Increasing Acid Tolerance of an Engineered Lactic Acid Bacterium Pediococcus acidilactici for L-Lactic Acid Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Reagents and Enzymes
2.3. Raw Material and Its Biorefinery Processing
2.4. Plasmid Construction
2.5. Adaptive Evolution
2.6. L-Lactic Acid Fermentation
2.7. Analysis
3. Results and Discussions
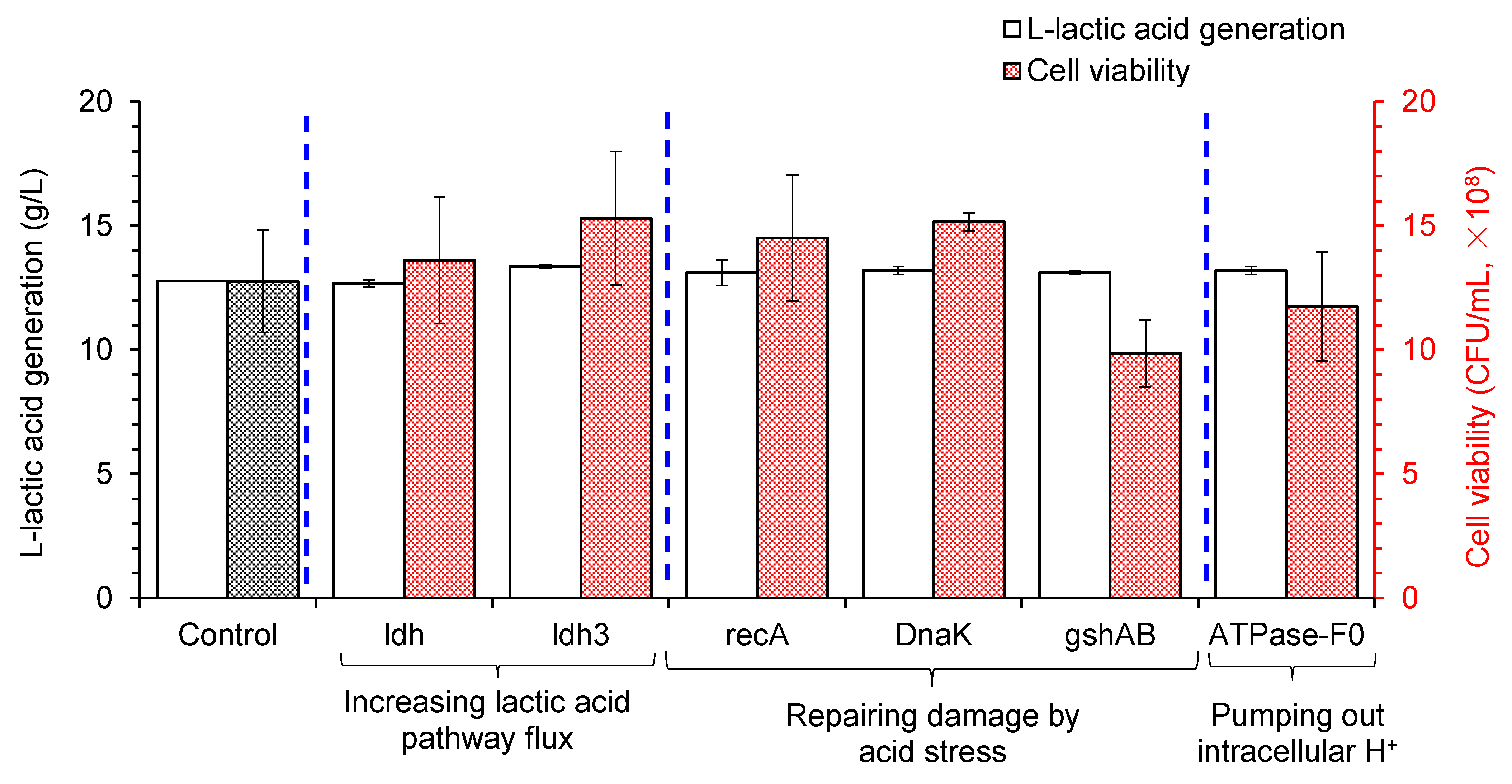
3.1. Metabolic Modification of LAB to Improve Acid Tolerance
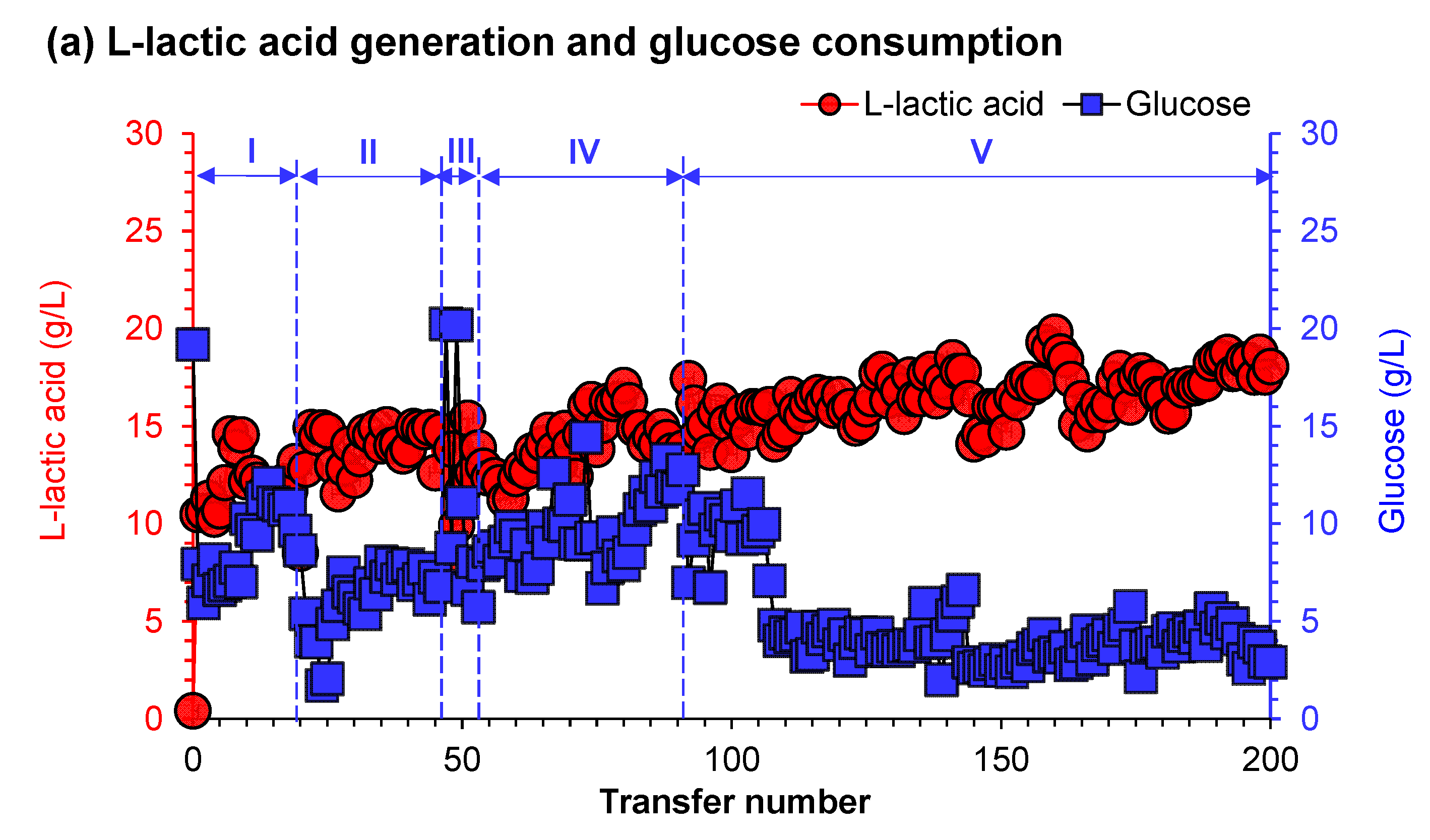
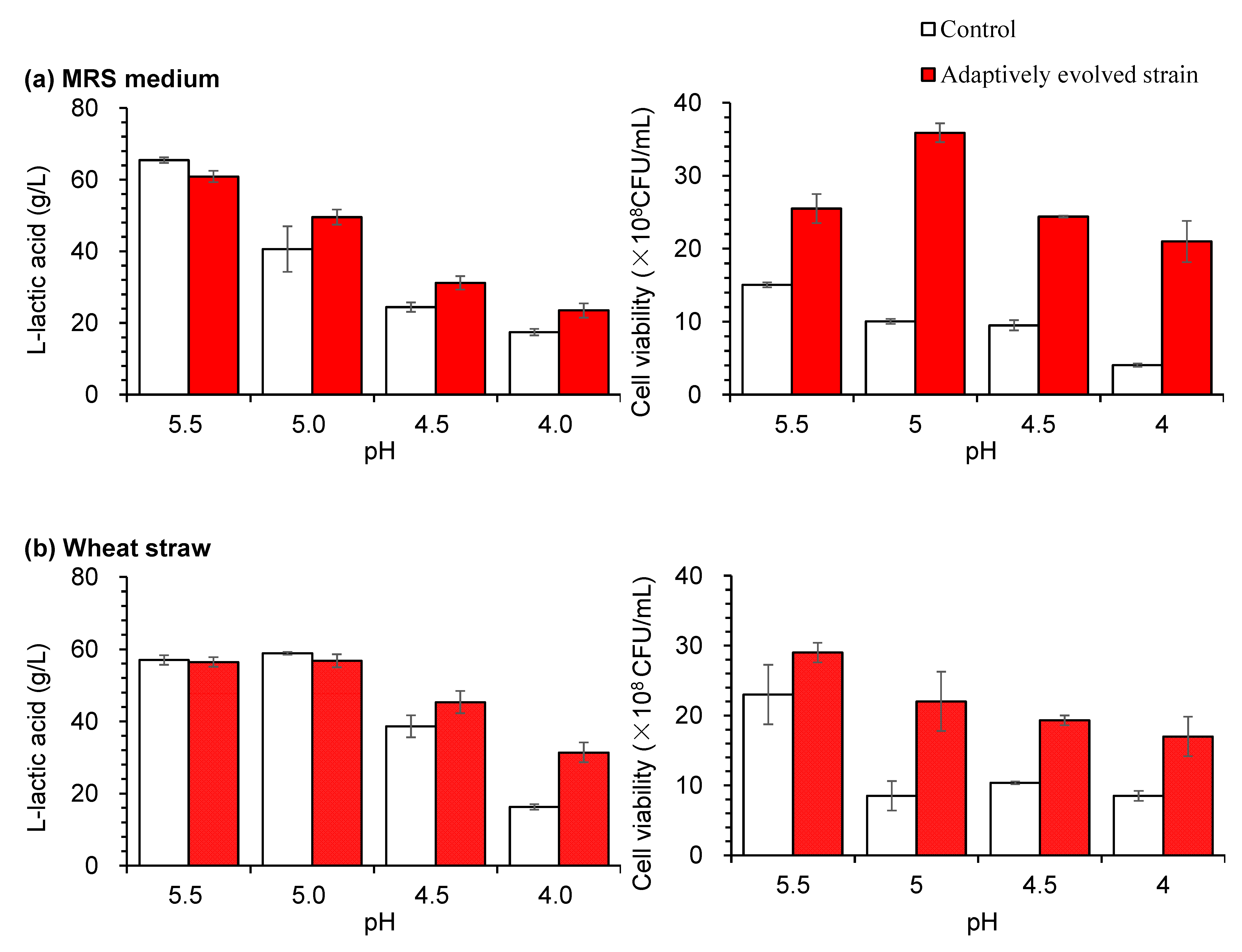
3.2. Low pH Adaptive Evolution of LAB and Fermentation Evaluation
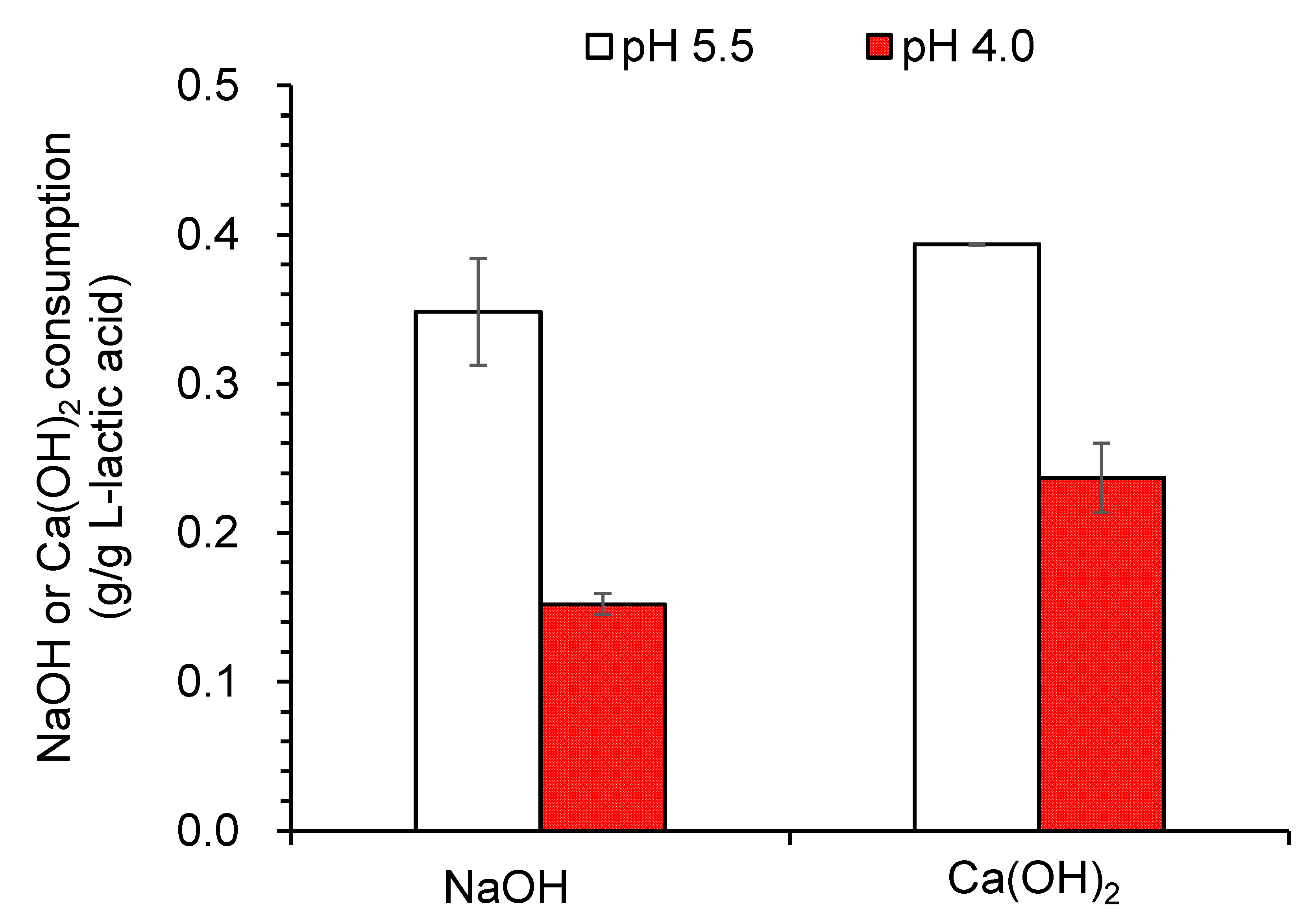
3.3. Neutralizing Agents Reduction for L-Lactic Acid Fermentation under Low pH
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reddy, G.; Altaf, M.; Naveena, B.J.; Venkateshwar, M.; Kumar, E.V. Amylolytic bacterial lactic acid fermentation—A review. Biotechnol. Adv. 2008, 26, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, M.A.; Sonomoto, K. Opportunities to overcome the current limitations and challenges for efficient microbial production of optically pure lactic acid. J. Biotechnol. 2016, 236, 176–192. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, R.A.; Komesu, A.; Rossell, C.E.V.; Maciel, R. Challenges and opportunities in lactic acid bioprocess design—From economic to production aspects. Biochem. Eng. J. 2018, 133, 219–239. [Google Scholar] [CrossRef]
- Kumar, A.; Thakur, A.; Panesar, P.S. Lactic acid and its separation and purification techniques: A review. Rev. Environ. Sci. Biotechnol. 2019, 18, 823–853. [Google Scholar] [CrossRef]
- Komesu, A.; de Oliveira, J.A.R.; Martins, L.H.D.; Maciel, M.R.W.; Maciel, R. Lactic acid production to purification: A review. BioResources 2017, 12, 4364–4383. [Google Scholar] [CrossRef]
- Ye, L.D.; Zhao, H.; Li, Z.; Wu, J.C. Improved acid tolerance of Lactobacillus pentosus by error-prone whole genome amplification. Bioresour. Technol. 2013, 135, 459–463. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C. Surviving the acid test: Responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 2003, 67, 429–453. [Google Scholar] [CrossRef] [Green Version]
- van de Guchte, M.; Serror, P.; Chervaux, C.; Smokvina, T.; Ehrlich, S.D.; Maguin, E. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 2002, 82, 187–216. [Google Scholar] [CrossRef]
- Trip, H.; Mulder, N.L.; Lolkema, J.S. Improved acid stress survival of Lactococcus lactis expressing the histidine decarboxylation pathway of Streptococcus thermophilus CHCC1524. J. Biol. Chem. 2012, 287, 11195–11204. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, A.L.; Cardoso, F.S.; Bohn, A.; Neves, A.R.; Santos, H. Engineering trehalose synthesis in Lactococcus lactis for improved stress tolerance. Appl. Environ. Microbiol. 2011, 77, 4189–4199. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wu, C.D.; Du, G.C.; Chen, J. Enhanced acid tolerance in Lactobacillus casei by adaptive evolution and compared stress response during acid stress. Biotechnol. Bioprocess Eng. 2012, 17, 283–289. [Google Scholar] [CrossRef]
- Cubas-Cano, E.; González-Fernández, C.; Tomás-Pejó, E. Evolutionary engineering of Lactobacillus pentosus improves lactic acid productivity from xylose-rich media at low pH. Bioresour. Technol. 2019, 288, 121540. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Zhang, P.; Sun, J.E.; Tu, Y.; Gao, Q.Q.; Zhang, J.; Bao, J. Engineering wild-type robust Pediococcus acidilactici strain for high titer L- and D-lactic acid production from corn stover feedstock. J. Biotechnol. 2016, 217, 112–121. [Google Scholar] [CrossRef]
- Zhao, K.; Qiao, Q.A.; Chu, D.Q.; Gu, H.Q.; Dao, T.H.; Zhang, J.; Bao, J. Simultaneous saccharification and high titer lactic acid fermentation of corn stover using a newly isolated lactic acid bacterium Pediococcus acidilactici DQ2. Bioresour. Technol. 2013, 135, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.Y.; Fang, C.; He, N.L.; Bao, J. An oxidoreductase gene ZMO1116 enhances the p-benzoquinone biodegradation and chiral lactic acid fermentability of Pediococcus acidilactici. J. Biotechnol. 2020, 323, 231–237. [Google Scholar] [CrossRef]
- Qiu, Z.Y.; Gao, Q.Q.; Bao, J. Engineering Pediococcus acidilactici with xylose assimilation pathway for high titer cellulosic L-lactic acid fermentation. Bioresour. Technol. 2018, 249, 9–15. [Google Scholar] [CrossRef]
- He, Y.Q.; Zhang, J.; Bao, J. Acceleration of biodetoxification on dilute acid pretreated lignocellulose feedstock by aeration and the consequent ethanol fermentation evaluation. Biotechnol. Biofuels 2016, 9, 19. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhu, Z.N.; Wang, X.F.; Wang, N.; Wang, W.; Bao, J. Biodetoxification of toxins generated from lignocellulose pretreatment using a newly isolated fungus Amorphotheca resinae ZN1 and the consequent ethanol fermentation. Biotechnol. Biofuels 2010, 3, 26. [Google Scholar] [CrossRef] [Green Version]
- Adney, B.; Baker, J. Measurement of Cellulase Activities; AP-006. NREL. Analytical Procedure; National Renewable Energy Laboratory: Golden, CO, USA, 1996.
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–256. [Google Scholar] [CrossRef]
- Ghose, T. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Chu, D.; He, Y.; Bao, J. Dry pretreatment of lignocellulose with extremely low steam and water usage for bioethanol production. Bioresour. Technol. 2011, 102, 4480–4488. [Google Scholar] [CrossRef] [PubMed]
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.; Crocker, D. Determination of Structural Carbohydrates and Lignin in Biomass; Technical Report NREL/TP-510-42618; National Renewable Energy Laboratory: Golden, CO, USA, 2012.
- Zhu, Z.M.; Ji, X.M.; Wu, Z.M.; Zhang, J.; Du, J.C. Improved acid-stress tolerance of Lactococcus lactis NZ9000 and Escherichia coli BL21 by overexpression of the anti-acid component recT. J. Ind. Microbiol. Biotechnol. 2018, 45, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fu, R.Y.; Hugenholtz, J.; Li, Y.; Chen, J. Glutathione protects Lactococcus lactis against acid stress. Appl. Environ. Microbiol. 2007, 73, 5268–5275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, M.; Ohishi, H.; Benno, Y. H+-ATPase activity in Bifidobacterium with special reference to acid tolerance. Int. J. Food Microbiol. 2004, 93, 109–113. [Google Scholar] [CrossRef]
- Miwa, T.; Abe, T.; Fukuda, S.; Ohkawara, S.; Hino, T. Effect of reduced H+-ATPase activity on acid tolerance in Streptococcus bovis mutants. Anaerobe 2000, 6, 197–203. [Google Scholar] [CrossRef]
- Lee, S.; Kim, P. Current status and applications of adaptive laboratory evolution in industrial microorganisms. J. Microbiol. Biotechnol. 2020, 30, 793–803. [Google Scholar] [CrossRef]
- Li, C.L.; Gao, M.; Zhu, W.B.; Wang, N.H.; Ma, X.Y.; Wu, C.F.; Wang, Q.H. Recent advances in the separation and purification of lactic acid from fermentation broth. Process Biochem. 2021, 104, 142–151. [Google Scholar] [CrossRef]

| Strains | Characteristic |
| E. coli XL1-blue | Host for plasmid construction |
| P. acidilactici ZY271 | Parental strain for L-lactic acid fermentation |
| P. acidilactici ZY271 (pZY36e) | P. acidilactici ZY271 harboring empty plasmid pZY36e |
| P. acidilactici ZY271 (pZY36e-ldh) | P. acidilactici ZY271 harboring expression plasmid pZY36e-ldh |
| P. acidilactici ZY271 (pZY36e-ldh3) | P. acidilactici ZY271 harboring expression plasmid pZY36e-ldh3 |
| P. acidilactici ZY271 (pZY36e-ATPase-F0) | P. acidilactici ZY271 harboring expression plasmid pZY36e-ATPase-F0 |
| P. acidilactici ZY271 (pZY36e-recA) | P. acidilactici ZY271 harboring expression plasmid pZY36e-recA |
| P. acidilactici ZY271 (pZY36e-DnaK) | P. acidilactici ZY271 harboring expression plasmid pZY36e-DnaK |
| P. acidilactici ZY271 (pZY36e-gshAB) | P. acidilactici ZY271 harboring expression plasmid pZY36e-gshAB |
| Plasmids | Characteristic |
| pZY36e | Expression plasmid by PldhD promotor |
| pZY36e-ldh | Plasmid for expression of ldh by PldhD promotor |
| pZY36e-ldh3 | Plasmid for expression of ldh by PldhD promotor |
| pZY36e-ATPase-F0 | Plasmid for expression of ATPase-F0 by PldhD promotor |
| pZY36e-recA | Plasmid for expression of recA by PldhD promotor |
| pZY36e-DnaK | Plasmid for expression of DnaK by PldhD promotor |
| pZY36e-gshAB | Plasmid for expression of gshAB by PldhD promotor |
| Primers | Sequence (5′-3′) |
| ldh-F | TGCTCTAGAATGTCTAATATTCAAAATCATCAAAAAGTTGT |
| ldh-R | AAAACTGCAGTTATTTGTCTTGTTTTTCAGCAAGAG |
| ldh3-F | CTAGTCTAGAATGGCAAGAGTTGAAAAACCTCGT |
| ldh3-R | ACGCGTCGACTTATTGACGAACCTTAACGCCA |
| ATPase-F0-F | TGCTCTAGAGTGGGTGGTGAATCAATTTCA |
| ATPase-F0-R | AAAACTGCAGTCATTTACTCTCACCTAAACCTTCAAT |
| DnaK-F | CTAGTCTAGAATGGCAAGTAATAAAATTATTGGTATTGAC |
| DnaK-R | ACGCGTCGACTTATTTGTTGTCTTTGTCAGGATCG |
| gshA-F | CTAGTCTAGATGCTCTGGTGTGCAGACCAGAC |
| gshA-R | ACGCGTCGACTCAGGCGTGTTTTTCCAGCCACA |
| gshB-F | ACGCGTCGACATTTGGCGATTTGGGCTAAC |
| gshB-R | AACTGCAGTTACTGCTGCTGTAAACGTG |
| recA-F | ACGCGTCGACGTGGCAGATGAAAGAAAAGAAG |
| recA-R | AAAACTGCAGTTATTTCAAGTCTAATTCAGCTTGGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Z.; Chen, M.; Jia, J.; Bao, J. Increasing Acid Tolerance of an Engineered Lactic Acid Bacterium Pediococcus acidilactici for L-Lactic Acid Production. Fermentation 2022, 8, 96. https://doi.org/10.3390/fermentation8030096
Yan Z, Chen M, Jia J, Bao J. Increasing Acid Tolerance of an Engineered Lactic Acid Bacterium Pediococcus acidilactici for L-Lactic Acid Production. Fermentation. 2022; 8(3):96. https://doi.org/10.3390/fermentation8030096
Chicago/Turabian StyleYan, Zhao, Mingxing Chen, Jia Jia, and Jie Bao. 2022. "Increasing Acid Tolerance of an Engineered Lactic Acid Bacterium Pediococcus acidilactici for L-Lactic Acid Production" Fermentation 8, no. 3: 96. https://doi.org/10.3390/fermentation8030096
APA StyleYan, Z., Chen, M., Jia, J., & Bao, J. (2022). Increasing Acid Tolerance of an Engineered Lactic Acid Bacterium Pediococcus acidilactici for L-Lactic Acid Production. Fermentation, 8(3), 96. https://doi.org/10.3390/fermentation8030096







