incubation at 33 °C, only the electroporated cells containing the Tn5 transposon produced colonies on tetracycline-containing plates. A total of 100 colonies were randomly selected from 3 independent transformations to verify Tn5 transposon integration, and their DNA were isolated using QIAprep Spin Miniprep Kit. PCR identification revealed that 88 of the 100 strains contained fully integrated Tn5 transposons in their genomes. Next, cultured the 88 PCR-positive strains in rich medium (pH 7.0) with 230 g/L glucose and 10 g/L yeast extract at 33 °C for 60 h under anaerobic conditions to test the new operon’s role in ethanol production. The strain with the highest ethanol titer was named Zm-PMXFWTY and stored at −80 °C in rich medium containing 20% glycerol for further fermentation studies.
2.4. Assay of Ethanol, Biomass, and Soluble Residual
The ethanol concentration in fermentation liquid was measured by gas chromatography. Samples (1 mL) were obtained by filtration, and 1 μL per sample was injected into a DBFFAP 30 cm × 0.53 mm × 1.5 μm capillary column of type-6890 gas chromatography (GC, Agilent, Santa Clara, CA, USA) for quantification. The column was initially maintained at 80 °C and gradually heated to 180 °C at 10 °C/min. The detector and injector temperatures were 250 °C and 200 °C, respectively, with a 1 min retention time after injection. The carrier gas was N2 and was maintained at a flow rate of 5 mL/min. In particular, when observing the dynamic ethanol concentration changes during fermentation, real-time ethanol concentrations were calculated from the production of CO2 as described by Zhang et al. [1]. Specifically, in ED and EMP pathways, in which 1 molecule of pyruvate generates 1 molecule of ethanol and releases 1 molecule of CO2 simultaneously (Figure 1). The CO2 produced was vented through an S-shaped plastic valve via bubbling; thus, the production of CO2 was represented by the weight loss of the fermentation flask. For the initial 100 g of fermentation medium, the ethanol concentration is predicted as follows:
and the ethanol yield is computed as follows [29]:
EtOH predicted (g/kg) = Total CO2 (mol) × 46.07 (g/mol) × 1000 (g/kg)/[100 g−total CO2 (mol) × 44.01 (g/mol)
EtOH yield (%) = EtOH production (g)/[0.51× total sugar consumption (g)] × 100%
An amount of 50 μL cell broth fermented for 24 h was mixed with 950 μL sterile distilled water, and 10 μL 1% methylene blue staining solution and 30 μL distilled water were added to 10 μL of the mixture. Then, 10 μL of the above sample was taken on a 16 × 16 cell counting plate and counted by microscopy (×5000). The colony forming units (CFU) were calculated as follows:
CFU/mL = Number of cells observed × 256 × 5000 × dilution times
To evaluate the biomass (dry cell weight) accumulated during fermentation, the cell pellet obtained by centrifugation at 10,000 rpm was resuspended in sterile distilled water, then heated the suspension at 130 °C using a LICHEN DHS-10A electronic moisture analyzer until the mass no longer decreased. In addition, the supernatant was subjected using the same instrument and heating program to detect soluble residual. Particularly in the mixed sugar fermentation part of the validation experiment, the various residual monosaccharides were measured separately by liquid chromatography (Shimadzu, Japan), the separation was conducted on an Aminex HPX-87H column (Bio-Rad, Heracles, CA, USA) at 55 °C, the eluent was 5 mM H2SO4, and maintained a flow rate of 0.5 mL/min.
2.5. GC/MS Analysis of Fatty Acids
An amount of 5 L fermentation broth was removed by rapid centrifugation (2000 rpm, 10 min) to remove the supernatant. After washing the cell pellet 3 times with sterile water, the cell pellet was suspended in 3 times the mass of HCl solution (4 mol/L), and the mixture was heated to 100 °C and held for 3 min before being immediately cooled to isolate the intact cell membrane structure [30]. Overloaded vigorously with 1.5 times the volume of CHCl3/CH3OH (2:1, v/v) for 2 min. The lower (CHCl3) phase was separated and subsequently evaporated to dryness by means of a LICHEN DHS-10A electronic moisture analyzer. The resulting extract was redissolved in CHCl3/CH3OH (2:1, v/v) and subjected to GC/MS assay on type-6890 GC installed with a 5973 EI MSD mass selection checker (Hewlett-Packard) using a BP21 chromatographic column. Internal standards were incorporated to adjust for variations in derivatization efficiency and sample volume upon heating (Scheme 2). The carrier gas was He gas (maintained a flow of 0.6 mL/min), while injector and detector temperatures were 250 and 240 °C, respectively. Data were assessed through the NIST Mass Spectral Search procedure.
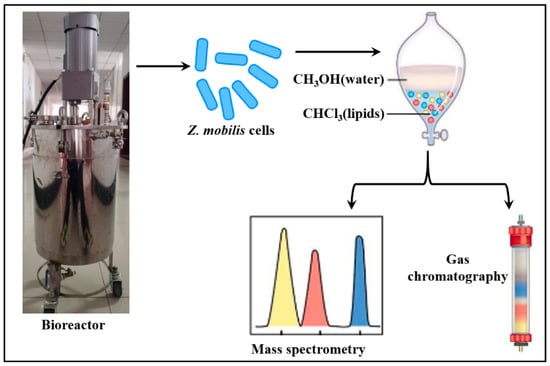
Scheme 2.
Procedures for the isolation, extraction, and identification of Z. mobilis lipids.
2.6. Response Surface Experimental Design for Optimizing Ethanol Production
Design Expert 10 software was utilized to design response surface experiments. The first part of the experiment aimed to determine the optimal conditions for ethanol production, and it was carried out by central composite design (CCD). The independent variables were glucose concentration (X1, g/L), temperature (X2, °C), and initial pH (X3), while the dependent variable (predicted response) was the amount of ethanol production (Y1, g/kg of broth). The CCD design involved a total of 17 experiments, with each variable having 5 levels. A quadratic polynomial was used to predict the response of the dependent variable Y1:
In Equation (4), a0 is an offset item, ai, aii, aji are linear items, square items, and interaction items, respectively. Based on the above experimental results, the optimal temperature and initial pH were identified for use in the xylose–hexose coupling fermentation experiment. Furthermore, the appropriate xylose ratio was determined by varying the total sugar concentration (X4, g/L) and xylose concentration (X5, g/L) as independent variables. To investigate the ethanol fermentability of the recombinant strain Zm-PMXFWTY under different ratios of mixed sugar medium, a CCD design consisting of 11 experiments was conducted.
2.7. Genetic Stability Testing and Verification Experiments
Stability testing of Zm-PMXFWTY was performed in shaker flasks containing 50 g medium (60 g/L glucose, 30 g/L xylose, 10 g/L mannose, 10 g/L yeast extract, 2 g/L KH2PO4, and 100 mg/L ampicillin, initial pH 7.0). Continuous transfers (5 mL to 50 g) were conducted every 24 h for 100 days. After 100 generations, the optimized conditions determined in Section 2.6 were employed for ethanol production fermentation to assess the stability of Zm-PMXFWTY and validate the accuracy of the model described in Section 2.6 in forecasting the max ethanol concentration. Verification experiments were performed in 3 replicates with the optimization conditions.
3. Results and Discussion
3.1. Less Residual Sugar and More Complete Fermentation under High-Gravity Condition
HG fermentation is a prevalent process for bioethanol production, usually carried out in the fermentation medium with a sugar content greater than 250 g/L [31]. Here, the fermentation performance of Zm-PMXFWTY, Z. mobilis CP4, and angel yeast (S. cerevisiae S288C) in a medium (pH 7.0) containing 250 g/L glucose and 10 g/L yeast extract was compared. The relationship between the predicted ethanol concentration (x) calculated from the shaker flask weight loss (CO2 release) and the ethanol (y) measured by gas chromatography satisfies the following regression equation (Figure 2B):
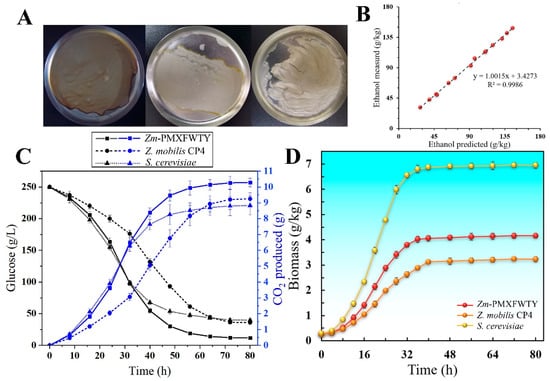
Figure 2.
Predicted ethanol concentration from CO2 weight loss vs. ethanol measured by GC (B), dynamic process of CO2 weight loss (C) and biomass accumulation (D), biomass is expressed in dry cell weight, the dry cell weights from left to right correspond to Zm-PMXFWTY, Z. mobilis CP4, and S. cerevisiae S288C, respectively (A).
The linearity coefficient and the regression coefficient are both close to 1, indicating that weight loss due to CO2 release can accurately represent dynamic ethanol production. Moreover, the longitudinal intercept of 3.4273 (g/kg of broth) is caused by the saturated CO2 dissolved in the shaker.
Figure 2C shows that Z. mobilis CP4 achieved higher ethanol concentrations. Although Z. mobilis CP4 and S. cerevisiae consumed similar glucose throughout the fermentation cycle, Z. mobilis CP4 accumulated 3.24 g/kg of biomass, which was lower than yeast’s 6.96 g/kg (Figure 2D), and which allowed Z. mobilis CP4 to use up to 98% of consumed sugars for ethanol production, up from 95.7% in S. cerevisiae. However, Z. mobilis CP4 had a slower ethanol production rate due to its low biomass density in the logarithmic growth period, leading to a prolonged fermentation time and higher potential for contamination [32]. Compared to the 60 h fermentation period of S. cerevisiae, Z. mobilis CP4 reached stable ethanol concentrations after 70 h. With the integration of exogenous genes, the Zm-PMXFWTY strain not only reduced the lag period of ethanol production but also had a significantly higher sugar consumption rate (95.4%) compared to the original Z. mobilis CP4 (85.6%) and S. cerevisiae (84.1%) (Figure 2C). This suggests that at high sugar concentrations, Zm-PMXFWTY is able to achieve more thorough fermentation in shorter cycles.
3.2. Utilization of Xylose and Mannose by Recombinant Z. mobilis-PMXFWTY
WT Z. mobilis is unable to utilize xylose and mannose, which are crucial ingredients in lignocellulosic saccharification products. By incorporating the phosphomannoisomerase (PMI) gene from Bacillus atrophaeus UCMB-5137 [33], Z. mobilis can convert mannose-6-phosphate to fructose-6-phosphate, which is able to enter the ED pathway (Figure 1). For xylose utilization, coupling with the pentose phosphate pathway is achieved by integrating xylose metabolism genes from E. coli K12 (xylA, xylB, tktA, and talB), resulting in the production of glyceraldehyde-3-phosphate and fructose-6-phosphate, both of which enter the glycolytic pathway of the ethanol metabolism [18]. The utilization rates of different sugars by Zm-PMXFWTY were evaluated at various total sugar concentrations (80 g/L, 150 g/L, and 230 g/L). The outcomes showed that Zm-PMXFWTY could convert mannose at a rate similar to that of glucose, with both above 94% (Figure 3A).
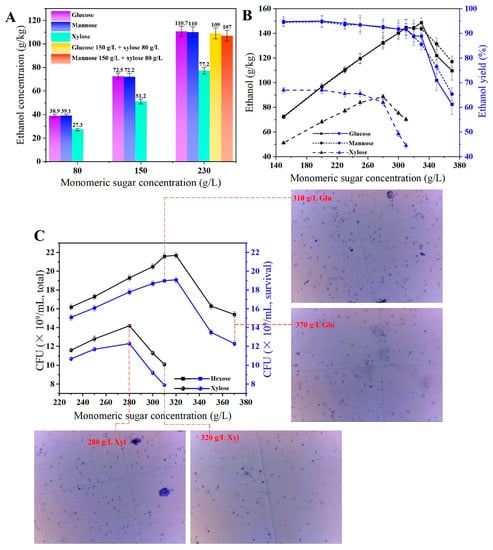
Figure 3.
Utilization of various monosaccharides by Zm-PMXFWTY (A,B) and CFU and survival rate under different sugar degrees (C). Colony-forming units were counted by methylene blue staining. Live cells with intact membrane structure excluded methylene blue and appeared transparent, and cells that lost activity showed blue color due to increased membrane permeability and nucleic acid binding to methylene blue [34], from which the survival rate of Zm-PMXFWTY under high sugar stress was calculated (C).
However, for xylose alone, the ethanol concentrations were 27.3, 51.2, and 77.2 g/kg, respectively, which were only equivalent to about 67% of the theoretical yield. Interestingly, in a mixed medium containing 230 g/L hexose–xylose (150 g/L glucose or mannose + 80 g/L xylose), the ethanol production was 109 g/kg and 107 g/kg, respectively, which was higher than the sum of 150 g/L hexose alone and 80 g/L xylose alone, and is close to the case of 230 g/L hexose. The addition of hexose increases the ethanol conversion of xylose to be obvious; this is shown in Figure 1, where the hexose phosphate shunt couples with the xylose metabolic pathway. To explain the reasons for low ethanol conversion due to xylose-alone fermentation, research by Li et al. [18] shows that may be due to the lower expression level of epimerase responsible for the interconversion of xylulose 5-phosphate and ribose 5-phosphate in Z. mobilis.
The ability of Zm-PMXFWTY to utilize glucose, mannose, and xylose, respectively, for ethanol production in a high-gravity environment was further tested. The findings indicated that the utilization rate of mannose by Zm-PMXFWTY was very close to that of glucose at almost all concentrations, and significantly higher than that of xylose (Figure 3B, Table S5). For all kinds of monosaccharides, the ethanol production of Zm-PMXFWTY generally increased with increasing the initial sugar concentration, but beyond a certain threshold, further increases in the sugar concentration resulted in a significant decrease in ethanol production. This is due to the fact that at high sugar concentrations, increased external osmotic pressure will lead to the dehydration of the cells and collapse of the plasma membrane ion gradient (Figure 3C) [25]. The threshold for glucose and mannose was approximately 320 g/L, while for xylose, it was approximately 280 g/L. This difference in threshold can be attributed to xylose’s smaller molecular weight (150.13) compared to glucose and mannose (180.16), resulting in a higher molar concentration at the same mass concentration and, therefore, a higher osmotic pressure.
3.3. Fatty Acid Overproduction Increases the Stress Tolerance under HG Conditions
The osmosis of the cell membrane of Z. mobilis by high sugar stress was evident (Figure 3C). Since lipids are key membrane components that counteract the inhibitory effects of heat and organic solvents, this study aimed to enhance fatty acid flux by introducing thioesterase (TesA) from A. baylyi to release fatty acyl groups from fatty acyl carrier proteins into free fatty acids by introducing FAR from Simmondsia chinensis and WAX/DGAT from A. baylyi to synthesize fatty alcohols and wax esters. Zm-PMXFWTY and Z. mobilis CP4 were both fermented in a medium containing 230 g/L total sugar and 10 g/L yeast extract (pH 6.5) at 33 °C for 60 h. After extracting and methylating the total fatty acids, the fatty acid composition of both strains was assayed by GC/MS (Table 2).

Table 2.
Comparison of fatty acid production and composition between Zm-PMXFWTY and Z. mobilis CP4 and their effects on ethanol production.
WT Z. mobilis CP4 mainly produced Cis-9-octadecenoic acids (C18:1, 63.3%), along with hexadecanoic acids (C16:0, 18.7%) and myristic acids (C14:0, 13.7%). By overexpressing the TesA/FAR/WS/DGAT operon, Zm-PMXFWTY mainly produced C18:1 acids (47.8%) and C16:0 acids (47.7%), with other components including C22:4 acids (1.7%), C14:0 acids (1.8%), and C12:0 acids (1.0%). Notably, the UFAs level and SFAs level of Zm-PMXFWTY were 10.5 times and 18.3 times higher than those of Z. mobilis CP4, respectively.
Fatty acid production and composition have been proposed as potential indicators for assessing microorganisms’ tolerance to ethanol [29]. These acids are vital resources for a diverse range of bioproducts, such as surfactants, lubricants, and solvents, and their importance has increased in recent years [30]. This study demonstrated that overexpressing the TesA/FAR/WS/DGAT operon boosted fatty acid production in Zm-PMXFWTY by 12-fold compared to Z. mobilis CP4, reaching 0.4 g/L as a percentage of bacterial dry biomass 11.1 wt%. Furthermore, integration of the fatty acid metabolism pathway also increased the ethanol production of Zm-PMXFWTY by 11 g/kg. The ethanol to fatty acid ratio dropped from 3316:1 in Z. mobilis CP4 to 275:1 in Zm-PMXFWTY (Table 2). Since ethanol is the world’s most produced biofuel, the coupled synthesis of fatty acids with ethanol may enable the production of vast amounts of fatty acids at low cost. The HG fermentation performance of Zm-PMXFWTY and Z. mobilis CP4 was further tested, with S. cerevisiae serving as the control. Specifically, they were subjected to tolerance experiments involving a high sugar concentration, acetate, and initial ethanol (Figure 4).
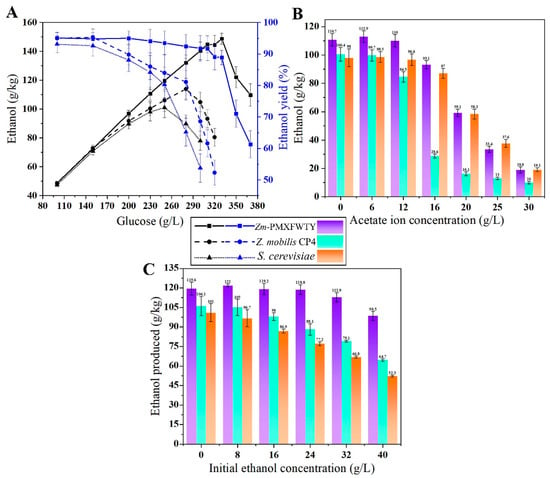
Figure 4.
Comparison of high-gravity fermentation performance (A), acetate tolerance (B), and initial ethanol tolerance (C) of the three strains.
These experiments were conducted in a medium containing 10 g/L yeast extract (pH 6.5) at 33 °C for 72 h. When the glucose concentration was low, the ethanol yield of the three strains did not differ distinctly. However, when the glucose concentration surpassed 200 g/L, they showed a loss of viability. When the glucose concentration exceeded 250 g/L and 280 g/L, respectively, the ethanol production of S. cerevisiae and CP4 declined significantly (Figure 4A), and their fermentative capacity was severely damaged under very high osmotic pressure conditions. On the other hand, Zm-PMXFWTY generated up to 144.9 g/kg of ethanol in a medium containing 310 g/L glucose with a theoretical yield of 91.7%, which was clearly superior to Z. mobilis CP4 and S. cerevisiae. Even when the medium contained 16 g/L acetate ion (230 g/L glucose), Zm-PMXFWTY maintained about 84% of its original ethanol productivity, with ethanol production 7% higher than that of S. cerevisiae (Figure 4B). In contrast, Z. mobilis CP4 could hardly produce ethanol under the same conditions.
The tolerance of the 3 strains to the initial ethanol was tested in a 250 g/L glucose medium, and the results showed that Zm-PMXFWTY had an 18% decrease in ethanol productivity under 40 g/L initial ethanol (Figure 4C). In contrast, Z. mobilis CP4 and S. cerevisiae had reductions of 39% and 48%, respectively. Typically, even after a downstream purification process, a small amount of ethanol remained in the previous batch of fermentation broth. This poses a challenge since additional ethanol recovery systems require high costs due to equipment investment and energy supply [35]. While this study demonstrated that Zm-PMXFWTY maintained a stable fermentation performance in a medium with 0–30 g/L initial ethanol (Figure 4C). Therefore, the previous batch of fermentation broth can be directed to the next fermentation process without the need for an additional ethanol recovery system.
3.4. Reduce Nutrient Requirements for the Fermentation Process
Compared to traditional starchy feedstocks, cellulosic ethanol production is often carried out under high solids loading and poor nutritional conditions. Yeast extracts consisting of amino acids, vitamins, and carbohydrates are essential to overcome the inhibitory effect of lignocellulosic hydrolysates in fermentation, but they are also the most expensive component of the medium. Previous studies have shown that in addition to the lysine pathway gene (yfdZ) and the methionine pathway gene (metB), WT Z. mobilis CP4 possesses a relatively complete amino acid synthesis gene [36], and the integration of the yfdZ and metB genes of E. coli K12 into the genome of Z. mobilis could reduce its nutritional requirements during fermentation. Additionally, since acetate inhibits methionine synthesis, overexpression of the metB operon may also play a role in the tolerance of Zm-PMXFWTY to acetate [37]. To investigate the effect of the yeast extract content on ethanol synthesis, fermentation was performed at 33 °C for 72 h in a medium containing 250 g/L glucose (initial pH 7.0). Figure 5A shows that in the high-nutrient medium, the differences in ethanol production of the three strains were small. However, ethanol synthesis by Zm-PMXFWTY was more efficient when the yeast extract content was below 4 g/L, while Z. mobilis CP4 and S. cerevisiae were significantly inhibited. Notably, in chemically defined media without amino acids and vitamins (0 g/L yeast extract), Zm-PMXFWTY had a final ethanol titer of 112.1 g/kg, which was 93.7% of the case of the 10 g/L yeast extract and 87.7% of the theoretical yield, significantly higher than that of the original strain CP4 and S. cerevisiae.
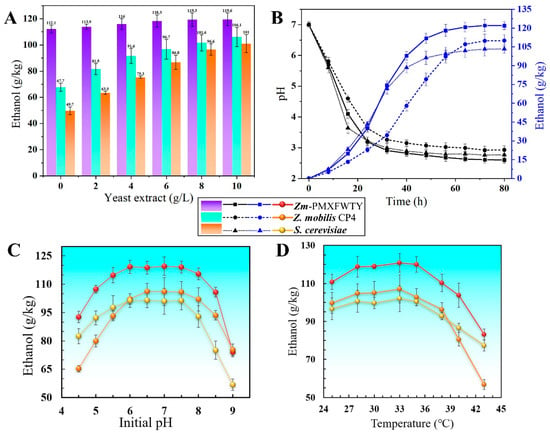
Figure 5.
Comparison of the tolerance of three strains to low nutrient stress (A), acid stress (B,C), and high temperature stress (D).
3.5. Single-Factor Experiments on the Effects of Ethanol Yield
In the bioethanol industry, a high acid environment at the end of the fermentation batch is prevalent, as microorganisms acidify the growth medium throughout fermentation. Protonated substances will inhibit bacterial growth and reduce ethanol productivity. A 24-amino acid proton buffer peptide (PBP, MGVKPTDDLKTIKTAYRRLARKYH) from the E. coli K12 cbpA gene (an acid-inducing gene) was selected to improve acid-stress resistance in Z. mobilis. This peptide can theoretically occupy more than 9 protons when the pH inside the cell is lower than the minimum pKa (R-group) of 6.0, because 33.4% of amino acids are capable of occupying 1 or more protons [38]. Fermentation was then conducted for 72 h at 33 °C in a medium containing 250 g/L glucose and 10 g/L yeast extract (initial pH 4.5–9.0) to compare the acid tolerance of the 3 strains. The findings revealed that in acidic environments, Zm-PMXFWTY had smaller reductions in fermentation performance compared to Z. mobilis CP4 (Figure 5C). The dynamic pH–ethanol production changes showed that Zm-PMXFWTY survived at lower pH levels and produced more ethanol than the original strain CP4 and S. cerevisiae in the late stages of fermentation (Figure 5B). These results suggest that the proton buffer peptide helps protect Z. mobilis from low pH and maintain its ethanol-producing activity. Furthermore, the growth environment with a lower pH level also contributes to lower sterility demands of fermentation, leading to enhanced cost-effectiveness in the fermentation process [19].
During fermentation, microorganisms produce heat as a result of metabolic activity. Failure to provide adequate cooling can lead to overheating, resulting in growth and fermentation problems. Temperature is a commonly studied physical factor affecting ethanol fermentation. The temperature applicable to Z. mobilis CP4 was relatively low, as indicated in Figure 5D, where the ethanol content was highest at 33 °C (250 g/L glucose, 10 g/L yeast extract, initial pH 7.0), reaching 107.1 g/kg, and decreased with rising temperatures. At 38 °C, CP4 ethanol concentration fell by approximately 10% compared to 33 °C, and the ethanol production capacity significantly declined at temperatures above 38 °C. The construction of heat-stable strains is of immense worth to the bioethanol process due to the reduced cost of cooling. The small heat-shock protein of P. furiosus (Pfu-sHSP) is a molecular chaperone, a homolog of α-crystallin that prevents protein aggregation caused by thermal stress or chemotreatment, leading to maintaining the vitality of Z. mobilis [19]. When the temperature increased from 33 °C to 40 °C, Zm-PMXFWTY and Z. mobilis CP4 ethanol production decreased by 14% and 25%, respectively. That is, Zm-PMXFWTY containing the Pfu-sHSP gene displayed better heat resistance than Z. mobilis CP4.
3.6. Response Surface Experiments for Optimal Fermentation Conditions and Hexose–Xylose Coupling
Based on prior experimental results, it was determined that medium sugar concentration, temperature (X2, °C), and initial pH (X3) are critical process parameters affecting ethanol production (Y1, g/kg). Since Zm-PMXFWTY demonstrates similar fermentability to both glucose and mannose, hexose concentration (X1, g/L) was used to express it. The central composite design (CCD) experimental was applied to assess the impact of the independent versus the dependent variable while minimizing the number of experimental runs. The design included 17 experiments across 5 levels with 3 replicates at the center point. The center point treatments (5, 14, and 17) had the highest ethanol concentrations (141.6, 142.1, and 141.3 g/kg, respectively, Table S1), indicating that Zm-PMXFWTY exhibited higher fermentative activity at higher sugar concentrations and neutral pH. Response surface methodology (RSM) produced the following regression equation (expressed as actual values):
In Figure 6A–C, the response surface plots are displayed for the optimization of ethanol fermentation conditions. At 312 g/L hexose, 33.5 °C, and initial pH 6.75, a maximum concentration of 143.8 g/kg of ethanol was observed. The effect of the xylose ratio on the ethanol yield was then studied at the determined optimal temperature and initial pH. Figure 6D shows the operating conditions and experimental results for a total of 11 experiments designed using CCD, with the total sugar content (X4, g/L) and xylose content (X5, g/L) as independent variables. It can be seen that ethanol production increases with the increasing hexose concentration and decreases with the increasing xylose concentration (Figure 6D, Table S3). RSM analysis led to the derivation of the following regression equation:
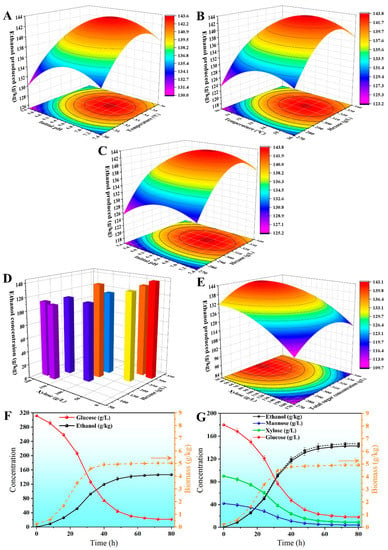
Figure 6.
Response surface diagram of optimal fermentation conditions (A–C) and hexose–xylose coupling relationship (E). The results of hexose–xylose-coupled CCD experiments (D) and validation experiments (F,G). The black dashed line in (G) is the ethanol curve for glucose-alone fermentation (same as in (F)) for comparison only.
The response surface plots of the hexose–xylose-coupled fermentation experiments clearly showed that ethanol production increased with the increasing sugar concentration at lower total sugar concentrations, and this trend was reversed with further increases in the sugar concentration. The model also predicted that at any ratio level, the ethanol production from the xylose–hexose mixed fermentation was lower than that of the single hexose fermentation at the same concentration (Figure 6E). This is consistent with the conclusions from Section 3.2 that the xylose utilization pathway requires the coupling of a hexose phosphate shunt to achieve higher ethanol production, and the higher osmotic pressure of xylose in relation to hexose also influences the activity of Zm-PMXFWTY. Fortunately, the suboptimal contour (139.8 g/kg) was observed at the coordinates of about 310 g/L total sugar and 90 g/L xylose (Figure 6E), reaching 97.7% of the optimal value.
The analysis of variance (ANOVA) for the fitted equations (Tables S2 and S4) calculated F-values which exceeded the analogous values in the statistics tables (<0.0001), indicating the model’s significance. The model’s accuracy and generalizability were confirmed with coefficients of determination at 0.9967 and 0.9987, respectively, rendering the analysis of relevant response trends reasonable. Additionally, the lack-of-fit tests were not significant, demonstrating the model’s accurate representation of the data in the experimental area.
3.7. Genetic Stability Testing and Verification Experiments
Researchers commonly employ shuttle plasmids for bacterial transformation, but such plasmids tend to be unstable in Z. mobilis when grown without antibiotic selection pressure [39]. This instability can worsen in mixed cultures because Z. mobilis must compete with other microorganisms. Additionally, the use of antibiotics to maintain plasmids is not ideal for industrial applications due to the risk of antibiotic resistance. To address these issues, this study utilized a Tn5-based random mutagenesis system to integrate the cloned genes into the genome of Z. mobilis, achieving comparable stability to genomic DNA. After 100 passages, PCR analysis confirmed that the genetic makeup of Zm-PMXFWTY remained unchanged from its original isolation. Fermentation experiments were conducted on glucose (312 g/L) and mixed sugar (180 g/L glucose, 90 g/L xylose, 42 g/L mannose) using the 100th generation recombinant strain, under optimal conditions (33.5 °C, initial pH 6.75) outlined in Section 3.7. The ethanol concentration of Zm-PMXFWTY in the glucose medium was 146.7 g/kg (Figure 6F) with a positive deviation of 2.01%, demonstrating the stability of the bacterial strain and validating the model’s accuracy. The ethanol yield was 92.2%, very close to the theoretical level. In the mixed sugar medium, the ethanol concentration of Zm-PMXFWTY was 142.8 g/kg with a positive deviation of 2.07%, only 2.66% lower than in the glucose alone (Figure 6G). Based on the preliminary literature research, this is the first report of reaching more than 14 wt% ethanol in broth containing 3 monosaccharides. For most lignocellulose hydrolysates, xylose accounted for about 30% of the total sugars. Therefore, this result highlights the potential of Zm-PMXFWTY to produce ethanol from lignocellulosic carbon sources.
4. Conclusions
This study successfully integrated a series of functional genes into the genome of Z. mobilis CP4. Because of the coupling of the ED pathway, hexose phosphate shunt pathway, and fatty acid pathway, the recombinant strain Zm-PMXFWTY was able to use glucose, mannose, and xylose simultaneously for ethanol and fatty acid production under low nutrient conditions. When xylose accounted for less than 30% of the total sugar, Zm-PMXFWTY achieved ethanol concentrations of more than 14 wt%. Additionally, it displayed a strong tolerance to heat and acidic environments. These findings create a new bridge for the future microbial conversion of lignocellulosic biomass to ethanol.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9060569/s1, Table S1: CCD experiment to find the optimum fermentation conditions; Table S2: analysis of variance (ANOVA) of the CCD experiment on optimum fermentation conditions; Table S3: CCD experiment of hexose–xylose-coupled fermentation at 33.5 °C and initial pH 6.75; Table S4: analysis of variance (ANOVA) of the CCD experiment on hexose–xylose-coupled fermentation; Table S5: ethanol fermentation results of Z. mobilis CP4, Zm-PMXFWTY and angel yeast.
Author Contributions
Conceptualization, H.W.; methodology, J.H. and H.W.; software, W.W.; validation, J.H., F.Z. and X.J.; formal analysis, F.Z.; investigation, X.J.; resources, L.P.; data curation, Y.F.; writing—original draft preparation, J.H.; writing—review and editing, W.W.; visualization, W.W.; supervision, H.W.; project administration, H.W.; funding acquisition, H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the State Key Laboratory of Biochemical Engineering of the Chinese Academy of Sciences (KF-011) and Nanjing 321 Leading Science and Technology Entrepreneurial Talents (1351100).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are included in the Supplementary Materials.
Acknowledgments
The authors thank Kai Wang (Hubei University of Technology) for participating in the discussion of the experimental part of the response surface design.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, K.; Wells, P.; Liang, Y.; Love, J.; Parker, D.A.; Botella, C. Effect of diluted hydrolysate as yeast propagation medium on ethanol production. Bioresour. Technol. 2019, 271, 1–8. [Google Scholar] [CrossRef]
- Ghalavand, Y.; Nikkhah, H.; Nikkhah, A. Heat pump assisted divided wall column for ethanol azeotropic purification. J. Taiwan Inst. Chem. E 2021, 123, 206–218. [Google Scholar] [CrossRef]
- Avilés Martínez, A.; Saucedo-Luna, J.; Segovia-Hernandez, J.G.; Hernandez, S.; Gomez-Castro, F.I.; Castro-Montoya, A.J. Dehydration of Bioethanol by Hybrid Process Liquid–Liquid Extraction/Extractive Distillation. Ind. Eng. Chem. Res. 2011, 51, 5847–5855. [Google Scholar] [CrossRef]
- Khalid, A.; Aslam, M.; Qyyum, M.A.; Faisal, A.; Khan, A.L.; Ahmed, F.; Lee, M.; Kim, J.; Jang, N.; Chang, I.S.; et al. Membrane separation processes for dehydration of bioethanol from fermentation broths: Recent developments, challenges, and prospects. Renew. Sustain. Energy Rev. 2019, 105, 427–443. [Google Scholar] [CrossRef]
- Saini, S.; Chandel, A.K.; Sharma, K.K. Past practices and current trends in the recovery and purification of first generation ethanol: A learning curve for lignocellulosic ethanol. J. Clean. Prod. 2020, 268, 122357. [Google Scholar] [CrossRef]
- Lee, S.C.; Woo, H.C.; Kim, Y.H. Energy-efficient ethanol recovery process using 2-methyl pentanol extraction. Fuel 2022, 310, 122393. [Google Scholar] [CrossRef]
- Grisales Díaz, V.H.; Willis, M.J. Ethanol production using Zymomonas mobilis: Development of a kinetic model describing glucose and xylose co-fermentation. Biomass Bioenergy 2019, 123, 41–50. [Google Scholar] [CrossRef]
- Phwan, C.K.; Chew, K.W.; Sebayang, A.H.; Ong, H.C.; Ling, T.C.; Malek, M.A.; Ho, Y.C.; Show, P.L. Effects of acids pre-treatment on the microbial fermentation process for bioethanol production from microalgae. Biotechnol. Biofuels 2019, 12, 191. [Google Scholar] [CrossRef]
- Yu, K.L.; Chen, W.-H.; Sheen, H.-K.; Chang, J.-S.; Lin, C.-S.; Ong, H.C.; Show, P.L.; Ling, T.C. Bioethanol production from acid pretreated microalgal hydrolysate using microwave-assisted heating wet torrefaction. Fuel 2020, 279, 118435. [Google Scholar] [CrossRef]
- Sun, C.; Song, G.; Pan, Z.; Tu, M.; Kharaziha, M.; Zhang, X.; Show, P.L.; Sun, F. Advances in organosolv modified components occurring during the organosolv pretreatment of lignocellulosic biomass. Bioresour. Technol. 2023, 368, 128356. [Google Scholar] [CrossRef]
- Chen, X.; Kuhn, E.; Jennings, E.W.; Nelson, R.; Tao, L.; Zhang, M.; Tucker, M.P. DMR (deacetylation and mechanical refining) processing of corn stover achieves high monomeric sugar concentrations (230 g·L−1) during enzymatic hydrolysis and high ethanol concentrations (>10% v/v) during fermentation without hydrolysate purification or concentration. Energy Environ. Sci. 2016, 9, 1237–1245. [Google Scholar]
- Koppram, R.; Tomas-Pejo, E.; Xiros, C.; Olsson, L. Lignocellulosic ethanol production at high-gravity: Challenges and perspectives. Trends Biotechnol. 2014, 32, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Van Aalst, A.C.A.; de Valk, S.C.; van Gulik, W.M.; Jansen, M.L.A.; Pronk, J.T.; Mans, R. Pathway engineering strategies for improved product yield in yeast-based industrial ethanol production. Synth. Syst. Biotechnol. 2022, 7, 554–566. [Google Scholar] [CrossRef]
- Zhao, N.; Bai, Y.; Liu, C.G.; Zhao, X.Q.; Xu, J.F.; Bai, F.W. Flocculating Zymomonas mobilis is a promising host to be engineered for fuel ethanol production from lignocellulosic biomass. Biotechnol. J. 2014, 9, 362–371. [Google Scholar] [CrossRef]
- Li, R.; Jin, M.; Du, J.; Li, M.; Chen, S.; Yang, S. The Magnesium Concentration in Yeast Extracts Is a Major Determinant Affecting Ethanol Fermentation Performance of Zymomonas mobilis. Front. Bioeng. Biotechnol. 2020, 8, 957. [Google Scholar] [CrossRef]
- Xia, J.; Yang, Y.; Liu, C.G.; Yang, S.; Bai, F.W. Engineering Zymomonas mobilis for Robust Cellulosic Ethanol Production. Trends Biotechnol. 2019, 37, 960–972. [Google Scholar] [CrossRef]
- Olivieri, G.; Wijffels, R.H.; Marzocchella, A.; Russo, M.E. Bioreactor and Bioprocess Design Issues in Enzymatic Hydrolysis of Lignocellulosic Biomass. Catalysts 2021, 11, 680. [Google Scholar] [CrossRef]
- Li, X.; Chen, Y.; Nielsen, J. Harnessing xylose pathways for biofuels production. Curr. Opin. Biotechnol. 2019, 57, 56–65. [Google Scholar] [CrossRef]
- Samir Ali, S.; Al-Tohamy, R.; Khalil, M.A.; Ho, S.H.; Fu, Y.; Sun, J. Exploring the potential of a newly constructed manganese peroxidase-producing yeast consortium for tolerating lignin degradation inhibitors while simultaneously decolorizing and detoxifying textile azo dye wastewater. Bioresour. Technol. 2022, 351, 126861. [Google Scholar] [CrossRef] [PubMed]
- Lam, F.H.; Ghaderi, A.; Fink, G.R.; Stephanopoulos, G. Biofuels. Engineering alcohol tolerance in yeast. Science 2014, 346, 71–75. [Google Scholar] [CrossRef]
- Sturtevant, D.; Lu, S.; Zhou, Z.W.; Shen, Y.; Wang, S.; Song, J.M.; Zhong, J.; Burks, D.J.; Yang, Z.Q.; Yang, Q.Y.; et al. The genome of jojoba (Simmondsia chinensis): A taxonomically isolated species that directs wax ester accumulation in its seeds. Sci. Adv. 2020, 6, eaay3240. [Google Scholar] [CrossRef] [PubMed]
- Kalscheuer, R.; Stoveken, T.; Malkus, U.; Reichelt, R.; Golyshin, P.N.; Sabirova, J.S.; Ferrer, M.; Timmis, K.N.; Steinbuchel, A. Analysis of storage lipid accumulation in Alcanivorax borkumensis: Evidence for alternative triacylglycerol biosynthesis routes in bacteria. J. Bacteriol. 2007, 189, 918–928. [Google Scholar] [CrossRef]
- Rottig, A.; Wenning, L.; Broker, D.; Steinbuchel, A. Fatty acid alkyl esters: Perspectives for production of alternative biofuels. Appl. Microbiol. Biotechnol. 2010, 85, 1713–1733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, T.; Hong, Z.; Ma, C.; Fang, X.; Zheng, F.; Teng, W.; Zhang, C.; Si, T. Biosynthesis of Hybrid Neutral Lipids with Archaeal and Eukaryotic Characteristics in Engineered Saccharomyces cerevisiae. Angew. Chem. Int. Ed. 2023, 62, e202214344. [Google Scholar]
- He, M.X.; Wu, B.; Shui, Z.X.; Hu, Q.C.; Wang, W.G.; Tan, F.R.; Tang, X.Y.; Zhu, Q.L.; Pan, K.; Li, Q.; et al. Transcriptome profiling of Zymomonas mobilis under ethanol stress. Biotechnol. Biofuels 2012, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Pan, C.; Tschaplinski, T.J.; Hurst, G.B.; Engle, N.L.; Zhou, W.; Dam, P.; Xu, Y.; Rodriguez, M., Jr.; Dice, L.; et al. Systems biology analysis of Zymomonas mobilis ZM4 ethanol stress responses. PLoS ONE 2013, 8, e68886. [Google Scholar]
- Lee, D.D.; Galera-Laporta, L.; Bialecka-Fornal, M.; Moon, E.C.; Shen, Z.; Briggs, S.P.; Garcia-Ojalvo, J.; Suel, G.M. Magnesium Flux Modulates Ribosomes to Increase Bacterial Survival. Cell 2019, 177, 352–360.e13. [Google Scholar] [CrossRef]
- Jia, X.; Wei, N.; Wang, T.; Wang, H. Use of an EZ-Tn5-based random mutagenesis system to create a Zymomonas mobilis with significant tolerance to heat stress and malnutrition. J. Ind. Microbiol. Biotechnol. 2013, 40, 811–822. [Google Scholar] [CrossRef]
- Qin, L.; Dong, S.; Yu, J.; Ning, X.; Xu, K.; Zhang, S.J.; Xu, L.; Li, B.Z.; Li, J.; Yuan, Y.J.; et al. Stress-driven dynamic regulation of multiple tolerance genes improves robustness and productive capacity of Saccharomyces cerevisiae in industrial lignocellulose fermentation. Metab. Eng. 2020, 61, 160–170. [Google Scholar] [CrossRef]
- Wei, S.; He, Y.; Yang, J.; Li, Y.; Liu, Z.; Wang, W. Effects of exogenous ascorbic acid on yields of citrinin and pigments, antioxidant capacities, and fatty acid composition of Monascus ruber. LWT 2021, 154, 112800. [Google Scholar] [CrossRef]
- Burphan, T.; Tatip, S.; Limcharoensuk, T.; Kangboonruang, K.; Boonchird, C.; Auesukaree, C. Enhancement of ethanol production in very high gravity fermentation by reducing fermentation-induced oxidative stress in Saccharomyces cerevisiae. Sci. Rep. 2018, 8, 13069. [Google Scholar] [CrossRef]
- Yuan, X.; Shen, G.; Chen, S.; Chen, X.; Zhang, C.; Liu, S.; Jin, M. Modified simultaneous saccharification and co-fermentation of DLC pretreated corn stover for high-titer cellulosic ethanol production without water washing or detoxifying pretreated biomass. Energy 2022, 247, 123488. [Google Scholar] [CrossRef]
- Chan, W.Y.; Dietel, K.; Lapa, S.V.; Avdeeva, L.V.; Borriss, R.; Reva, O.N. Draft Genome Sequence of Bacillus atrophaeus UCMB-5137, a Plant Growth-Promoting Rhizobacterium. Genome Announc. 2013, 1, e00233-13. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, R.H.; Adler, H.; Pickhardt, M.; Mandelkow, E. Lest we forget you--methylene blue…. Neurobiol. Aging 2011, 32, 2325.e7–2325.e16. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.; Li, Y.; Nan, J.; Shi, C.; Li, S. Combined Vapor Permeation and Continuous Solid-State Distillation for Energy-Efficient Bioethanol Production. Energies 2021, 14, 2266. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, T.; Zhou, W.; Jia, X.; Wang, H. Use of a Tn5-based transposon system to create a cost-effective Zymomonas mobilis for ethanol production from lignocelluloses. Microb. Cell Fact. 2013, 12, 41. [Google Scholar] [CrossRef]
- Mordukhova, E.A.; Pan, J.G. Evolved cobalamin-independent methionine synthase (MetE) improves the acetate and thermal tolerance of Escherichia coli. Appl. Environ. Microbiol. 2013, 79, 7905–7915. [Google Scholar] [CrossRef]
- Baumler, D.J.; Hung, K.F.; Bose, J.L.; Vykhodets, B.M.; Cheng, C.M.; Jeong, K.C.; Kaspar, C.W. Enhancement of acid tolerance in Zymomonas mobilis by a proton-buffering peptide. Appl. Biochem. Biotechnol. 2006, 134, 15–26. [Google Scholar] [CrossRef]
- Li, R.; Shen, W.; Yang, Y.; Du, J.; Li, M.; Yang, S. Investigation of the impact of a broad range of temperatures on the physiological and transcriptional profiles of Zymomonas mobilis ZM4 for high-temperature-tolerant recombinant strain development. Biotechnol. Biofuels 2021, 14, 146. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).