For this study, the alcoholic fermentation experimental processes have been conducted in a bioreactor (EVO, 10 L, software NEPTUNE, Pierre Guerin, France) in the research laboratory and have been divided into four distinct control situations. These concern the initial concentration of the substrate (210 and 180 g/L), the initial concentration of the biomass (0.1 and 0.2 g/L), the temperature values (20, 22, 24, and 26 °C), and the fermentation medium characteristics: mash malted, mash malt enriched with B1 vitamin (thiamine), white grape juice (Sauvignon Blanc must) and white grape must (Sauvignon Blanc must) enriched with a B1 vitamin (thiamine). For the experiments, the wine yeasts
Saccharomyces oviformis and
Saccharomyces ellipsoideus were used, these being seeded on a culture medium [
15]. During the process’s evolution, the cell concentration has been calculated on the basis of three different measurements: optical density, dry substance, and the total number of cells. The ethanol concentration has been determined using HPLC-MS (Agilent, USA, Agilrom, Bucharest, Romania) equipment and the substrate concentration has been determined with a spectrophotometric technique.
The bioreactor has been equipped with sensors and analyzers to monitor pH, temperature, level and stirred speed control, dissolved O2, and the released CO2 and O2. From the obtained experimental data, based on the domain knowledge of the authors, 30 “representative” sets were used, considering the following contexts obtained from a combination of variables: 210 and 180 g/L as the initial concentration of the substrate, 0.1 g/L as the initial concentration of the biomass, the four mediums, and the four temperatures.
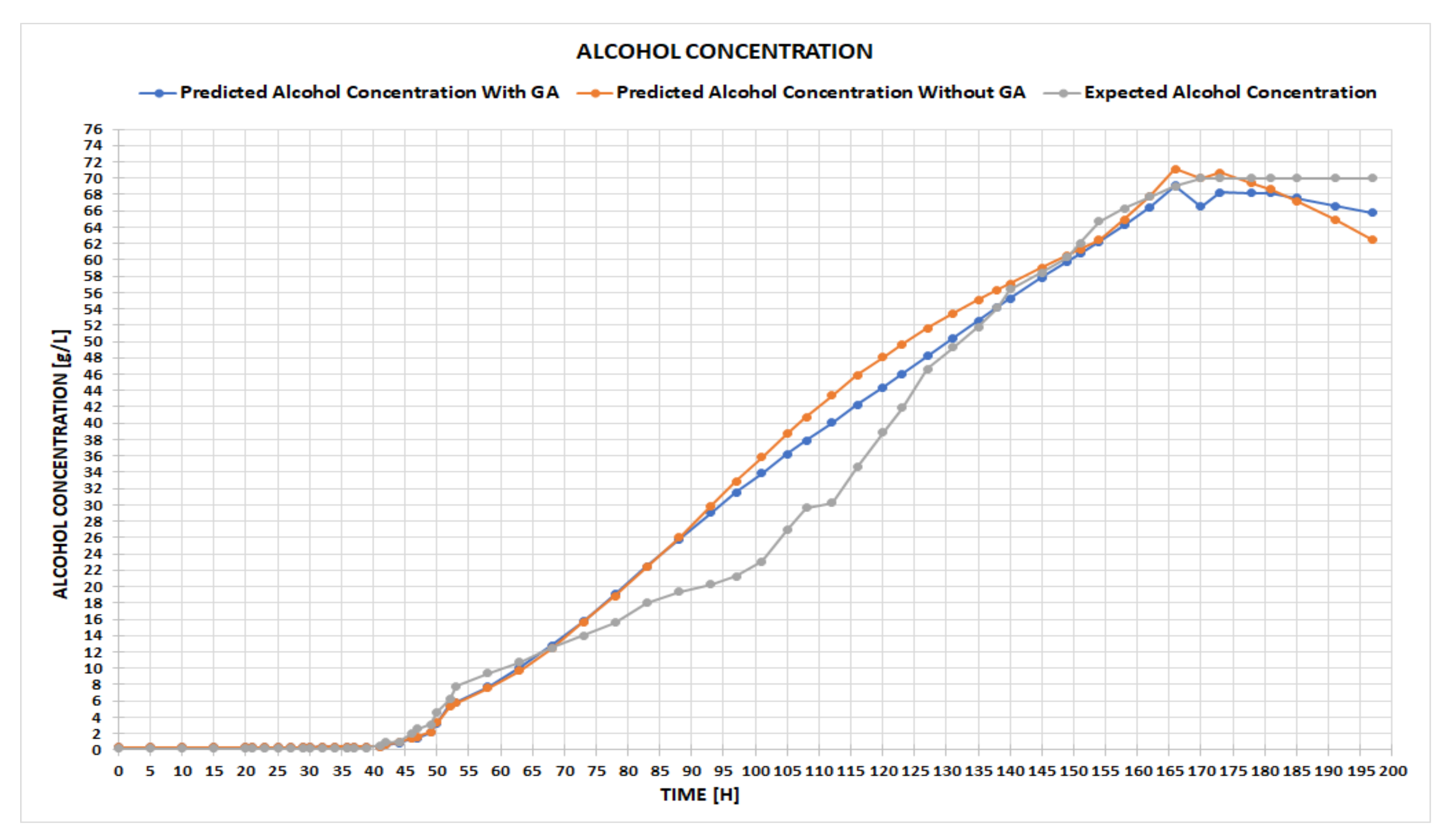
Although neural networks have previously been successfully applied in control processes [
16], from our experience, we can say that there is no single solution (in terms of NN configuration) to suit all problems. For this reason, we believe that cross-fertilizing ideas belonging to evolutionary computing (such as automatic design space exploration, based on genetic algorithms) with artificial intelligence (neural networks) can lead to better results. Thus, we tried to improve and enrich the current solution by replacing the manual design -space exploration for the architecture of the NN with a basic, automatic design space exploration based on a genetic algorithm. The idea started from the fact that several parameters of the NN are chosen randomly at the beginning of the process, such as the weights, the number of hidden layers, the number of neurons on a hidden layer, and the learning rate. More precisely, the aim is to replace the initial random weights of the NN with some that are generated by the genetic algorithm, run for between 30 and 50 generations, in order to increase prediction accuracy. To achieve this goal, an NN has been developed in Visual Studio C# and trained using the experimental data for an alcoholic fermentation process from white winemaking; then, based on this NN, we have been predicted the desired variables of the process.
The neural network was trained using experimental data obtained from the bioreactor wherein the fermentation occurred. The data were collected either by samples acquisition and analysis or from the transductors. Different datasets were used in the training and testing steps, which contain some or all of the following process variables:
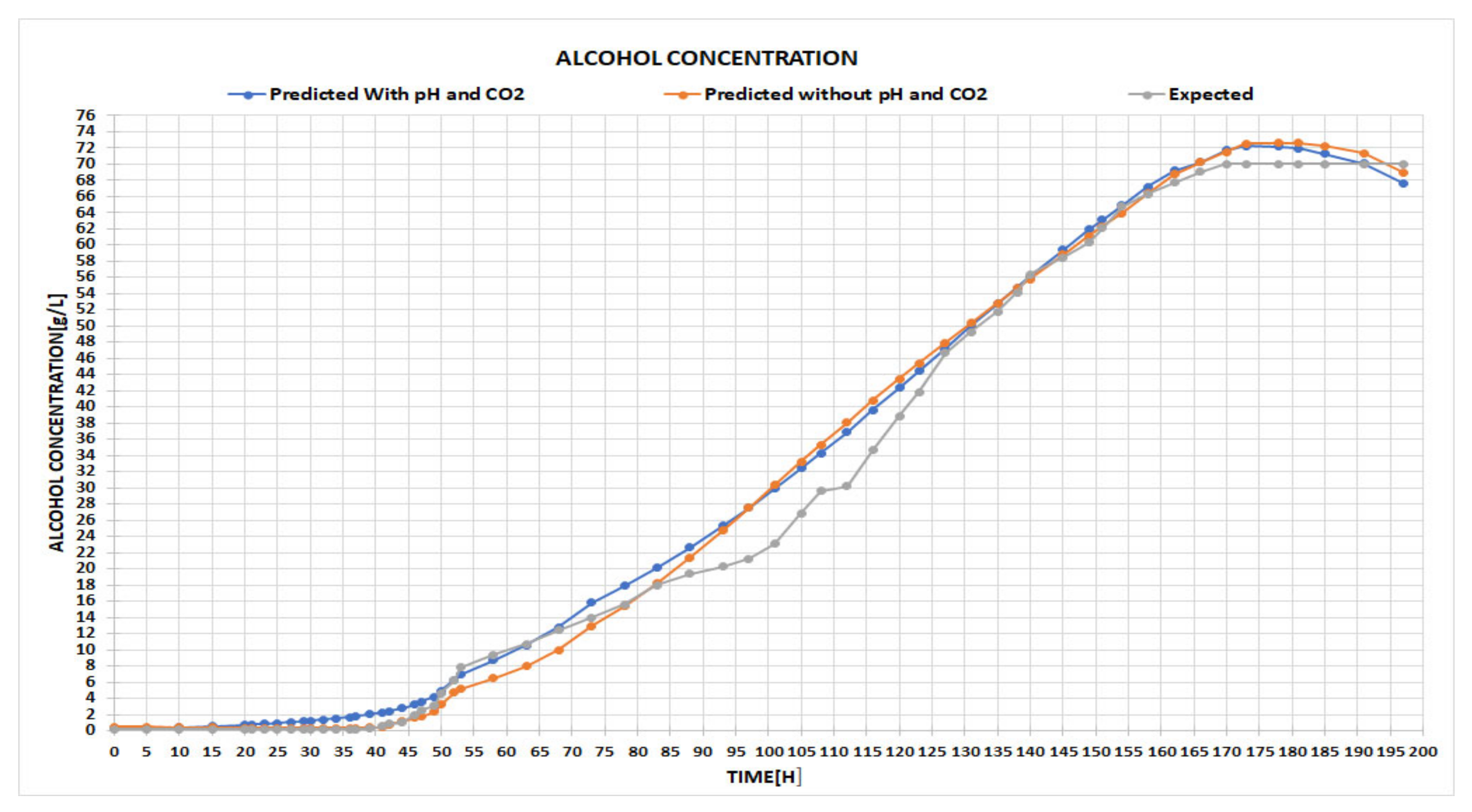
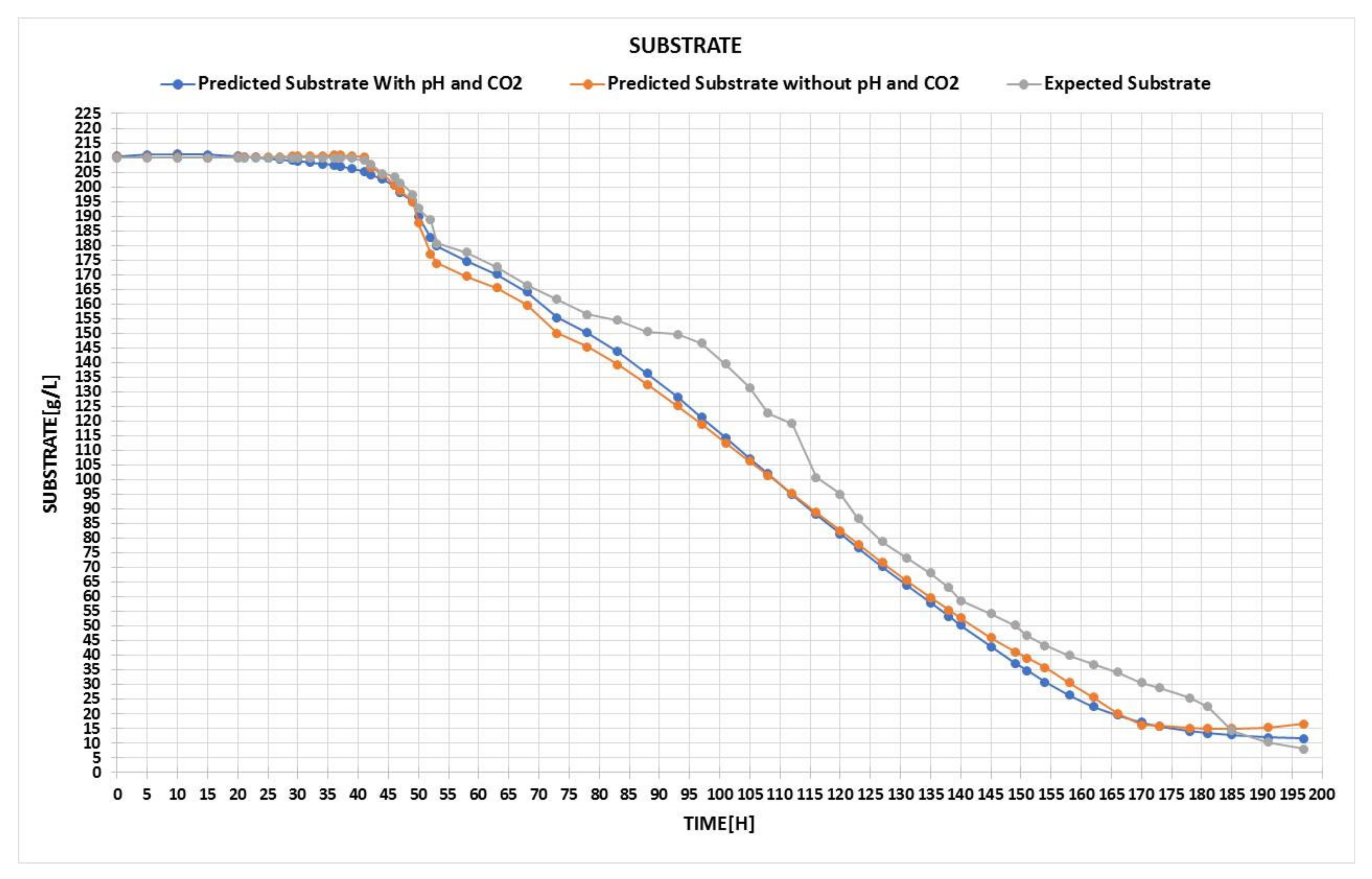
Based on the input, two different configurations were outlined: the first one used only 4 parameters, such as temperature, biomass, time, and initial substrate, while the second one used 2 additional inputs of CO2 concentration and pH.
2.1. User Guide and System Requirements
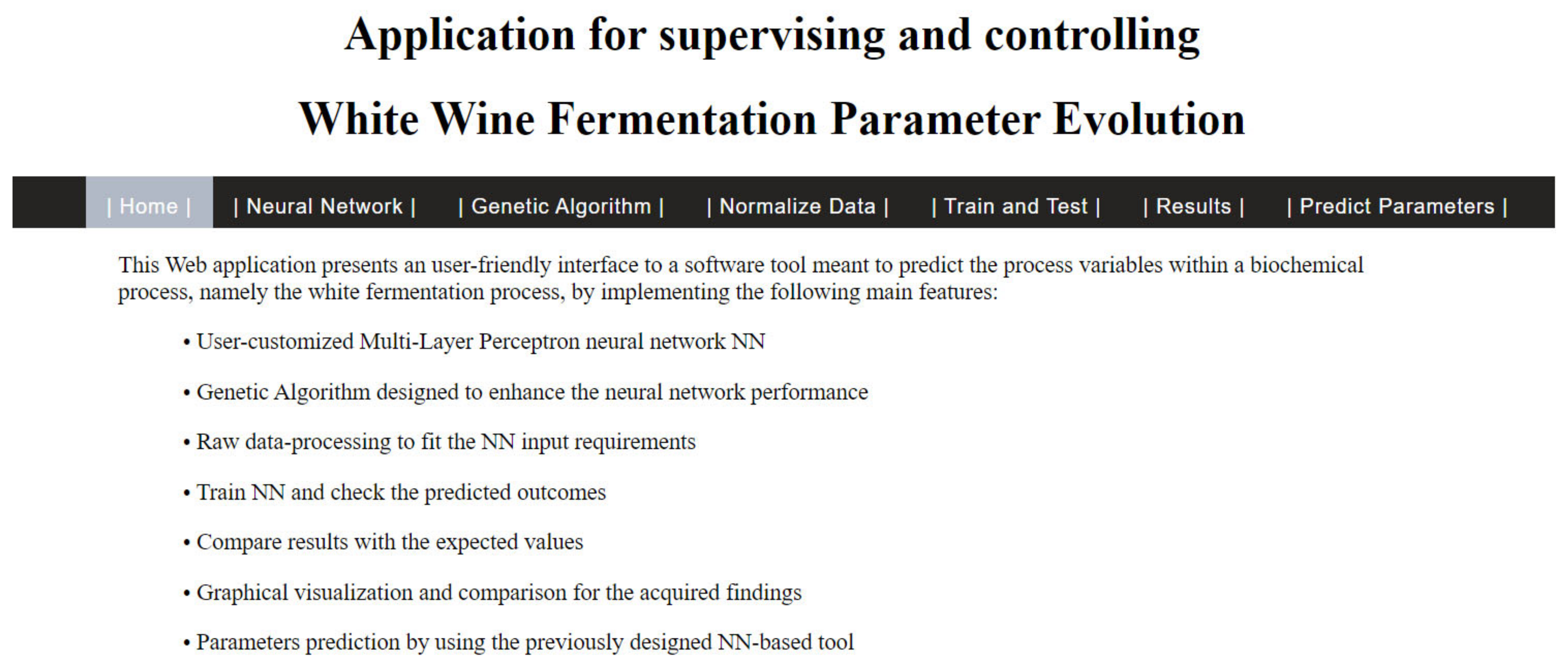
The software application was developed in Visual Studio IDE with C# in 2 different phases: the so-called “back-end” implementation, wherein the architecture of the overall project was designed, and the “front-end” implementation, wherein a user-friendly interface (UI) was created based on an ASP.NET Web Forms Application in order to remotely monitor the fermentation process. This way, we could ensure that the software solution that is proposed is easily portable and platform-independent; thus, migrating it to a Linux/Android/iOS machine would require only a new UI, instead of having to create everything from scratch.
The graphical interface, or the front end, was developed as a website, with one menu bar and several dedicated web pages so that it could facilitate remote access by an expert. The menu bar allows the end-user to easily navigate throughout the different stages of the prediction process, as can be seen in
Figure 2.
Through this software application, the user can accomplish the following tasks:
Choose an Excel (*.xlsx) file in which the training data of the network are structured.
Choose an Excel (*.xlsx) file for generating a normalized data set.
Choose the type of network to predict the variables.
Set up the iteration number that the network will execute in the training process.
Set up the neuron number for the hidden layer of the network.
Set up the number of hidden layers of the network.
Comparatively visualize the graphics of the desired output of the network and the real output.
Predict the desired variable functions of several variables, defined as the input network.
From a hardware point of view, to run properly, the application needs systems with quad-core processors, 8 GB RAM, and a minimum of 850 MB up to 20 GB of HDD available space, depending on the Visual Studio features installed. We created an archive using the sources of the applications, which can be accessed at the following web address (
http://webspace.ulbsibiu.ro/adrian.florea/html/simulatoare/WineFermentationPrediction.zip (accessed on 20 February 2022)) and downloaded to local computers by any interested parties. We recommend using Windows 10 or a later version. Further details about the software’s use are provided by the README.txt file in the archive. This software application was developed using the full Visual Studio license provided for academic use, free of charge, by the Lucian Blaga University of Sibiu, with the full support of the Hasso Plattner Foundation in Germany. For this reason, those who use our source must do so only for educational purposes. Besides it being free and easy to use, our tool provides the following advantages: flexibility, extensibility, interactivity, and performance.
2.2. Architecture and Design
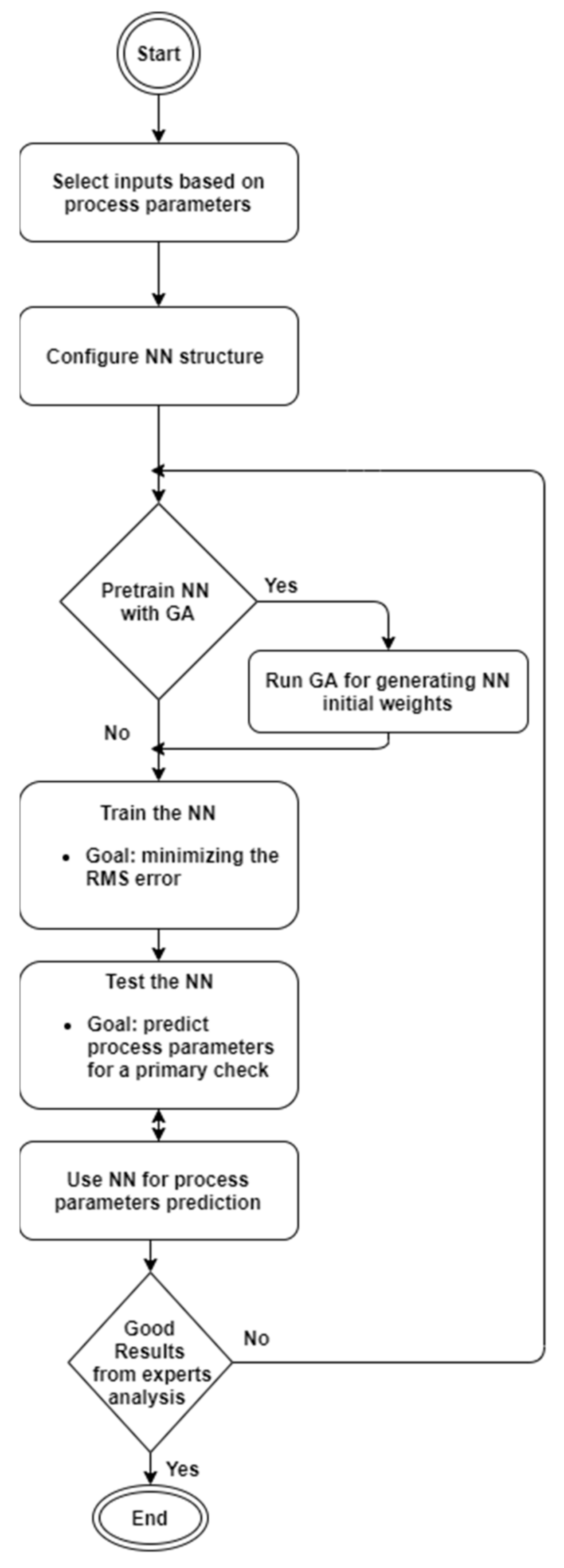
The menu bar displays all the steps required for the prediction process in a logical and chronological order, as shown in the diagram in
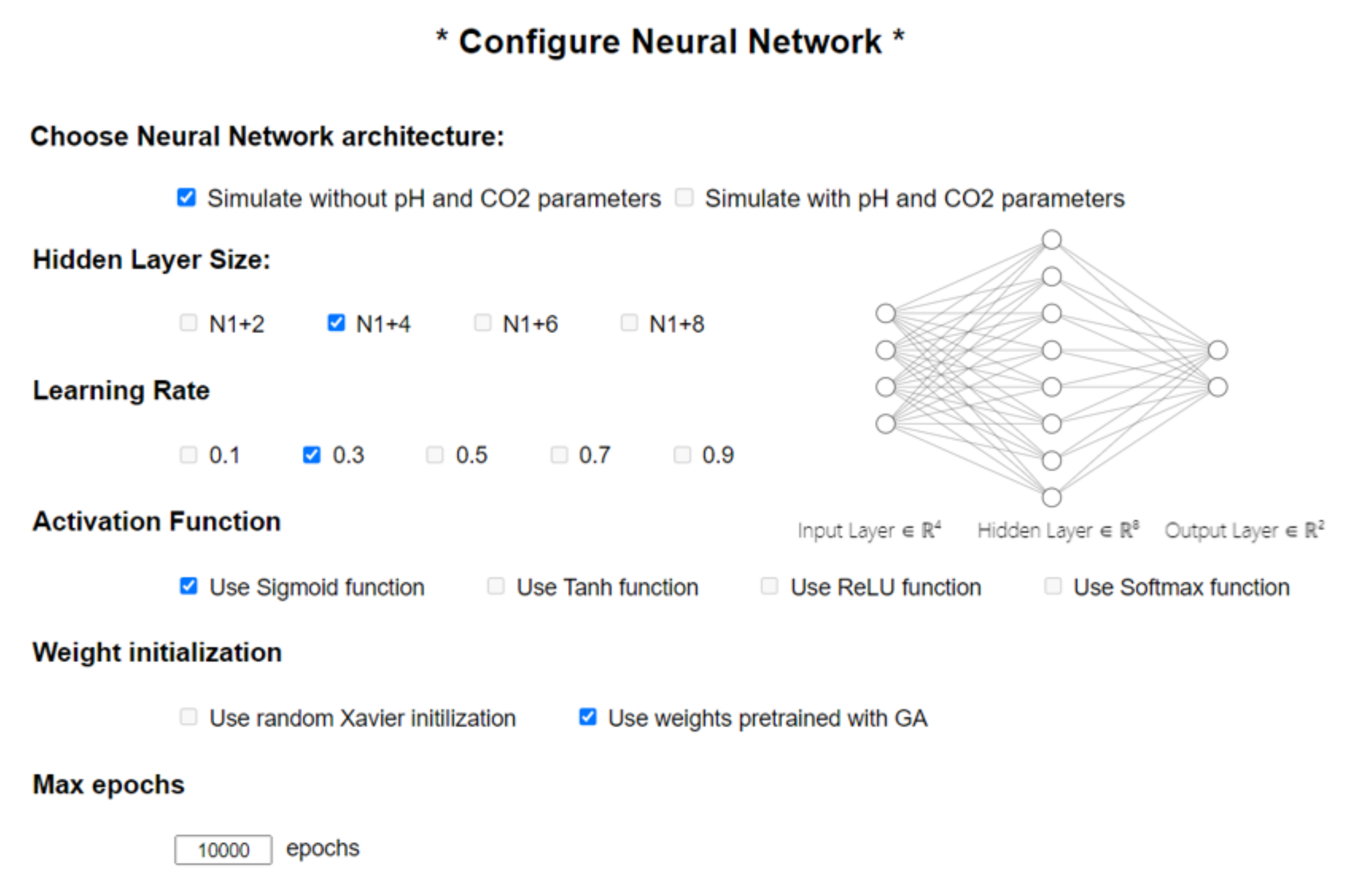
Figure 1. The first step was to configure the neural network that fits the input dataset. We could perform this intuitively and easily from the graphical interface by using checkboxes and text boxes, as highlighted in
Figure A1 in
Appendix A. The user interface (UI) allows the user to select the type of data they want to use for the simulation from between the two possible choices previously outlined in
Section 2. Their selection would influence the entire network architecture by changing the number of neurons on the input layer, denoted henceforth as N1. The application simulates a feed-forward multi-layer perceptron (MLP) network with a backpropagation (BP) algorithm. The backpropagation learning model is a learning algorithm used in feed-forward networks. BP comprises two steps: the first step (forward) is where information is passed from input to output, followed by a step from output to input. The forward step propagates the input vector to the first level of the network; the outputs of this level produce a new vector that will be the input for the next level until it reaches the last level, where the outputs are the network outputs. The second step backward (backward) is similar to the forward step, except that errors are propagated backward through the network to cause the weights to adjust. Based on gradient descent for weight adjustment, the BP algorithm uses the chain rule to compute the gradient of the error for each unit with respect to its weights. There are several other parameters that must be customized, such as: the number of neurons in the hidden layer, the learning rate, which in the literature is usually given the notation β, the activation function, the weights initialization, and the maximum number of epochs for which the network has to be trained.
For the activation functions, four of the most well-known functions in feed-forward neural networks were taken into consideration. Each one has its own advantages and disadvantages; it is up to the user to choose the function that best suits the input data. Although the sigmoid (Equation (1)) and hyperbolic tangent (Equation (2)) are two of the most widely used functions, they both face the problem of vanishing gradients. As an alternative, there is the ReLU function (Equation (3)), which stands for the rectified linear unit; this is meant to activate only those neurons that have a significant output (higher than 0). Another option to the well-known sigmoid function would be SoftMax (Equation (4)).
For the weight initialization step, there are two different approaches we considered that best suited our dataset. The first approach is a pseudo-random initialization that generates sub-unitary values, called the Xavier initialization [
17]. Instead of generating completely random weights in the (0, 1) domain, this initialization chooses values from a random uniform distribution, bounded by the following interval:
where
is the number of incomings in terms of network connections and
is the number of outgoing network connections from that layer. Assuming that the configuration selected for the neural network in
Figure A1 represents a 3-layer MLP neural network (Input–Hidden–Output), for the hidden layer, the value of
would be the number of the neurons on the input layer and
the number of neurons on the output layer.
The other approach is the one that brings innovation to our application, i.e., finding the initial weighting configuration that yields the best results by using a heuristic algorithm, such as a genetic algorithm. The original premise was that finding an optimal layout for our network can be narrowed down to a searching problem within an infinite space of solutions. It has previously been established that an exhaustive search would be time-consuming and pointless, which is what led us to use heuristic methods, among which the best-known one is the genetic algorithm.
For this to be possible, we had to exclude the easy and intuitive approach of using the neural network libraries proposed by MATLAB (also used by us in a previous work [
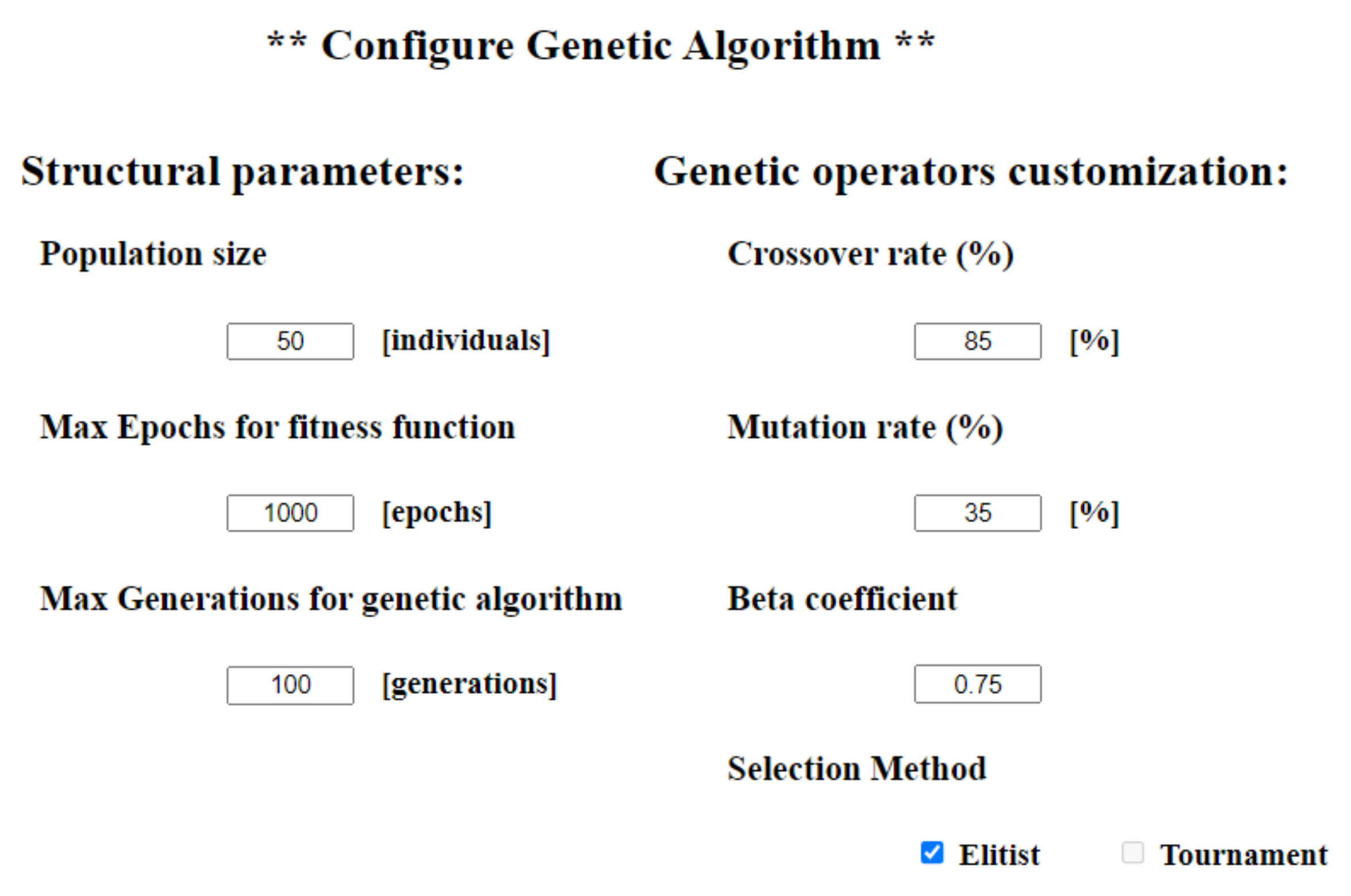
16]) and create our own user-customizable solution, with the help of C# and the ASP.NET framework. Akin to implementation for the neural network design, the expert user is given the opportunity to fully customize the genetic algorithm operators using the controls offered by the ASP web pages, such as text boxes and checkboxes (
Figure A2).
The process starts with a population of randomly generated individuals. In our approach, each individual represents a different neural network, with its own generated weights. To narrow it down to a simple approach, the individual can be represented as a linear matrix of weights, as follows:
Here, each weight is generated with the above-mentioned Xavier initialization. In
Figure 3, we are using the NN selected in
Figure A1 (from
Appendix A), wherein we set 4 neurons on the input layer, 1 single hidden layer with 8 neurons, and 2 neurons on the output layer. The fitness function used to evaluate each individual is the backpropagation algorithm itself, while the fitness score is the output of this algorithm, i.e., the network error that resulted after training the network for the specific number of epochs selected by the user. The number of individuals selected for the initial population and the maximum number of generations is again selected by the user; this must be a trade-off between having an optimal solution and a proper execution time.
Each individual has a finite number of genes, which in our case are represented by the linear matrix of weights, where one weight corresponds to one gene. Each string of genes, also called the chromosome, is a possible solution to the search problem. In order to find the optimal configuration, we applied the three genetic operators to the initial population. As previously mentioned, the application is fully customizable, as this is its main purpose and, although there are many proposals for this type of crossover, mutation, or selection, we implemented only the ones that best suited our dataset and that provided acceptable results. Hence, as a parent selection method, either the elitist method, which chooses the two individuals with the best fitness score, or the tournament method, which selects the two best-evaluated individuals among several n of randomly selected individuals, can be chosen. The latter choice follows the principle that it is not always the first, best, 2 parents that generate the best offspring, giving the chance for weaker evaluated individuals to participate in the crossover process.
As for the crossover process, there are several well-known methods, such as the k-point crossover, uniform crossover, or order-based crossover. For our solution, we opted for a probability-based uniform crossover.
Here, W
1i,j and W
2i,j are the genes of the parents, arising from the selection process. More precisely, for each gene of the selected chromosome, a random probability P is generated; if the generated P is less than the crossover probability, denoted as P
crossover in
Figure 4, or the crossover rate in
Figure A2 (in
Appendix A), the value of this gene is exchanged with the value of the corresponding gene of the second selected chromosome. It is advisable that in all cases, no matter the data set used, the crossover rate needs to be high enough to ensure that the future generation is somehow distinct and can bring improvements compared with the previous generation.
After crossover, the mutation operator will be used in order to bring diversity to the population with its best-known types: bit-string mutation, flip bit, boundary, uniform, or Gaussian. Conversely to the high rate of the crossover, the mutation probability must be low enough not to generate completely new random offspring while still bringing diversity to the population. We have chosen a probability-based uniform mutation (
Figure 5), but it is dynamically handled this time; we started with a user-defined mutation probability and adjusted it with each generation, as follows:
where
Beta is a sub-unitary coefficient selected by the expert user in the configuration box (
Figure A2).
After applying these operators, we have a group of individuals comprising the initial population and the newly generated offspring. The latter are evaluated using the same fitness function and, based on the fitness score, we replace the worst-evaluated individuals with the two derived descendants if they perform better than these individuals. Consequently, the survival of the individuals in the new generation is based on an elitist approach.
These operators are applied repeatedly to the group of individuals until the population converges, meaning that it does not produce significantly better-scored offspring, or until the maximum number of generations has been reached. In the end, the outcome is an individual representing the best or at least the optimal configuration of initial weights for our neural network.
2.4. Prediction Tool
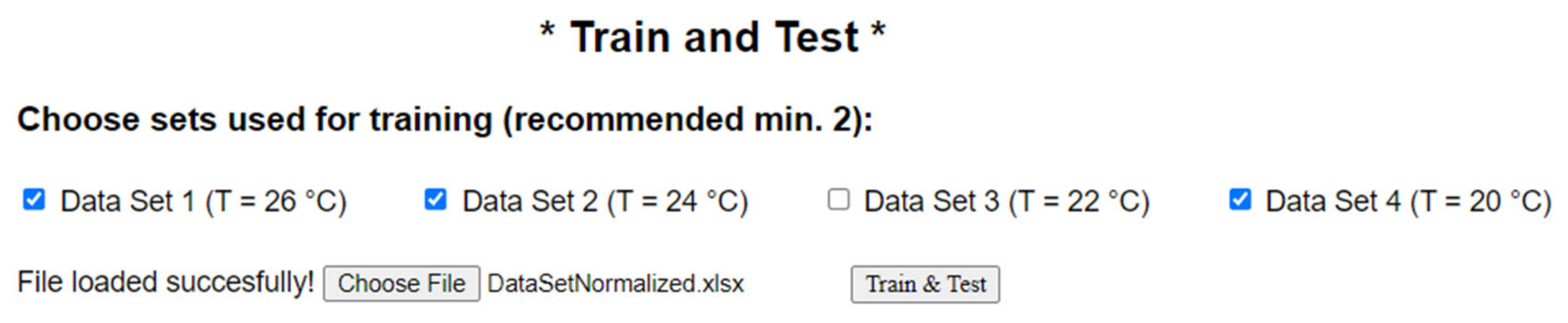
The neural network architecture selection is now complete, ready for training, and, finally, for testing. In order to do that, the spreadsheet format datasets will be used for both training and testing, as follows: the user must select the sets used for training by checking the corresponding boxes in the application. In our experiments, the training/testing ratio is 75/25 because we used 3 data sets for training and one for testing (see the configuration from
Figure A4 in
Appendix A). However, if we vary the number of sets on which we perform, the practice test, we also change the training/testing ratio of 75/25. Our application is flexible and allows having many configurations (for example, 50/50, or even 25/75. Considering
n (
benchmarks (data sets) and that we use
k benchmarks (
for training, we must have at least one set for training and the rest for testing, with at least one set for testing, although it is recommended for better results that more than 50% of the data is reserved for training. The total number of configurations that can be simulated is:
For example, selecting only one benchmark for training the total number of configurations that can be simulated is , because we may select anyone from all n benchmarks for training, and for testing, one from all n − 1 remaining data sets or select two from all n − 1 for testing, etc. Taking all these into consideration, we consider that the training/testing sets are representative of all combinations of the experimental conditions.
The expected output can be checked in the results section (
Figure 6), designed to accelerate the interpretation process by laying out both numerical and graphical results. The training and testing errors give important values for a first evaluation; there are situations where the training error is very low but the testing error is still high, so that we can safely conclude immediately that we are dealing with an overfitting problem (the model learns the detail and noise in the training data, to the extent that it negatively impacts the performance of the model with new data). Still, for a deeper evaluation, the graphics containing the results from the test are more meaningful.
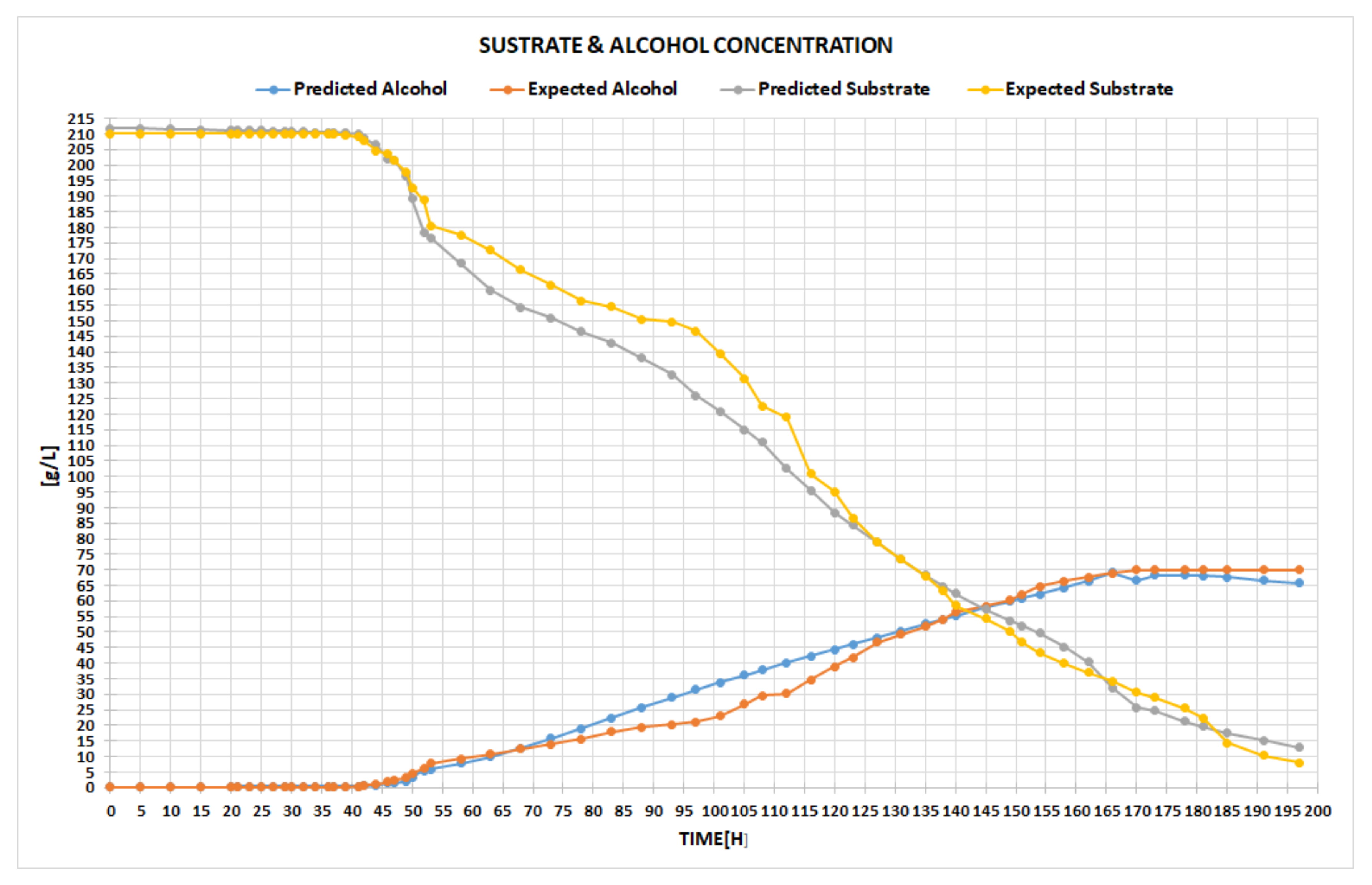
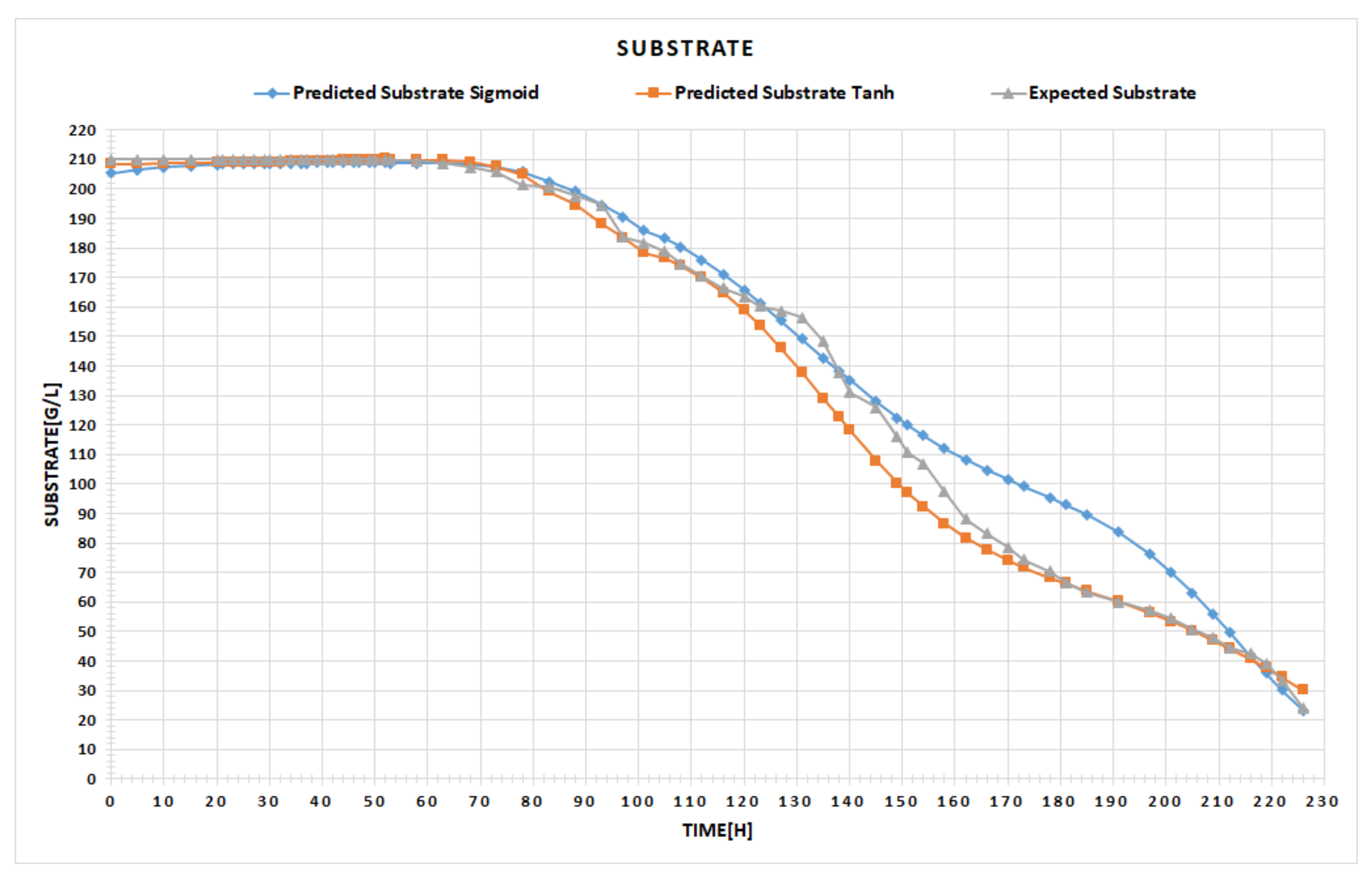
The left-hand chart in
Figure 6 highlights the expected ascending curve of alcohol generation, versus the one predicted by our network in the testing phase. The right-hand chart displays the expected quantity of the consumed substrate in time, compared to the one attained within the testing period. The user can switch between different chart types, for instance, by point, line, bar, column, or area, the main idea being to select the particular type of chart that better emphasizes the visual differences between the data. With all these facts being given, the expert can now conclude whether the results are satisfactory or not. If yes, the final step is to use the obtained tool for the prediction process of the fermentation parameters, as the example in
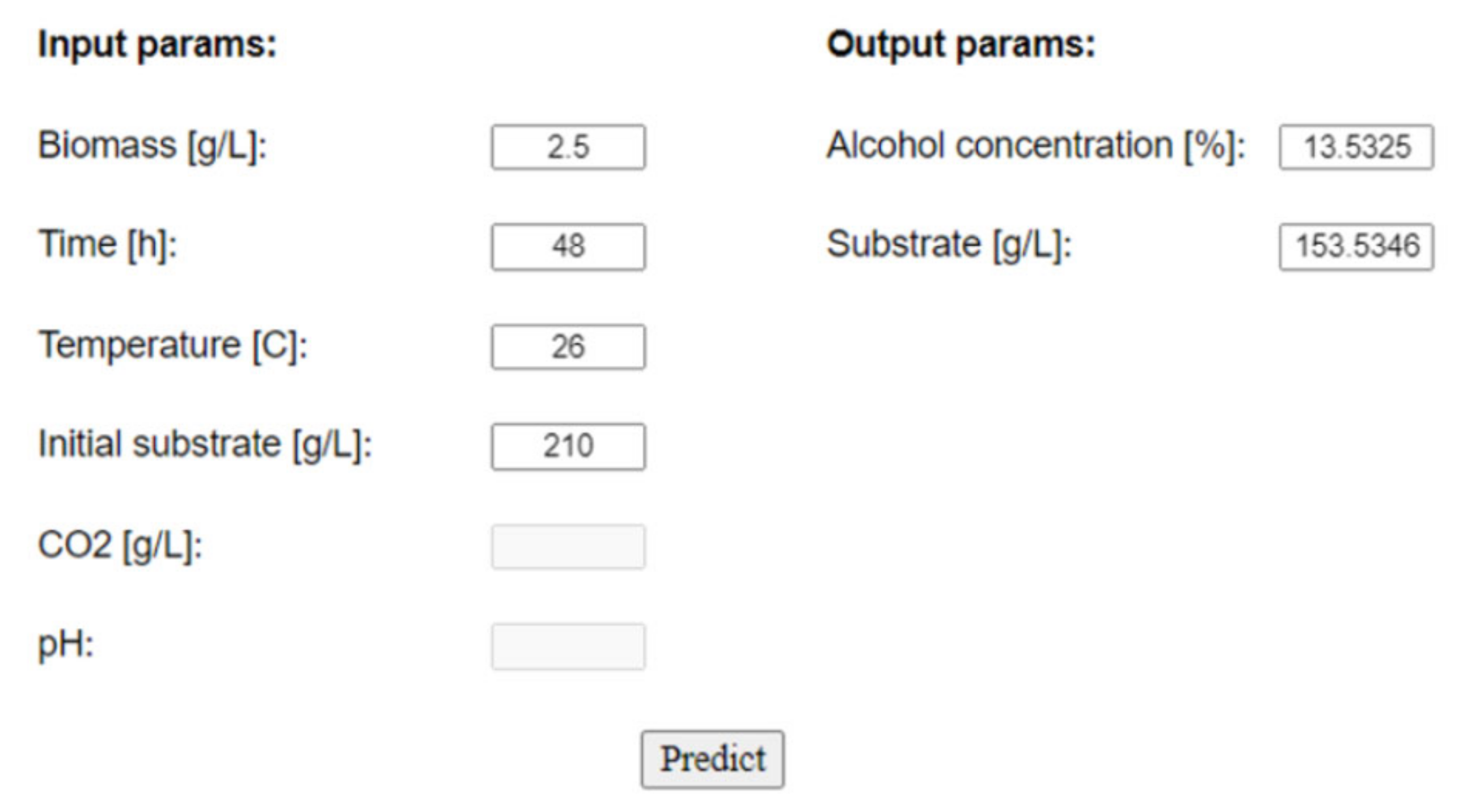
Figure 7 indicates.
In the left-hand input section, the user is required to fill in the boxes with the values of the process parameters, meaning the quantity of biomass and initial substrate at a certain time and temperature. Temperature is very important because even the slightest fluctuation can alter the oxidation of the wine and, therefore, significantly affect the quality. Additionally, information about CO
2 and pH can be used if the neural network architecture is designed and trained with this information from the beginning. This example outlines the usage of the tool for the neural network built in
Figure A1, with only 4 input parameters; thus, the last two boxes are disabled.
When pressing the “Predict” button in the output section, the numerical values for the output parameters can be read and further interpreted by the wine technology expert. Thus, the NN is able to determine the fermentation stage (providing real-time information regarding the fermentation process without reading the sensors) or predict wine quality based on the process variables. This can directly affect the productivity of the winery by saving time and money.