The Bacterial and Fungi Microbiota of Soy Sauce-Supplied Lactic Acid Bacteria Treated with High-Pressure Process
Abstract
:1. Introduction
2. Materials and Methods
2.1. Koji Fermentation
2.2. Microbiota Assay
2.3. Statistical Analysis
3. Results and Discussion
3.1. Bacterial Diversity in Soy Sauce after High-Salt (HS), Low-Salt (LS), and Low-Salt High Pressure (LS-HP)
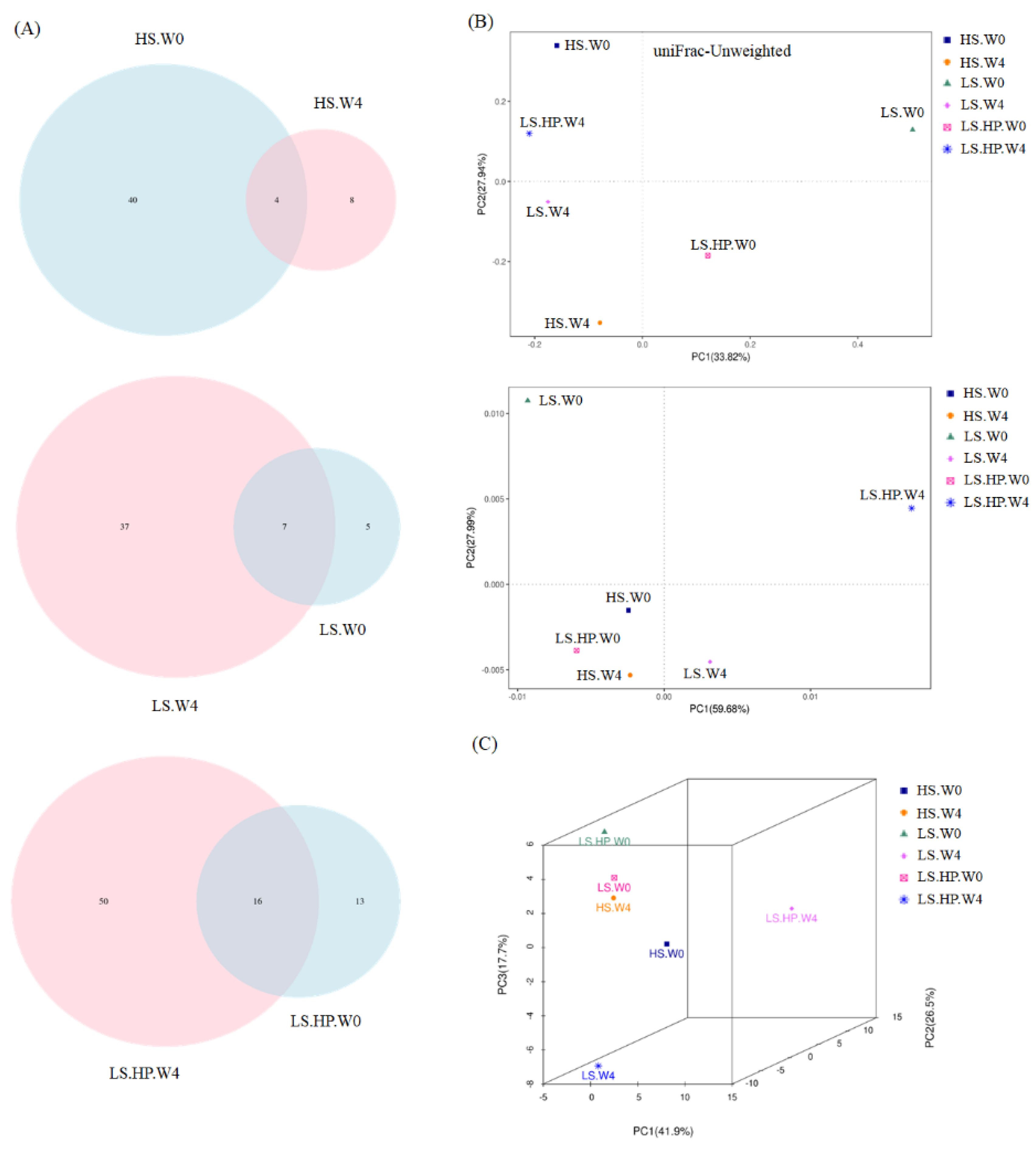
3.2. Bacterial Variation in HS, LS, and LS-HP-Treated Soy Sauce
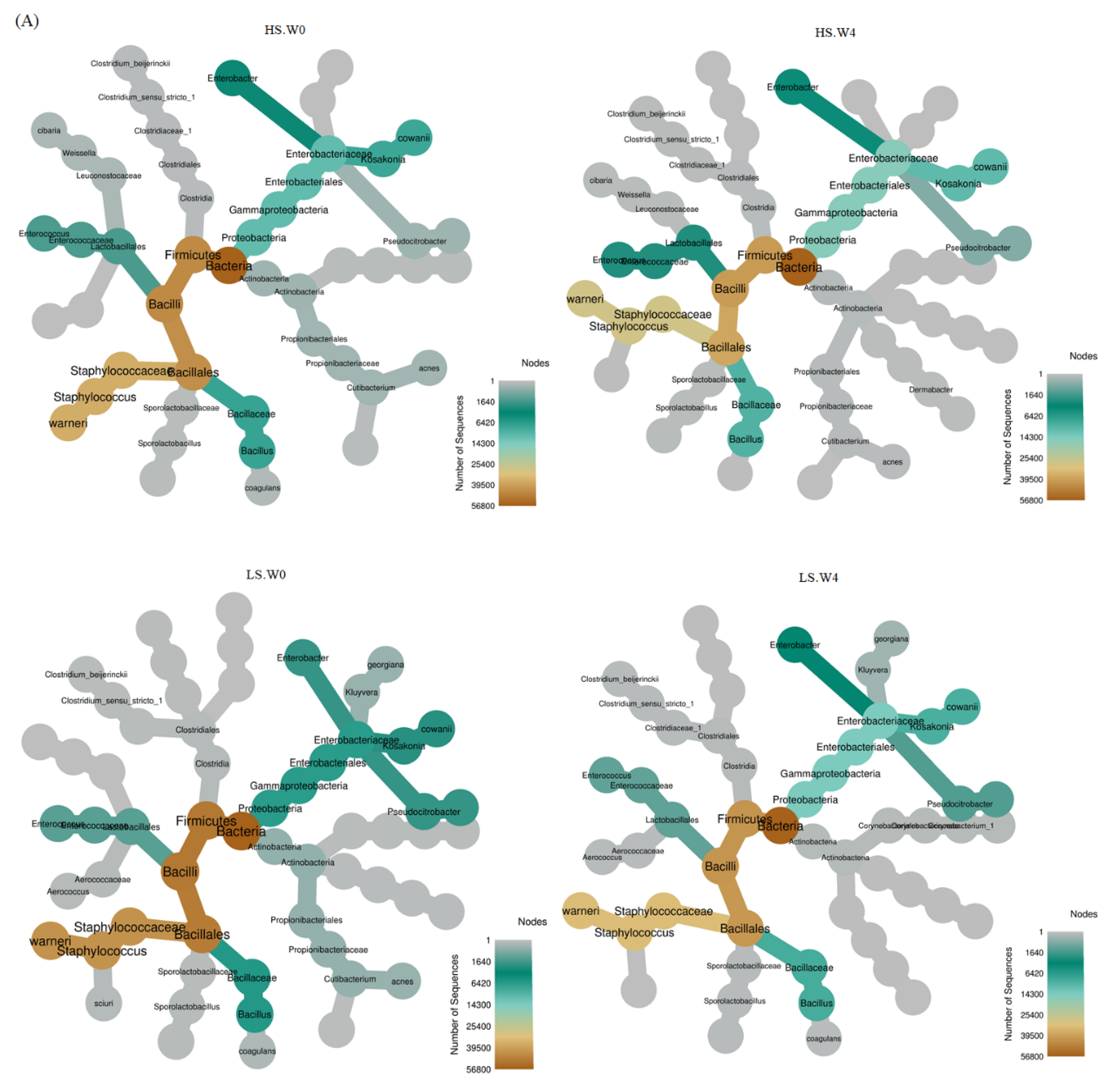
3.3. Bacterial Abundance in HS, LS, and LS-HP-Treated Soy Sauce
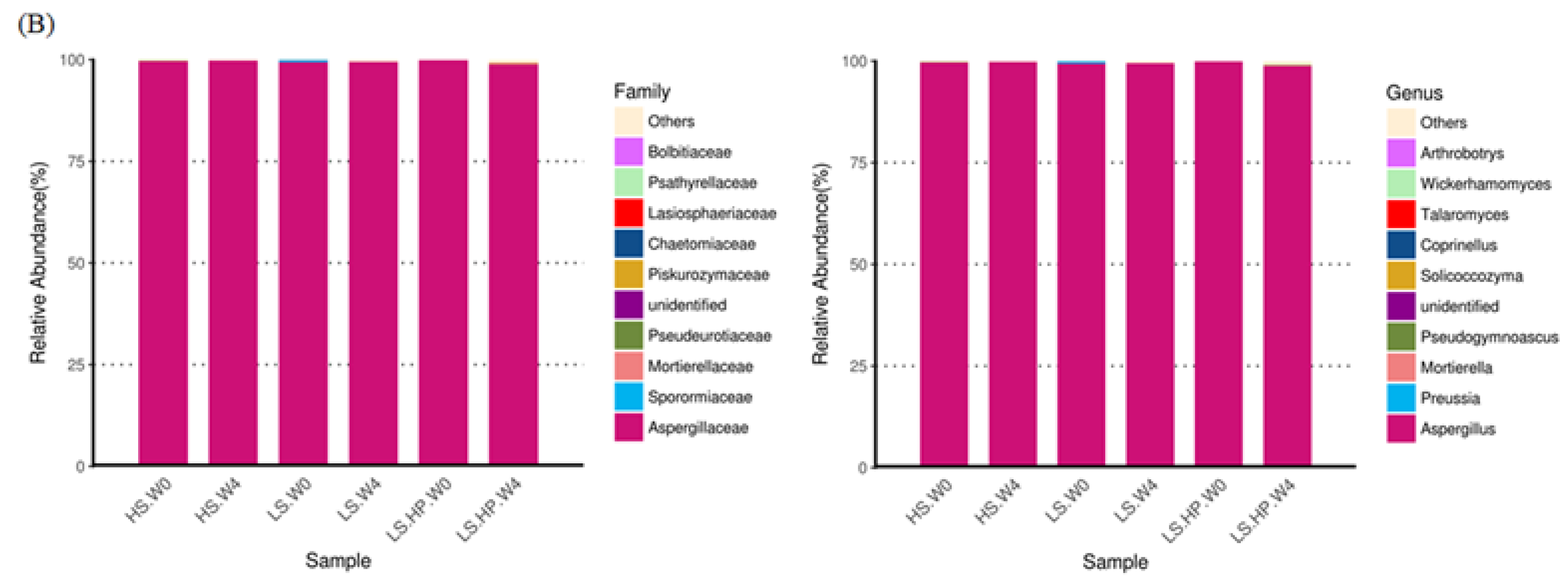
3.4. Fungal Diversity and Variation in HS, LS, and LS-HP-Treated Soil Sauce
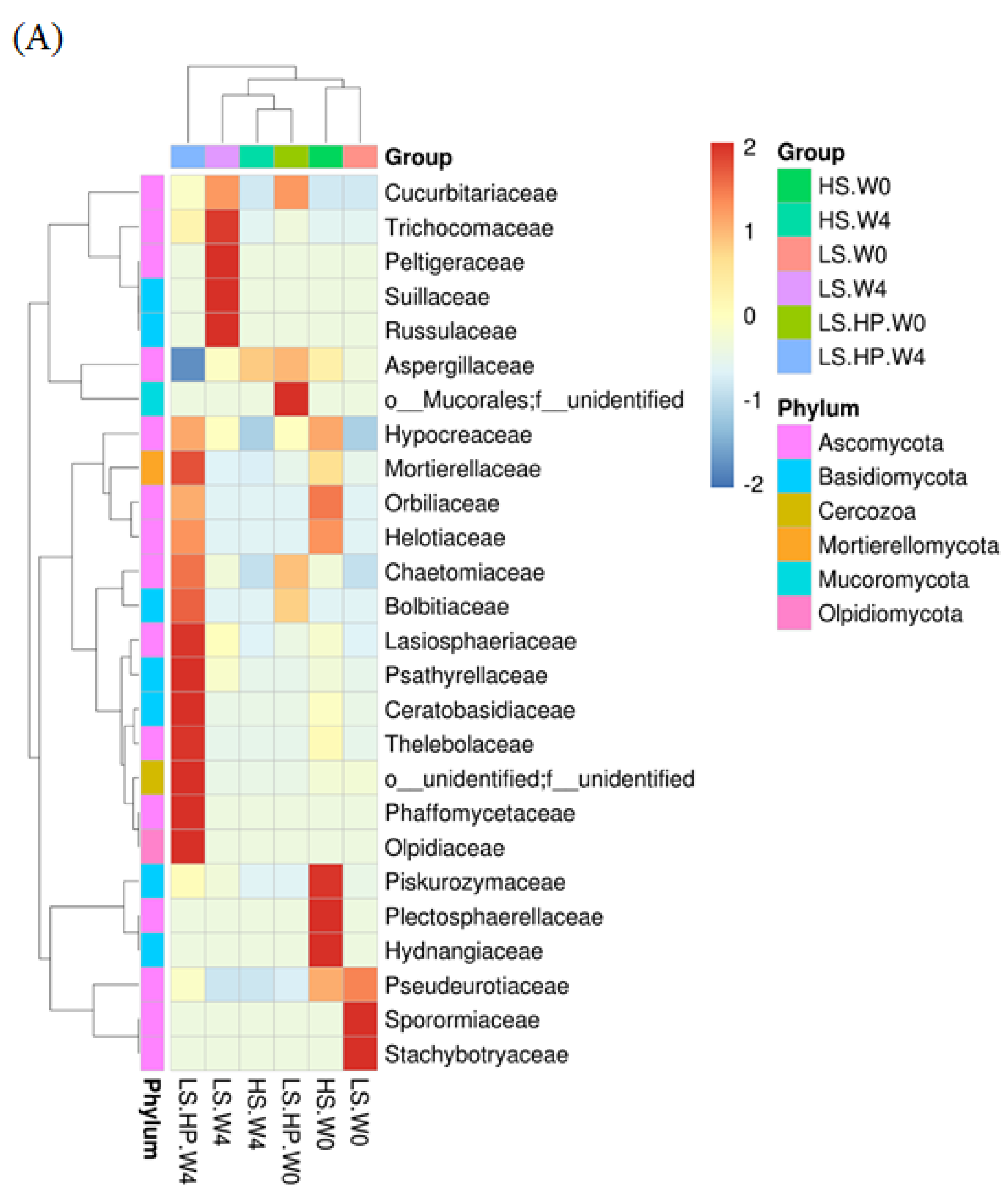
3.5. Heat Map of Fungal Community in HS, LS, and LS-HP Soy Sauce
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wicklow, D.T.; McAlpin, C.E.; Yeoh, Q.L. Diversity of Aspergillus oryzae genotypes (RFLP) isolated from traditional soy sauce production within Malaysia and Southeast Asia. Mycoscience 2007, 48, 373–380. [Google Scholar] [CrossRef]
- Luh, B.S. Industrial production of Soy sauce. J. Ind. Microbiol. 1995, 14, 467–471. [Google Scholar] [CrossRef]
- Yang, L.; Yang, H.L.; Tu, Z.C.; Wang, X.L. High-throughput sequencing of microbial community diversity and dynamics during douchi fermentation. PLoS ONE 2016, 11, e0168166. [Google Scholar] [CrossRef] [Green Version]
- Beland, F.A.; Benson, R.W.; Mellick, P.W.; Kovatch, R.M.; Roberts, D.W.; Fang, J.L.; Doerge, D.R. Effect of ethanol on the tumorigenicity of urethane (ethyl carbamate) in B6C3F1 mice. Food Chem. Toxicol. 2005, 43, 1–19. [Google Scholar] [CrossRef]
- Riachi, L.G.; Santos, A.; Moreira, R.F.; De Maria, C.A. A review of ethyl carbamate and polycyclic aromatic hydrocarbon contamination risk in cachaça and other Brazilian sugarcane spirits. Food Chem. 2014, 149, 159–169. [Google Scholar] [CrossRef]
- Chen, D.W.; Ren, Y.P.; Zhong, Q.D.; Shao, Y.; Zhao, Y.F.; Wu, Y.N. Ethyl carbamate in alcoholic beverages from China: Levels, dietary intake, and risk assessment. Food Control 2017, 72, 283–288. [Google Scholar] [CrossRef]
- Hasnip, S.; Crews, C.; Potter, N.; Christy, J.; Chan, D.; Bondu, T.; Matthews, W.; Walters, B.; Patel, K. Survey of ethyl carbamate in fermented foods sold in the United Kingdom in 2004. J. Agric. Food Chem. 2007, 55, 2755–2759. [Google Scholar] [CrossRef]
- Jiao, Z.; Dong, Y.; Chen, Q. Ethyl carbamate in fermented beverages: Presence, analytical chemistry, formation mechanism, and mitigation proposals. Compr. Rev. Food Sci. Food Saf. 2014, 13, 611–626. [Google Scholar] [CrossRef]
- Ryu, D.; Choi, B.; Kim, E.; Park, S.; Paeng, H.; Kim, C.I.; Lee, J.Y.; Yoon, H.J.; Koh, E. Determination of ethyl carbamate in alcoholic beverages and fermented foods sold in Korea. Toxicol. Res. 2015, 31, 289–297. [Google Scholar] [CrossRef]
- Weber, J.V.; Sharypov, V.I. Ethyl carbamate in foods and beverages—A review. In Climate Change, Intercropping, Pest Control and Beneficial Microorganisms; Lichtfouse, E., Ed.; Springer: Dordrecht, The Netherlands, 2009; pp. 429–452. [Google Scholar]
- Wu, P.G.; Pan, X.D.; Wang, L.Y.; Shen, X.H.; Yang, D.J. A survey of ethyl carbamate in fermented foods and beverages from Zhejiang, China. Food Control 2012, 23, 286–288. [Google Scholar] [CrossRef]
- Zhao, X.R.; Du, G.C.; Zou, H.J.; Fu, J.W.; Zhou, J.W.; Chen, J. Progress in preventing the accumulation of ethyl carbamate in alcoholic beverages. Trends Food Sci. Technol. 2013, 32, 97–107. [Google Scholar] [CrossRef]
- Matsudo, T.; Aoki, T.; Abe, K.; Fukuta, N.; Higuchi, T.; Sasaki, M.; Uchida, K. Determination of ethyl carbamate in soy-sauce and its possible precursor. J. Agric. Food Chem. 1993, 41, 352–356. [Google Scholar] [CrossRef]
- Zhang, J.R.; Fang, F.; Chen, J.; Du, G.C. The arginine deiminase pathway of koji bacteria is involved in ethyl carbamate precursor production in soy sauce. FEMS Microbiol. Lett. 2014, 358, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betts, G.D.; Linton, P.; Betteridge, R.J. Food spoilage yeasts: Effects of pH, NaCl and temperature on growth. Food Control 1999, 10, 27–33. [Google Scholar] [CrossRef]
- Tanaka, Y.; Watanabe, J.; Mogi, Y. Monitoring of the microbial communities involved in the soy sauce manufacturing process by PCR-denaturing gradient gel electrophoresis. Food Microbiol. 2012, 31, 100–106. [Google Scholar] [CrossRef]
- Wei, Q.; Wang, H.; Chen, Z.; Lv, Z.; Xie, Y.; Lu, F. Profiling of dynamic changes in the microbial community during the soy sauce fermentation process. Appl. Microbiol. Biotechnol. 2013, 97, 9111–9119. [Google Scholar] [CrossRef]
- Fang, F.; Zhang, J.; Zhou, J.; Zhou, Z.; Li, T.; Lu, L.; Zeng, W.; Du, G.; Chen, J. Accumulation of citrulline by microbial arginine metabolism during alcoholic fermentation of soy sauce. J. Agric. Food Chem. 2018, 66, 2108–2113. [Google Scholar] [CrossRef]
- Yamamoto, K. Food processing by high hydrostatic pressure. Biosci. Biotechnol. Biochem. 2017, 81, 672–679. [Google Scholar] [CrossRef] [Green Version]
- Del Arbol, J.T.; Pulido, R.P.; La Storia, A.; Burgos, M.J.G.; Lucas, R.; Ercolini, D.; Galvez, A. Changes in microbial diversity of brined green asparagus upon treatment with high hydrostatic pressure. Int. J. Food Microbiol. 2016, 216, 1–8. [Google Scholar] [CrossRef]
- Pulido, R.P.; Burgos, M.J.G.; Galvez, A.; Lucas, R. Changes in bacterial diversity of refrigerated mango pulp before and after treatment by high hydrostatic pressure. LWT Food Sci. Technol. 2017, 78, 289–295. [Google Scholar] [CrossRef]
- Liang, R.; Huang, J.; Wu, X.M.; Xu, Y.; Fan, J.; Wu, C.D.; Jin, Y.; Zhou, R. Characterizing the metabolites and the microbial communities of the soy sauce mash affected by temperature and hydrostatic pressure. Food Res. Int. 2019, 123, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Qian, M.; Shen, Y.; Qin, X.; Huang, H.; Yang, H.; He, Y.; Bai, W. A high-throughput sequencing approach to the preliminary analysis of bacterial communities associated with changes in amino acid nitrogen, organic acid and reducing sugar contents during soy sauce fermentation. Food Chem. 2021, 349, 129131. [Google Scholar] [CrossRef] [PubMed]
- Foster, Z.S.; Sharpton, T.J.; Grunwald, N.J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017, 13, e1005404. [Google Scholar] [CrossRef] [Green Version]
- Sergelidis, D.; Abrahim, A.; Papadopoulos, T.; Soultos, N.; Martziou, E.; Koulourida, V.; Govaris, A.; Pexara, A.; Zdragas, A.; Papa, A. Isolation of methicillin-resistant Staphylococcus spp. from ready-to-eat fish products. Lett. Appl. Microbiol. 2014, 59, 500–506. [Google Scholar] [CrossRef]
- Yan, Y.Z.; Qian, Y.L.; Ji, F.D.; Chen, J.Y.; Han, B.Z. Microbial composition during Chinese soy sauce koji-making based on culture dependent and independent methods. Food Microbiol. 2013, 34, 189–195. [Google Scholar] [CrossRef]
- Han, D.M.; Chun, B.H.; Feng, T.; Kim, H.M.; Jeon, C.O. Dynamics of microbial communities and metabolites in ganjang, a traditional Korean fermented soy sauce, during fermentation. Food Microbiol. 2020, 92, 103591. [Google Scholar] [CrossRef]
- Mota, M.J.; Lopes, R.P.; Sousa, S.; Gomes, A.M.; Delgadillo, I.; Saraiva, J.A. Lactobacillus reuteri growth and fermentation under high pressure towards the production of 1,3-propanediol. Food Res. Int. 2018, 113, 424–432. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, Y.; Zhao, L.; Wang, Y.T.; Rao, L.; Liao, X.J. Pressure-resistant acclimation of lactic acid bacteria from a natural fermentation product using high pressure. Innov. Food Sci. Emerg. Technol. 2021, 69, 102660. [Google Scholar] [CrossRef]
- Qi, Q.; Huang, J.; Zhou, R.Q.; Yang, M.L.; Zhang, L.; Peng, C.; Jin, Y.; Wu, C.D. Characterizing microbial community and metabolites of Cantonese soy sauce. Food Biosci. 2021, 40, 100872. [Google Scholar] [CrossRef]
- Wei, C.L.; Chao, S.H.; Tsai, W.B.; Lee, P.S.; Tsau, N.H.; Chen, J.S.; Lai, W.L.; Tu, J.C.Y.; Tsai, Y.C. Analysis of bacterial diversity during the fermentation of inyu, a high-temperature fermented soy sauce, using nested PCR-denaturing gradient gel electrophoresis and the plate count method. Food Microbiol. 2013, 33, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Del Arbol, J.T.; Pulido, R.P.; La Storia, A.; Burgos, M.J.G.; Lucas, R.; Ercolini, D.; Galvez, A. Microbial diversity in pitted sweet cherries (Prunus avium L.) as affected by high-hydrostatic pressure treatment. Food Res. Int. 2016, 89, 790–796. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.-C.; Lai, C.-Y.; Lee, B.-H.; Wu, S.-C. The Bacterial and Fungi Microbiota of Soy Sauce-Supplied Lactic Acid Bacteria Treated with High-Pressure Process. Fermentation 2022, 8, 97. https://doi.org/10.3390/fermentation8030097
Shi Y-C, Lai C-Y, Lee B-H, Wu S-C. The Bacterial and Fungi Microbiota of Soy Sauce-Supplied Lactic Acid Bacteria Treated with High-Pressure Process. Fermentation. 2022; 8(3):97. https://doi.org/10.3390/fermentation8030097
Chicago/Turabian StyleShi, Yeu-Ching, Chiung-Yu Lai, Bao-Hong Lee, and She-Ching Wu. 2022. "The Bacterial and Fungi Microbiota of Soy Sauce-Supplied Lactic Acid Bacteria Treated with High-Pressure Process" Fermentation 8, no. 3: 97. https://doi.org/10.3390/fermentation8030097
APA StyleShi, Y.-C., Lai, C.-Y., Lee, B.-H., & Wu, S.-C. (2022). The Bacterial and Fungi Microbiota of Soy Sauce-Supplied Lactic Acid Bacteria Treated with High-Pressure Process. Fermentation, 8(3), 97. https://doi.org/10.3390/fermentation8030097






