Changes in the Microbial Community and Biogenic Amine Content in Rapeseed Meal during Fermentation with an Antimicrobial Combination of Lactic Acid Bacteria Strains
Abstract
:1. Introduction
2. Materials and Methods
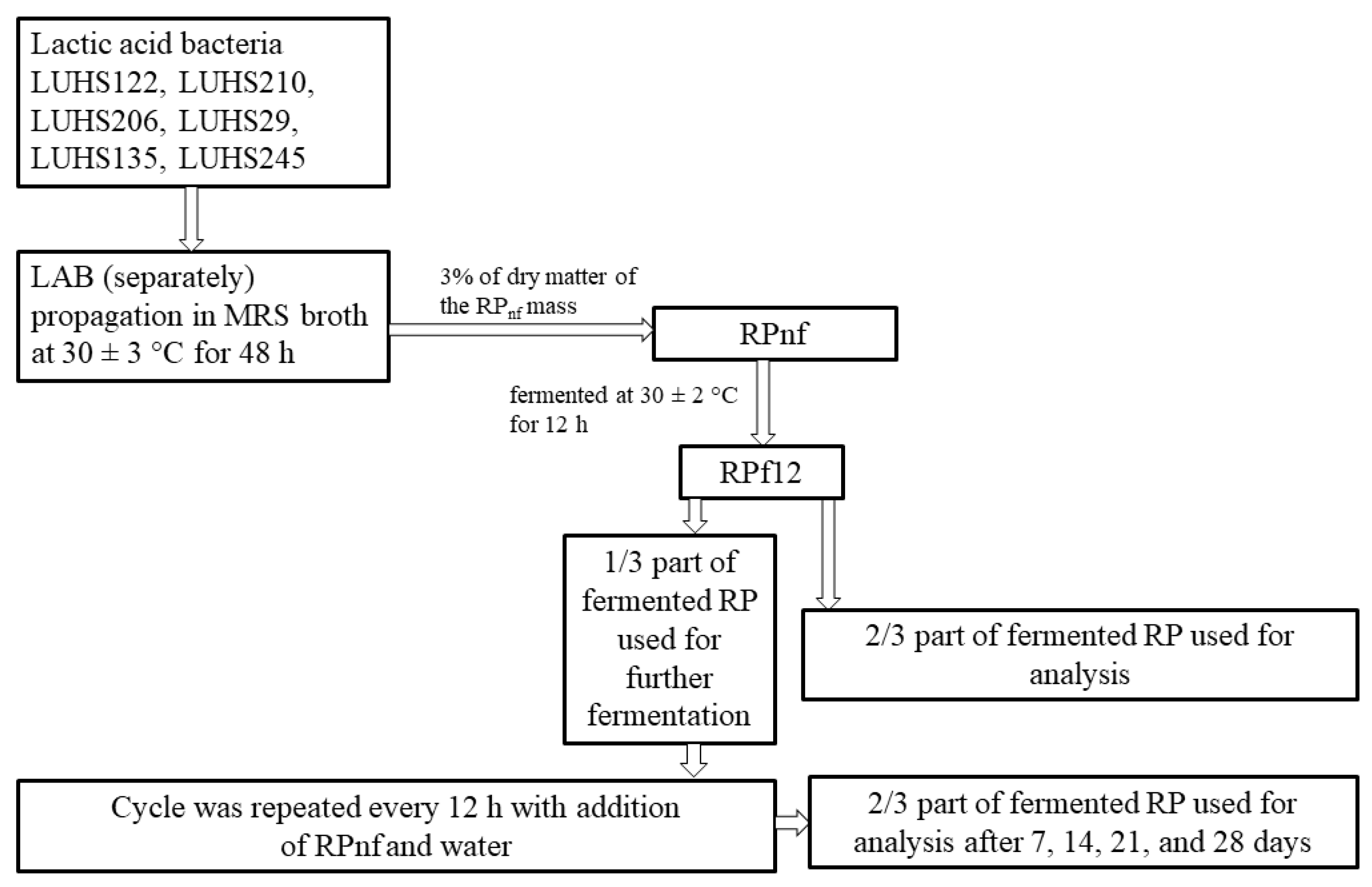
2.1. Lactic Acid Bacteria Strains and Rapeseed Meal Used in Experiments
2.2. Evaluation of Acidity and Microbiological Characteristics of Samples
2.3. Determination of Biogenic Amine Content in Samples
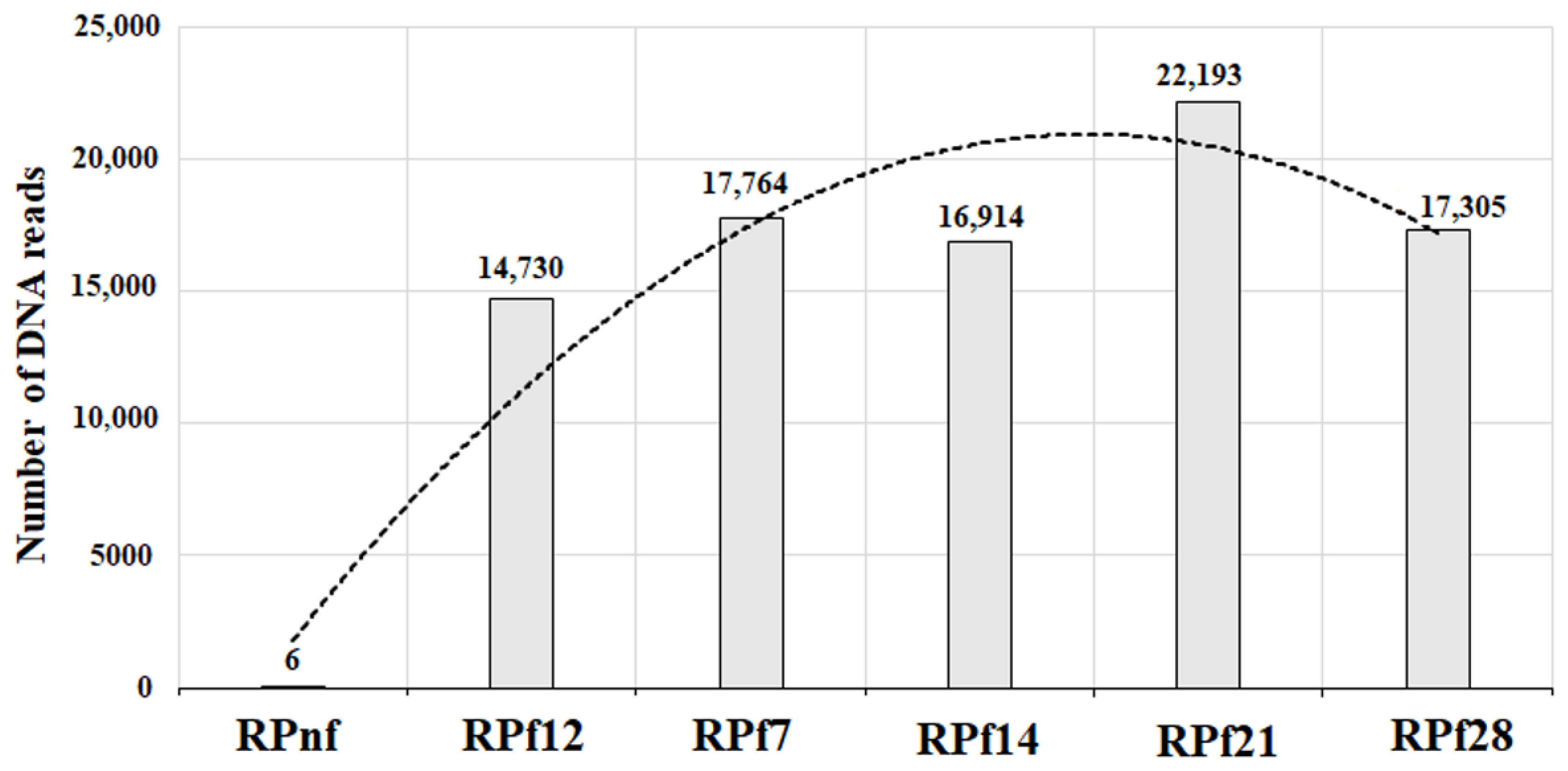
2.4. Metagenomic Analysis of Rapeseed Meal Samples
2.5. Statistical Analysis
3. Results and Discussion
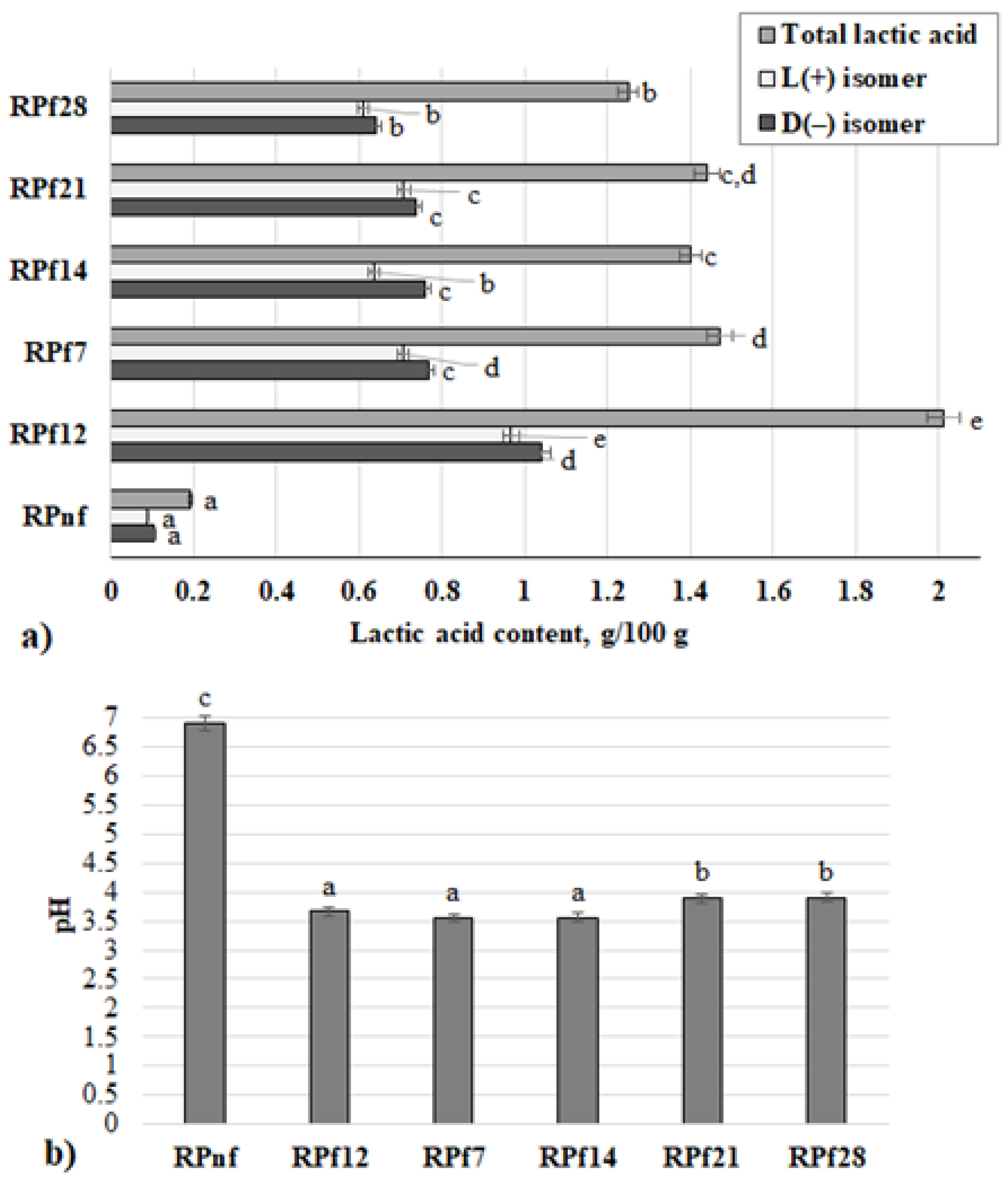
3.1. Acidity Parameters of Rapeseed Meal Samples
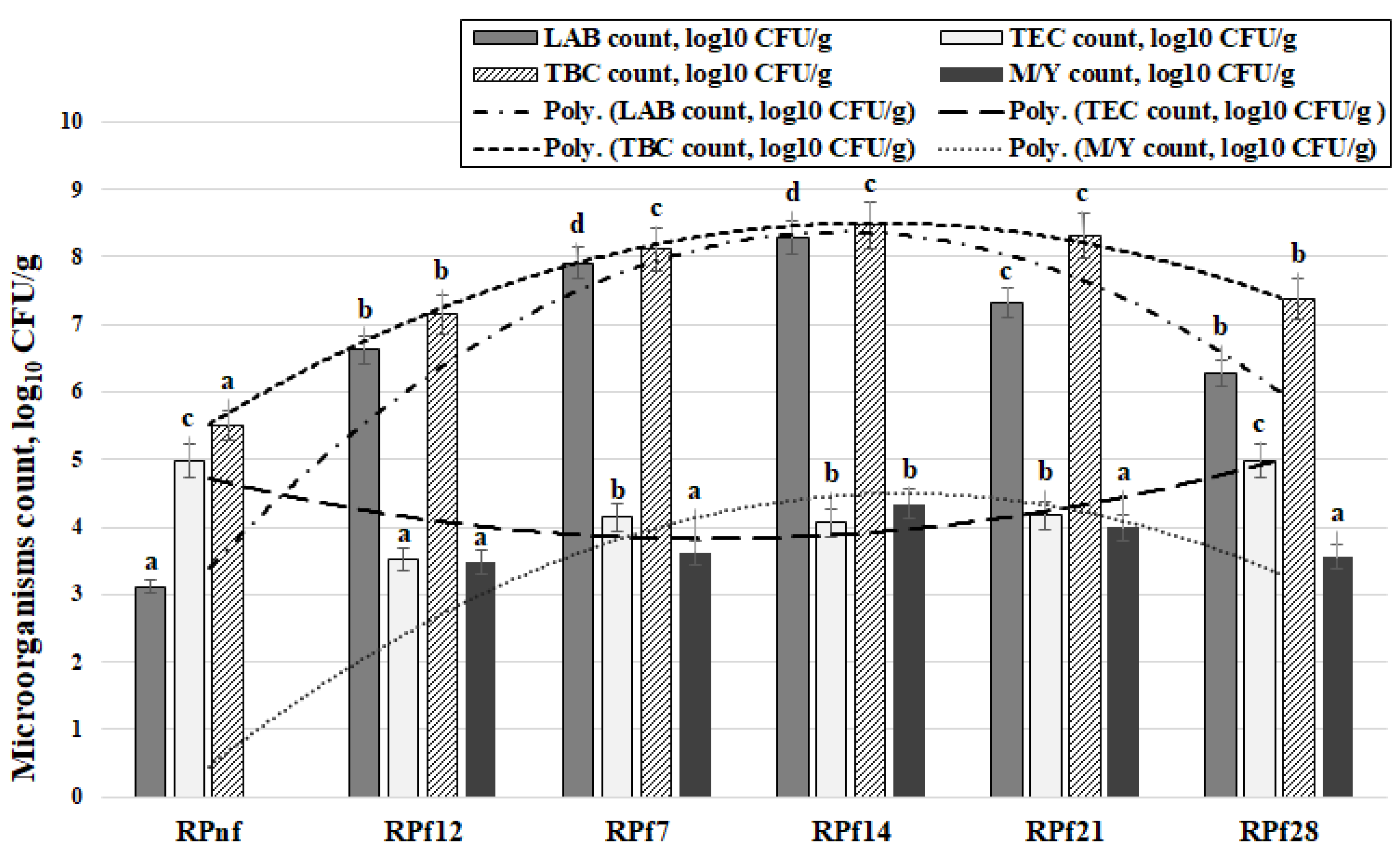
3.2. Microbiological Parameters of the Fermented Samples
3.3. Biogenic Amine Formation in Fermented Rapeseed Meal
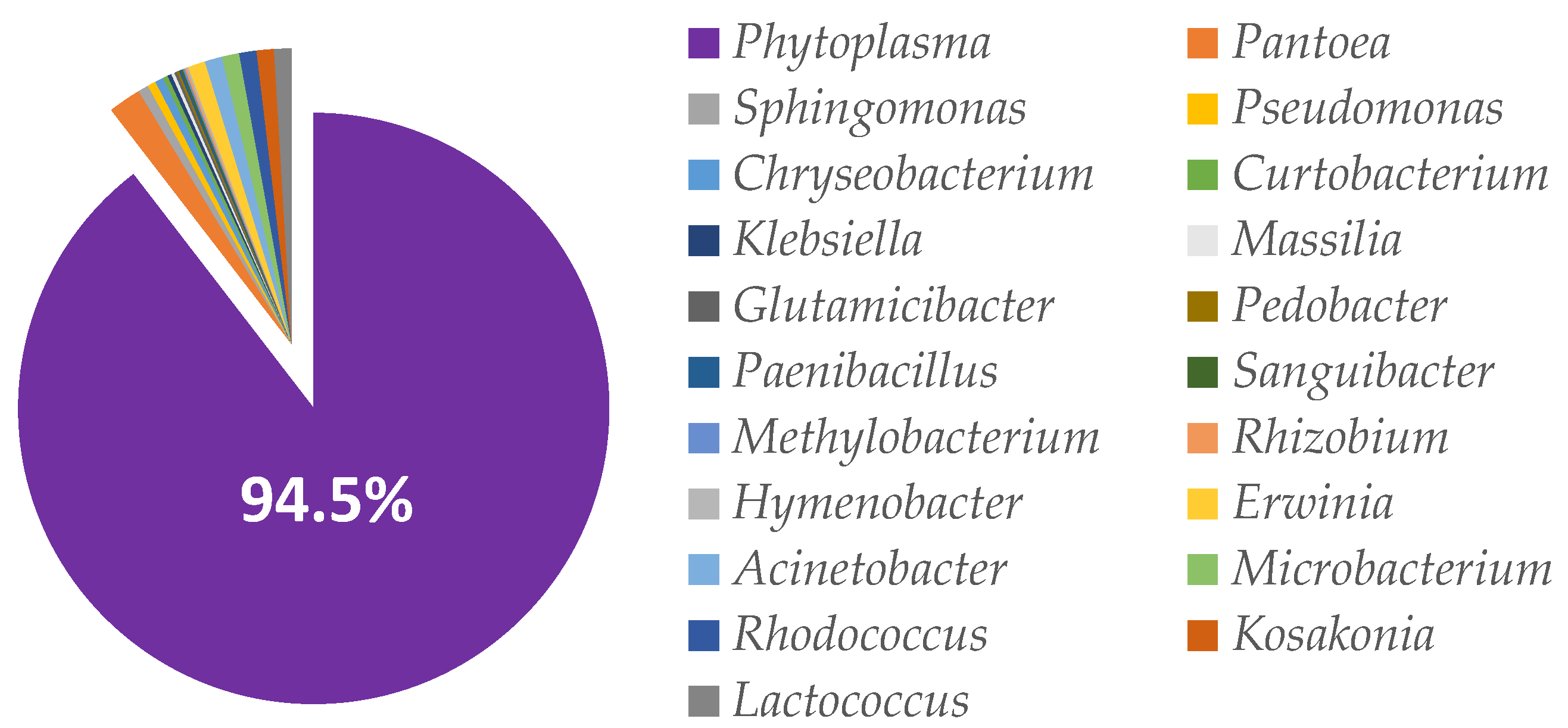
3.4. Microbial Profiles of the Non-Fermented and Fermented Rapeseed Meal
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ross, R.P.; Morgan, S.; Hill, C. Preservation and fermentation: Past, present and future. Int. J. Food Microbiol. 2002, 79, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Macori, G.; Cotter, P.D. Novel insights into the microbiology of fermented dairy foods. Curr. Opin. Biotechnol. 2018, 49, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Vadopalas, L.; Ruzauskas, M.; Lele, V.; Starkute, V.; Zavistanaviciute, P.; Zokaityte, E.; Bartkevics, V.; Pugajeva, I.; Reinolds, I.; Badaras, S.; et al. Combination of Antimicrobial Starters for Feed Fermentation: Influence on Piglet Feces Microbiota and Health and Growth Performance, Including Mycotoxin Biotransformation in vivo. Front. Vet. Sci. 2020, 7, 528990. [Google Scholar] [CrossRef]
- Vadopalas, L.; Ruzauskas, M.; Lele, V.; Starkute, V.; Zavistanaviciute, P.; Zokaityte, E.; Bartkevics, V.; Badaras, S.; Klupsaite, D.; Mozuriene, E.; et al. Pigs’ Feed Fermentation Model with Antimicrobial Lactic Acid Bacteria Strains Combination by Changing Extruded Soya to Biomodified Local Feed Stock. Animals 2020, 10, 783. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, P.M.; Kuniyoshi, T.M.; Oliveira, R.P.; Hill, C.; Ross, R.P.; Cotter, P.D. Antimicrobials for food and feed; a bacteriocin perspective. Curr. Opin. Biotechnol. 2020, 61, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Lele, V.; Sakiene, V.; Zavistanaviciute, P.; Ruzauskas, M.; Bernatoniene, J.; Jakstas, V.; Viskelis, P.; Zadeike, D.; Juodeikiene, G. Improvement of the antimicrobial activity of lactic acid bacteria in combination with berries/fruits and dairy industry by-products. J. Sci. Food Agric. 2019, 99, 3992–4002. [Google Scholar] [CrossRef]
- Bartkiene, E.; Lele, V.; Ruzauskas, M.; Domig, K.J.; Starkute, V.; Zavistanaviciute, P.; Bartkevics, V.; Pugajeva, I.; Klupsaite, D.; Juodeikiene, G.; et al. Lactic Acid Bacteria Isolation from Spontaneous Sourdough and Their Characterization Including Antimicrobial and Antifungal Properties Evaluation. Microorganisms 2020, 8, 64. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-J.; Choi, Y.-J.; Lee, M.J.; Park, S.J.; Oh, S.J.; Yun, Y.-R.; Min, S.G.; Seo, H.-Y.; Park, S.-H.; Lee, M.-A. Effects of combining two lactic acid bacteria as a starter culture on model kimchi fermentation. Food Res. Int. 2020, 136, 109591. [Google Scholar] [CrossRef]
- Hesseltine, C.W. Applications of Biotechnology in Traditional Fermented Foods; National Academies Press: Washington, DC, USA, 1992; ISBN 978-0-309-04685-5. [Google Scholar]
- Wójcik, W.; Łukasiewicz, M.; Puppel, K. Biogenic amines: Formation, action and toxicity—A review. J. Sci. Food Agric. 2021, 101, 2634–2640. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Kim, M.-J.; Moon, B. Various biogenic amines in Doenjang and changes in concentration depending on boiling and roasting. Appl. Biol. Chem. 2017, 60, 273–279. [Google Scholar] [CrossRef]
- Ly, D.; Mayrhofer, S.; Schmidt, J.-M.; Zitz, U.; Domig, K.J. Biogenic Amine Contents and Microbial Characteristics of Cambodian Fermented Foods. Foods 2020, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Bartkiene, E.; Bartkevics, V.; Krungleviciute, V.; Pugajeva, I.; Zadeike, D.; Juodeikiene, G. Lactic Acid Bacteria Combinations for Wheat Sourdough Preparation and Their Influence on Wheat Bread Quality and Acrylamide Formation. J. Food Sci. 2017, 82, 2371–2378. [Google Scholar] [CrossRef]
- Bartkiene, E.; Bartkevics, V.; Lele, V.; Pugajeva, I.; Zavistanaviciute, P.; Mickiene, R.; Zadeike, D.; Juodeikiene, G. A concept of mould spoilage prevention and acrylamide reduction in wheat bread: Application of lactobacilli in combination with a cranberry coating. Food Control 2018, 91, 284–293. [Google Scholar] [CrossRef]
- Bartkiene, E.; Zokaityte, E.; Lele, V.; Starkute, V.; Zavistanaviciute, P.; Klupsaite, D.; Cernauskas, D.; Ruzauskas, M.; Bartkevics, V.; Pugajeva, I.; et al. Combination of Extrusion and Fermentation with Lactobacillus plantarum and L. uvarum Strains for Improving the Safety Characteristics of Wheat Bran. Toxins 2021, 13, 163. [Google Scholar] [CrossRef]
- Vadopalas, L.; Badaras, S.; Ruzauskas, M.; Lele, V.; Starkute, V.; Zavistanaviciute, P.; Zokaityte, E.; Bartkevics, V.; Klupsaite, D.; Mozuriene, E.; et al. Influence of the Fermented Feed and Vaccination and Their Interaction on Parameters of Large White/Norwegian Landrace Piglets. Animals 2020, 10, 1201. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Ruzauskas, M.; Lele, V.; Zavistanaviciute, P.; Bernatoniene, J.; Jakstas, V.; Ivanauskas, L.; Zadeike, D.; Klupsaite, D.; Viskelis, P.; et al. Development of antimicrobial gummy candies with addition of bovine colostrum, essential oils and probiotics. Int. J. Food Sci. Technol. 2018, 53, 1227–1235. [Google Scholar] [CrossRef]
- Ben-Gigirey, B.; De Sousa, J.M.V.B.; Villa, T.G.; Velázquez, J.B. Histamine and Cadaverine Production by Bacteria Isolated from Fresh and Frozen Albacore (Thunnus alalunga). J. Food Prot. 1999, 62, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Bartkevics, V.; Rusko, J.; Starkute, V.; Bendoraitiene, E.; Zadeike, D.; Juodeikiene, G. The effect of Pediococcus acidilactici and Lactobacillus sakei on biogenic amines formation and free amino acid profile in different lupin during fermentation. LWT 2016, 74, 40–47. [Google Scholar] [CrossRef]
- Evans, T.G.; Reed, S.S.; Hibbs, J.B. Nitric Oxide Production in Murine Leishmaniasis: Correlation of Progressive Infection with Increasing Systemic Synthesis of Nitric Oxide. Am. J. Trop. Med. Hyg. 1996, 54, 486–489. [Google Scholar] [CrossRef]
- Kim, D.; Lee, K.D.; Choi, K.C. Role of LAB in silage fermentation: Effect on nutritional quality and organic acid production—An overview. AIMS Agric. Food 2021, 6, 216–234. [Google Scholar] [CrossRef]
- Ashayerizadeh, A.; Dastar, B.; Shargh, M.S.; Mahoonak, A.S.; Zerehdaran, S. Fermented rapeseed meal is effective in controlling Salmonella enterica serovar Typhimurium infection and improving growth performance in broiler chicks. Vet. Microbiol. 2017, 201, 93–102. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Ng, C.-C.; Su, H.; Tzeng, W.-S.; Shyu, Y.-T. Probiotic potential of noni juice fermented with lactic acid bacteria and bifidobacteria. Int. J. Food Sci. Nutr. 2009, 60, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Rawoof, S.A.A.; Kumar, P.S.; Vo, D.-V.N.; Devaraj, K.; Mani, Y.; Devaraj, T.; Subramanian, S. Production of optically pure lactic acid by microbial fermentation: A review. Environ. Chem. Lett. 2021, 19, 539–556. [Google Scholar] [CrossRef]
- Liptáková, D.; Matejčeková, Z.; Valík, Ľ. Lactic Acid Bacteria and Fermentation of Cereals and Pseudocereals. In Fermentation Processes; Jozala, A.F., Ed.; Intech Publisher: London, UK; Rijeka, Croatia, 2017; pp. 223–254. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Ding, Y.; Dong, H.; Hou, H.; Zhang, X. Distribution of Phenolic Acids and Antioxidant Activities of Different Bran Fractions from Three Pigmented Wheat Varieties. J. Chem. 2018, 2018, 6459243. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, A.; Narayanan, N.; Roychoudhury, P.K. L (+) lactic acid fermentation and its product polymerization. Electron. J. Biotechnol. 2004, 7, 167–178. [Google Scholar] [CrossRef]
- Kung, L., Jr.; Shaver, R.D.; Grant, R.J.; Schmidt, R.J. Silage review: Interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 2018, 101, 4020–4033. [Google Scholar] [CrossRef]
- Soundharrajan, I.; Park, H.S.; Rengasamy, S.; Sivanesan, R.; Choi, K.C. Application and Future Prospective of Lactic Acid Bacteria as Natural Additives for Silage Production—A Review. Appl. Sci. 2021, 11, 8127. [Google Scholar] [CrossRef]
- Agyirifo, D.S.; Wamalwa, M.; Otwe, E.P.; Galyuon, I.; Runo, S.; Takrama, J.; Ngeranwa, J. Metagenomics analysis of cocoa bean fermentation microbiome identifying species diversity and putative functional capabilities. Heliyon 2019, 5, e02170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pot, B.; Salvetti, E.; Mattarelli, P.; Felis, G.E. The potential impact of the Lactobacillus name change: The results of an expert meeting organised by the Lactic Acid Bacteria Industrial Platform (LABIP). Trends Food Sci. Technol. 2019, 94, 105–113. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Govea, F.E.; Muck, R.E.; Broderick, G.A.; Weimer, P.J. Lactobacillus plantarum effects on silage fermentation and in vitro microbial yield. Anim. Feed Sci. Technol. 2013, 179, 61–68. [Google Scholar] [CrossRef]
- Ekici, K.; Omer, A. Biogenic amines formation and their importance in fermented foods. BIO Web Conf. 2020, 17, 00232. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Capillas, C.; Herrero, A.M. Impact of Biogenic Amines on Food Quality and Safety. Foods 2019, 8, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özogul, F.; Hamed, I. The importance of lactic acid bacteria for the prevention of bacterial growth and their biogenic amines formation: A review. Crit. Rev. Food Sci. Nutr. 2018, 58, 1660–1670. [Google Scholar] [CrossRef] [PubMed]
- García, C.; Rendueles, M.; Díaz, M. Liquid-phase food fermentations with microbial consortia involving lactic acid bacteria: A review. Food Res. Int. 2019, 119, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef]
- Agarussi, M.C.N.; Pereira, O.G.; De Paula, R.A.; Da Silva, V.P.; Roseira, J.P.S.; Silva, F.F.E. Novel lactic acid bacteria strains as inoculants on alfalfa silage fermentation. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Vadopalas, L.; Zokaityte, E.; Zavistanaviciute, P.; Gruzauskas, R.; Starkute, V.; Mockus, E.; Klementaviciute, J.; Ruzauskas, M.; Lele, V.; Cernauskas, D.; et al. Supplement Based on Fermented Milk Permeate for Feeding Newborn Calves: Influence on Blood, Growth Performance, and Faecal Parameters, including Microbiota, Volatile Compounds, and Fatty and Organic Acid Profiles. Animals 2021, 11, 2544. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, T.; Tang, H.; Li, X.; Chen, Y.; Zhang, L.; Zhang, J. Probiotic potential and amylolytic properties of lactic acid bacteria isolated from Chinese fermented cereal foods. Food Control 2020, 111, 107057. [Google Scholar] [CrossRef]
- Gardini, F.; Özogul, Y.; Suzzi, G.; Tabanelli, G.; Özogul, F. Technological Factors Affecting Biogenic Amine Content in Foods: A Review. Front. Microbiol. 2016, 7, 1218. [Google Scholar] [CrossRef] [Green Version]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, J.; Skrede, A.; Mydland, L.T.; Øverland, M. Fractionation of rapeseed meal by milling, sieving and air classification—Effect on crude protein, amino acids and fiber content and digestibility. Anim. Feed Sci. Technol. 2017, 230, 143–153. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Esparza, N.C.; Latorre-Moratalla, M.L.; Comas-Basté, O.; Toro-Funes, N.; Veciana-Nogués, M.T.; Vidal-Carou, M.C. Polyamines in Food. Front. Nutr. 2019, 6, 108. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.; Hummel, J.; Kramer, E.; Hünerberg, M. Fermentation of liquid feed with lactic acid bacteria reduces dry matter losses, lysine breakdown, formation of biogenic amines, and phytate-phosphorus. Transl. Anim. Sci. 2022, 6, txac007. [Google Scholar] [CrossRef] [PubMed]
- Canibe, N.; Højberg, O.; Badsberg, J.H.; Jensen, B.B. Effect of feeding fermented liquid feed and fermented grain on gastrointestinal ecology and growth performance in piglets. J. Anim. Sci. 2007, 85, 2959–2971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olajugbagbe, T.E.; Odukoya, S.O.A.; Omafuvbe, B.O. Evaluation of the Effects of Pediococcus acidilactici Isolated from Wara, a Nigerian Milk Product, in the Prevention of Diarrhea and the Modulation of Intestinal Microflora in Wistar Rats. Asian J. Med. Health 2020, 18, 94–106. [Google Scholar] [CrossRef]


| Pearson Correlation (r) and Correlation Significance (p) | Tryptamine | Phenylethylamine | Putrescine | Cadaverine | Histamine | Tyramine | Spermidine | |
|---|---|---|---|---|---|---|---|---|
| D(–), g/100 g | r | −0.055 | 0.790 ** | 0.766 ** | −0.002 | 0.003 | 0.001 | −0.900 ** |
| p | 0.828 | 0.0001 | 0.0001 | 0.993 | 0.991 | 0.998 | 0.0001 | |
| L(+), g/100 g | r | −0.015 | 0.737 ** | 0.796 ** | 0.065 | 0.072 | 0.068 | −0.897 ** |
| p | 0.954 | 0.0001 | 0.0001 | 0.798 | 0.778 | 0.789 | 0.0001 | |
| Total lactic acid content, g/100 g | r | −0.034 | 0.757 ** | 0.772 ** | 0.029 | 0.036 | 0.036 | −0.888 ** |
| p | 0.893 | 0.0001 | 0.0001 | 0.908 | 0.888 | 0.888 | 0.0001 | |
| pH | r | −0.125 | −0.752 ** | −0.872 ** | −0.191 | −0.194 | −0.191 | 0.992 ** |
| p | 0.620 | 0.0001 | 0.0001 | 0.449 | 0.442 | 0.449 | 0.0001 | |
| LAB count, log10 CFU/g | r | −0.083 | 0.768 ** | 0.690 ** | 0.022 | 0.032 | 0.028 | −0.909 ** |
| p | 0.744 | 0.0001 | 0.002 | 0.930 | 0.899 | 0.913 | 0.0001 | |
| TEC count, log10 CFU/g | r | 0.565 * | −0.763 ** | −0.280 | 0.483 * | 0.471 * | 0.486 * | 0.559 * |
| p | 0.014 | 0.0001 | 0.261 | 0.042 | 0.048 | 0.041 | 0.016 | |
| TBC count, log10 CFU/g | r | −0.046 | 0.609 ** | 0.656 ** | 0.144 | 0.160 | 0.151 | −0.868 ** |
| p | 0.855 | 0.007 | 0.003 | 0.569 | 0.527 | 0.549 | 0.0001 | |
| M/Y count, log10 CFU/g | r | 0.120 | 0.649 ** | 0.821 ** | 0.250 | 0.259 | 0.255 | −0.973 ** |
| p | 0.635 | 0.004 | 0.0001 | 0.318 | 0.300 | 0.308 | 0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartkiene, E.; Gruzauskas, R.; Ruzauskas, M.; Zokaityte, E.; Starkute, V.; Klupsaite, D.; Vadopalas, L.; Badaras, S.; Özogul, F. Changes in the Microbial Community and Biogenic Amine Content in Rapeseed Meal during Fermentation with an Antimicrobial Combination of Lactic Acid Bacteria Strains. Fermentation 2022, 8, 136. https://doi.org/10.3390/fermentation8040136
Bartkiene E, Gruzauskas R, Ruzauskas M, Zokaityte E, Starkute V, Klupsaite D, Vadopalas L, Badaras S, Özogul F. Changes in the Microbial Community and Biogenic Amine Content in Rapeseed Meal during Fermentation with an Antimicrobial Combination of Lactic Acid Bacteria Strains. Fermentation. 2022; 8(4):136. https://doi.org/10.3390/fermentation8040136
Chicago/Turabian StyleBartkiene, Elena, Romas Gruzauskas, Modestas Ruzauskas, Egle Zokaityte, Vytaute Starkute, Dovile Klupsaite, Laurynas Vadopalas, Sarunas Badaras, and Fatih Özogul. 2022. "Changes in the Microbial Community and Biogenic Amine Content in Rapeseed Meal during Fermentation with an Antimicrobial Combination of Lactic Acid Bacteria Strains" Fermentation 8, no. 4: 136. https://doi.org/10.3390/fermentation8040136
APA StyleBartkiene, E., Gruzauskas, R., Ruzauskas, M., Zokaityte, E., Starkute, V., Klupsaite, D., Vadopalas, L., Badaras, S., & Özogul, F. (2022). Changes in the Microbial Community and Biogenic Amine Content in Rapeseed Meal during Fermentation with an Antimicrobial Combination of Lactic Acid Bacteria Strains. Fermentation, 8(4), 136. https://doi.org/10.3390/fermentation8040136










