Optimisation of Xylanase–Pectinase Cocktail Production with Bacillus amyloliquefaciens ADI2 Using a Low-Cost Substrate via Statistical Strategy
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Suspension Preparation
2.2. Production of Enzymes
2.3. Enzymes Assay
2.4. Effect of Culture Conditions on Bacillus amyloliquefaciens ADI2 Xylano-Pectinolytic Enzymes Production
2.5. Optimisation of Xylano-Pectinolytic Enzymes Production Using Central Composite Design (CCD)
2.6. Validation of the Model
3. Results and Discussion
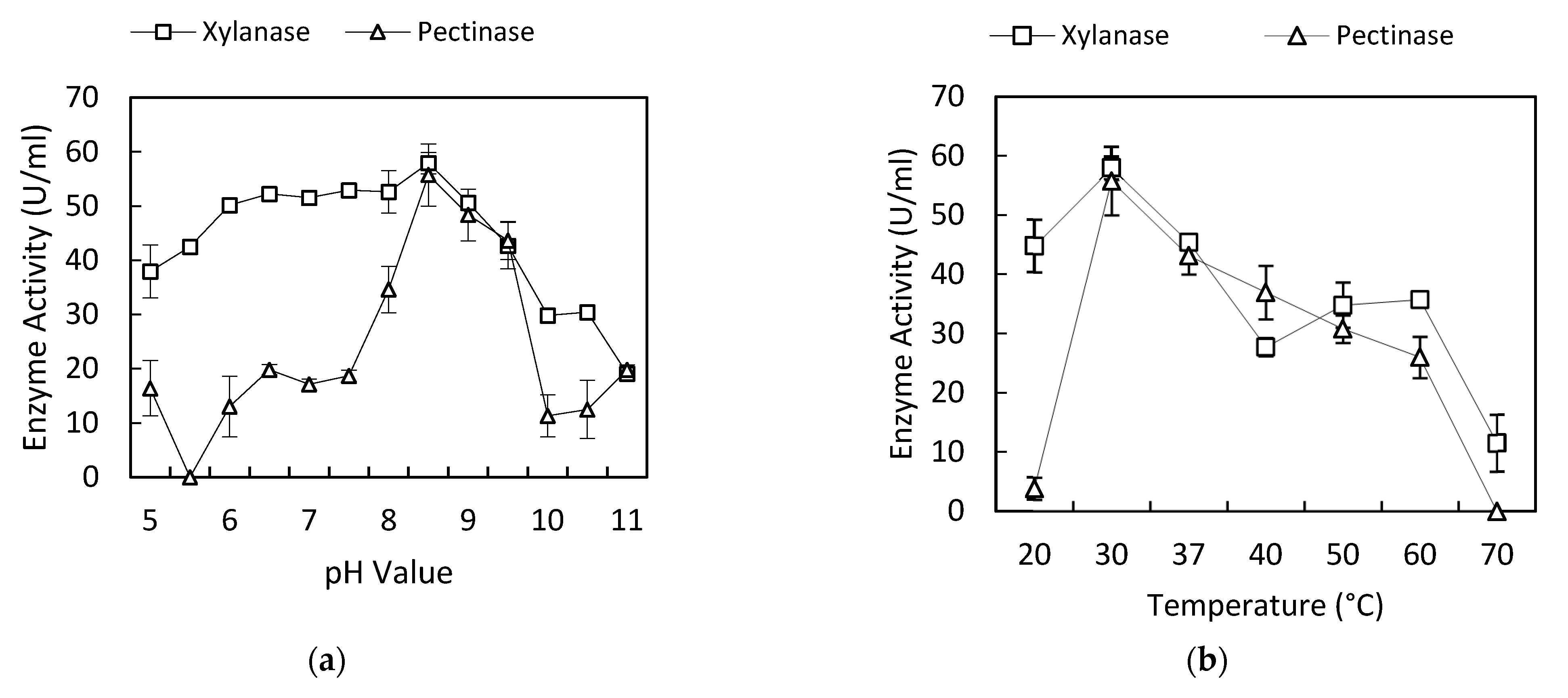
3.1. Effect of pH on Xylanase and Pectinase Production
3.2. Effect of Temperature on Xylanase and Pectinase
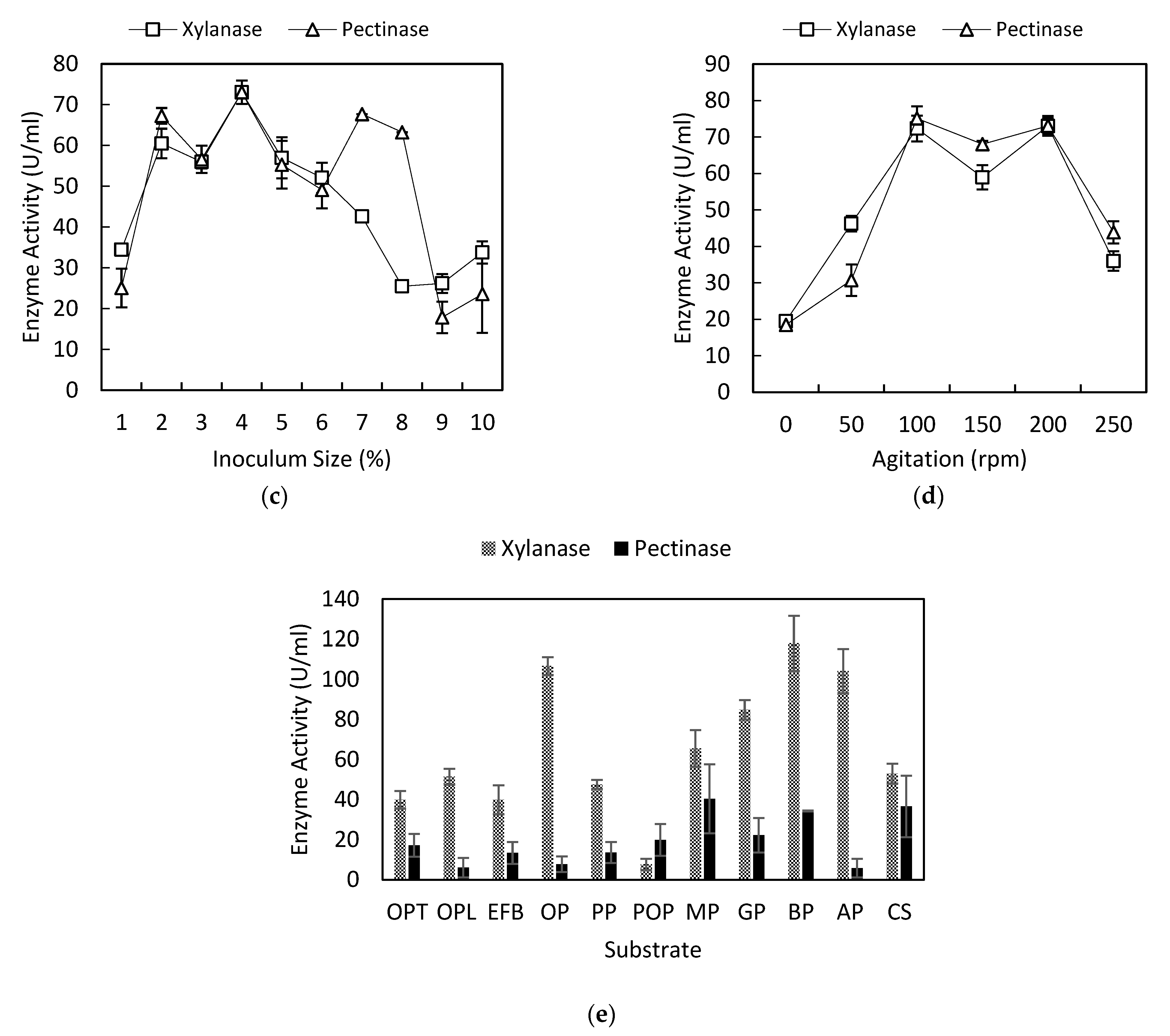
3.3. Effect of Inoculum Concentration on Xylanase and Pectinase Production
3.4. Effect of Agitation on Xylanase and Pectinase Production
3.5. Effect of the Substrate on Xylanase and Pectinase Production
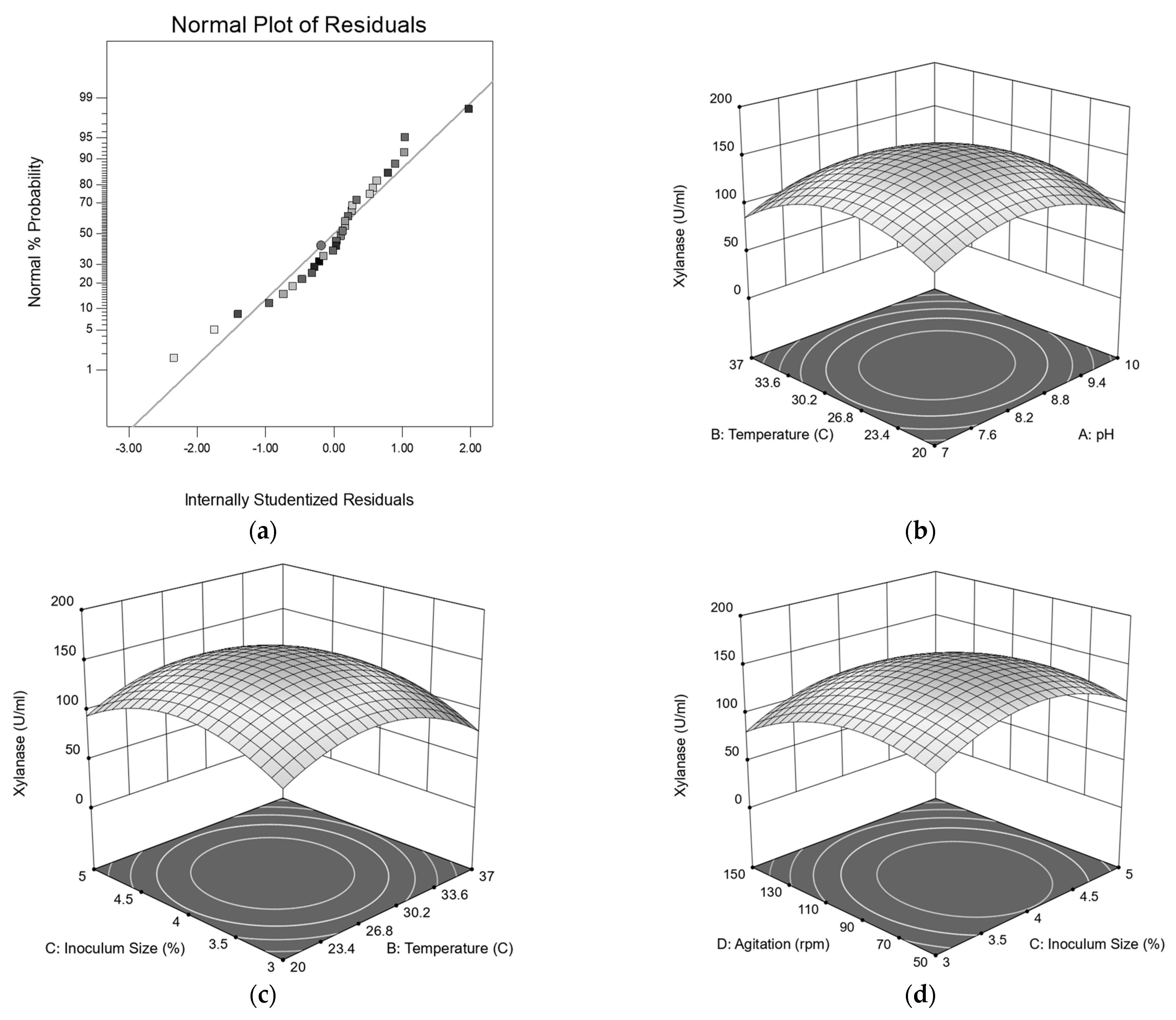
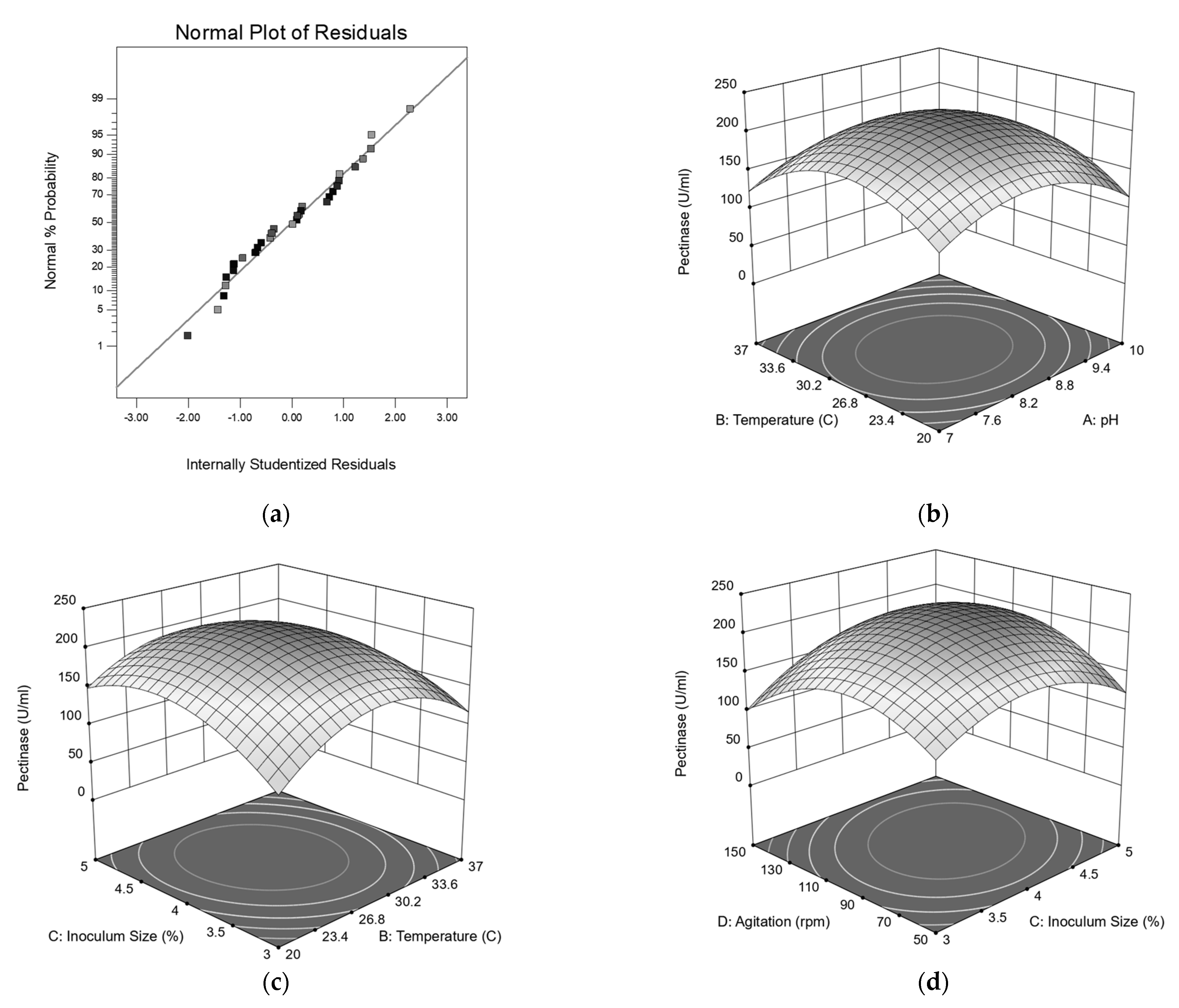
3.6. Central Composite Design Analysis
1.77AC + 5.80AD + 2.02BC + 0.21BD + 0.34CD − 30.11A2 − 33.92B2 − 31.86C2 −
25.56D2
12.15AC − 0.52AD − 10.72BC − 8.63BD + 13.66CD − 44.03A2 − 55.30B2 −
42.83C2 − 52.08D2
3.7. Validation of the Model
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of variance |
| CCD | Central composite design |
| CV | Coefficient of variation |
| OD | Optical density |
| OFAT | One factor at a time |
| RSM | Response surface methodology |
| SmF | Submerged fermentation |
| SSF | Solid-state fermentation |
References
- Silversides, F.G.; Scott, T.A.; Korver, D.R.; Afsharmanesh, M.; Hruby, M. A study on the interaction of xylanase and phytase enzymes in wheat-based diets fed to commercial white and brown egg laying hens. Poult. Sci. 2006, 85, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Romanowska, I.; Polak, J.; Bielecki, S. Isolation and properties of Aspergillus niger IBT-90 xylanase for bakery. Appl. Microbiol. Biotechnol. 2006, 69, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Selinheimo, E.; Kruus, K.; Buchert, J.; Hopia, A.; Autio, K. Effects of laccase, xylanase and their combination on the rheological properties of wheat doughs. J. Cereal Sci. 2006, 43, 152–159. [Google Scholar] [CrossRef]
- Reid, I.; Ricard, M. Pectinase in papermaking: Solving retention problems in mechanical pulps bleached with hydrogen peroxide. Enzyme Microb. Technol. 2000, 26, 115–123. [Google Scholar] [CrossRef]
- Wong, K.K.Y.; Nelson, S.L.; Saddler, J.N. Xylanase treatment for the peroxide bleaching of oxygen delignified kraft pulps derived from three softwood species. J. Biotechnol. 1996, 48, 137–145. [Google Scholar] [CrossRef]
- Garg, S. Xylanase: Applications in Biofuel Production. Curr. Metab. 2016, 4, 23–37. [Google Scholar] [CrossRef]
- Arora, N.; Banerjee, A.K.; Mutyala, S.; Murty, U.S. Comparative characterization of commercially important xylanase enzymes. Bioinformation 2009, 3, 446–453. [Google Scholar] [CrossRef]
- Iyer, P.V.; Ananthanarayan, L. Enzyme stability and stabilization-Aqueous and non-aqueous environment. Process Biochem. 2008, 43, 1019–1032. [Google Scholar] [CrossRef]
- Sujani, S.; Seresinhe, R.T. Exogenous enzymes in ruminant nutrition: A review. Asian J. Anim. Sci. 2015, 9, 85–99. [Google Scholar] [CrossRef]
- Pothiraj, C.; Kanmani, P.; Balaji, P. Bioconversion of Lignocellulose Materials. Mycobiology 2006, 34, 159. [Google Scholar] [CrossRef]
- Jecu, L. Solid state fermentation of agricultural wastes for endoglucanase production. Ind. Crops Prod. 2000, 11, 1–5. [Google Scholar] [CrossRef]
- Chahal, P.S.; Chahal, D.S.; Lê, G.B.B. Production of cellulase in solid-state fermentation with Trichoderma reesei MCG 80 on wheat straw. Appl. Biochem. Biotechnol. Part A Enzym. Eng. Biotechnol. 1996, 57, 433–442. [Google Scholar] [CrossRef]
- Haltrich, D.; Nidetzky, B.; Kulbe, K.D.; Steiner, W.; Župančič, S. Production of fungal xylanases. Bioresour. Technol. 1996, 58, 137–161. [Google Scholar] [CrossRef]
- Ertan, F.; Balkan, B.; Yarkın, Z. Determination of the effects of initial glucose on the production of α-amylase from Penicillium sp. under solid-state and submerged fermentation. Biotechnol. Biotechnol. Equip. 2014, 28, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Cunha, F.M.; Esperança, M.N.; Zangirolami, T.C.; Badino, A.C.; Farinas, C.S. Sequential solid-state and submerged cultivation of Aspergillus niger on sugarcane bagasse for the production of cellulase. Bioresour. Technol. 2012, 112, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.R.; Dayanand, A. Production of pectinase from deseeded sunflower head by Aspergillus niger in submerged and solid-state conditions. Bioresour. Technol. 2006, 97, 2054–2058. [Google Scholar] [CrossRef]
- Kar, S.; Sona Gauri, S.; Das, A.; Jana, A.; Maity, C.; Mandal, A.; Das Mohapatra, P.K.; Pati, B.R.; Mondal, K.C. Process optimization of xylanase production using cheap solid substrate by Trichoderma reesei SAF3 and study on the alteration of behavioral properties of enzyme obtained from SSF and SmF. Bioprocess Biosyst. Eng. 2013, 36, 57–68. [Google Scholar] [CrossRef]
- Hölker, U.; Höfer, M.; Lenz, J. Biotechnological advantages of laboratory-scale solid-state fermentation with fungi. Appl. Microbiol. Biotechnol. 2004, 64, 175–186. [Google Scholar] [CrossRef]
- Park, Y.S.; Kang, S.W.; Lee, J.S.; Hong, S.I.; Kim, S.W. Xylanase production in solid state fermentation by Aspergillus niger mutant using statistical experimental designs. Appl. Microbiol. Biotechnol. 2002, 58, 761–766. [Google Scholar] [CrossRef]
- Koch, A.L. Turbidity measurements of bacterial cultures in some available commercial instruments. Anal. Biochem. 1970, 38, 252–259. [Google Scholar] [CrossRef]
- Widyasti, E.; Shikata, A.; Hashim, R.; Sulaiman, O.; Sudesh, K.; Wahjono, E.; Kosugi, A. Biodegradation of fibrillated oil palm trunk fiber by a novel thermophilic, anaerobic, xylanolytic bacterium Caldicoprobacter sp. CL-2 isolated from compost. Enzym. Microb. Technol. 2018, 111, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Bibi, N.; Ali, S.; Tabassum, R. Statistical Optimization of Pectinase Biosynthesis from Orange Peel by Bacillus licheniformis Using Submerged Fermentation. Waste Biomass Valorization 2016, 7, 467–481. [Google Scholar] [CrossRef]
- Kaur, A.; Mahajan, R.; Singh, A.; Garg, G.; Sharma, J. Application of cellulase-free xylano-pectinolytic enzymes from the same bacterial isolate in biobleaching of kraft pulp. Bioresour. Technol. 2010, 101, 9150–9155. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Gunst, R.F.; Myers, R.H.; Montgomery, D.C. Response Surface Methodology: Process and Product Optimization Using Designed Experiments. Technometrics 1996, 38, 285–286. [Google Scholar] [CrossRef]
- Sethi, B.K.; Jana, A.; Nanda, P.K.; Das Mohapatra, P.K.; Sahoo, S.L. Thermostable acidic protease production in Aspergillus terreus NCFT 4269.10 using chickling vetch peels. J. Taibah Univ. Sci. 2016, 10, 571–583. [Google Scholar] [CrossRef]
- Moon, S.H.; Parulekar, S.J. A parametric study ot protease production in batch and fed-batch cultures of Bacillus firmus. Biotechnol. Bioeng. 1991, 37, 467–483. [Google Scholar] [CrossRef]
- Saar Dover, R.; Bitler, A.; Shimoni, E.; Trieu-Cuot, P.; Shai, Y. Multiparametric AFM reveals turgor-responsive net-like peptidoglycan architecture in live streptococci. Nat. Commun. 2015, 6, 7193. [Google Scholar] [CrossRef]
- Pillet, F.; Formosa-Dague, C.; Baaziz, H.; Dague, E.; Rols, M.P. Cell wall as a target for bacteria inactivation by pulsed electric fields. Sci. Rep. 2016, 6, 19778. [Google Scholar] [CrossRef]
- Ramírez-Nuñez, J.; Romero-Medrano, R.; Nevárez-Moorillón, G.V.; Gutiérrez-Méndez, N. Effect of pH and salt Gradient on the autolysis of Lactococcus lactis strains. Braz. J. Microbiol. 2011, 42, 1495–1499. [Google Scholar] [CrossRef][Green Version]
- Akhavan Sepahy, A.; Ghazi, S.; Akhavan Sepahy, M. Cost-Effective Production and Optimization of Alkaline Xylanase by Indigenous Bacillus mojavensis AG137 Fermented on Agricultural Waste. Enzyme Res. 2011, 2011, 593624. [Google Scholar] [CrossRef] [PubMed]
- Annamalai, N.; Thavasi, R.; Jayalakshmi, S.; Balasubramanian, T. Thermostable and alkaline tolerant xylanase production by Bacillus subtilis isolated form marine environment. Indian J. Biotechnol. 2009, 8, 291–297. [Google Scholar]
- Mei, Y.; Chen, Y.; Zhai, R.; Liu, Y. Cloning, purification and biochemical properties of a thermostable pectinase from Bacillus halodurans M29. J. Mol. Catal. B Enzym. 2013, 94, 77–81. [Google Scholar] [CrossRef]
- Sethi, S.; Datta, A.; Gupta, B.L.; Gupta, S. Optimization of Cellulase Production from Bacteria Isolated from Soil. ISRN Biotechnol. 2013, 2013, 985685. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.; Dafforn, T.R.; Fryer, P.J.; Cox, P.W. Kinetic study of the thermal denaturation of a hyperthermostable extracellular α-amylase from Pyrococcus furiosus. Biochim. Biophys. Acta—Proteins Proteom. 2013, 1834, 2600–2605. [Google Scholar] [CrossRef]
- Dubeau, H.; Chahal, D.S.; Ishaque, M. Xylanase ofChaetomiumcellulolyticum: Its nature of production and hydrolytic potential. Biotechnol. Lett. 1987, 9, 275–280. [Google Scholar] [CrossRef]
- Lee, J.; Seo, E.; Kweon, D.-H.; Park, K.; Jin, Y.-S. Fermentation of rice bran and defatted rice bran for butanol 5 production using clostridium beijerinckii NCIMB 8052. J. Microbiol. Biotechnol. 2009, 19, 482–490. [Google Scholar] [CrossRef]
- Meyrath, J.; Suchanek, G. Chapter III Inoculation Techniques—Effects Due to Quality and Quantity of Inoculum. Methods Microbiol. 1972, 7, 159–209. [Google Scholar] [CrossRef]
- Raj, A.; Kumar, S.; Singh, S.K. A highly thermostable xylanase from Stenotrophomonas maltophilia: Purification and partial characterization. Enzyme Res. 2013, 2013, 429305. [Google Scholar] [CrossRef]
- Lincoln, R.E. Control of stock culture preservation and inoculum build-up in bacterial fermentation. J. Biochem. Microbiol. Technol. Eng. 1960, 2, 481–500. [Google Scholar] [CrossRef]
- Irfan, M.; Asghar, U.; Nadeem, M.; Nelofer, R.; Syed, Q. Optimization of process parameters for xylanase production by Bacillus sp. in submerged fermentation. J. Radiat. Res. Appl. Sci. 2016, 9, 139–147. [Google Scholar] [CrossRef]
- Kaushal, R.; Sharma, N.; Dogra, V. Optimization of the production and molecular characterization of cellulase-free xylanase from an alkalophillic Bacillus subtilis SD8 isolated from paper mill effluent. Appl. Biochem. Microbiol. 2015, 51, 551–559. [Google Scholar] [CrossRef]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140. [Google Scholar] [CrossRef] [PubMed]
- Azzaz, H.H.; Murad, H.A.; Kholif, A.M.; Hanfy, M.A.; Abdel Gawad, M.H. Optimization of culture conditions affecting fungal cellulase production. Res. J. Microbiol. 2012, 7, 23. [Google Scholar] [CrossRef][Green Version]
- Srivastava, S.K.; Gopalkrishnan, K.S.; Ramachandran, K.B. The production of β-glucosidase in shake-flasks by Aspergillus wentii. J. Ferment. Technol. 1987, 65, 95–99. [Google Scholar] [CrossRef]
- Stanbury, P.F.; Whitaker, A.; Hall, S.J. Principles of Fermentation Technology, 3rd ed.; Elsevier Science & Technology: Amsterdam, The Netherlands, 2016; ISBN 9780080999531. [Google Scholar]
- Fritsche, E.; Paschos, A.; Beisel, H.G.; Böck, A.; Huber, R. Crystal structure of the hydrogenase maturating endopeptidase HYBD from Escherichia coli. J. Mol. Biol. 1999, 288, 989–998. [Google Scholar] [CrossRef]
- Kapoor, M.; Nair, L.M.; Kuhad, R.C. Cost-effective xylanase production from free and immobilized Bacillus pumilus strain MK001 and its application in saccharification of Prosopis juliflora. Biochem. Eng. J. 2008, 38, 88–97. [Google Scholar] [CrossRef]
- Geok, L.P.; Nyonya, C.; Razak, A.; Abd Rahman, R.N.Z.; Basri, M.; Salleh, A.B. Isolation and screening of an extracellular organic solvent-tolerant protease producer. Biochem. Eng. J. 2003, 13, 73–77. [Google Scholar] [CrossRef]
- Helianti, I.; Ulfah, M.; Nurhayati, N.; Suhendar, D.; Finalissari, A.K.; Wardani, A.K. Production of Xylanase by Recombinant Bacillus subtilis DB104 Cultivated in Agroindustrial Waste Medium. HAYATI J. Biosci. 2016, 23, 125–131. [Google Scholar] [CrossRef]
- Ghazala, I.; Sayari, N.; Romdhane, M.B.; Ellouz-Chaabouni, S.; Haddar, A. Assessment of pectinase production by Bacillus mojavensis I4 using an economical substrate and its potential application in oil sesame extraction. J. Food Sci. Technol. 2015, 52, 7710–7722. [Google Scholar] [CrossRef][Green Version]
- Juarez-Garcia, E.; Agama-Acevedo, E.; Sáyago-Ayerdi, S.G.; Rodríguez-Ambriz, S.L.; Bello-Pérez, L.A. Composition, digestibility and application in breadmaking of banana flour. Plant Foods Hum. Nutr. 2006, 61, 131–137. [Google Scholar] [CrossRef] [PubMed]
- González-Montelongo, R.; Gloria Lobo, M.; González, M. Antioxidant activity in banana peel extracts: Testing extraction conditions and related bioactive compounds. Food Chem. 2010, 119, 1030–1039. [Google Scholar] [CrossRef]
- Reddy, G.V.; Ravindra Babu, P.; Komaraiah, P.; Roy, K.R.R.M.; Kothari, I.L. Utilization of banana waste for the production of lignolytic and cellulolytic enzymes by solid substrate fermentation using two Pleurotus species (P. ostreatus and P. sajor-caju). Process Biochem. 2003, 38, 1457–1462. [Google Scholar] [CrossRef]
- Barman, S.; Sit, N.; Badwaik, L.S.; Deka, S.C. Pectinase production by Aspergillus niger using banana (Musa balbisiana) peel as substrate and its effect on clarification of banana juice. J. Food Sci. Technol. 2015, 52, 3579–3589. [Google Scholar] [CrossRef][Green Version]
- Sivasubramanian, S.; Namasivayam, S.K.R. Phenol degradation using Candida tropicalis SSK01 isolated from petroleum contaminated soil under optimized medium composition. J. Pure Appl. Microbiol. 2014, 8, 641–650. [Google Scholar]
- Zhou, J.; Yu, X.; Ding, C.; Wang, Z.; Zhou, Q.; Pao, H.; Cai, W. Optimization of phenol degradation by Candida tropicalis Z-04 using Plackett-Burman design and response surface methodology. J. Environ. Sci. 2011, 23, 22–30. [Google Scholar] [CrossRef]
- Tepe, O.; Dursun, A.Y. Exo-pectinase production by Bacillus pumilus using different agricultural wastes and optimizing of medium components using response surface methodology. Environ. Sci. Pollut. Res. 2014, 21, 9911–9920. [Google Scholar] [CrossRef]
- Deepak, V.; Kalishwaralal, K.; Ramkumarpandian, S.; Babu, S.V.; Senthilkumar, S.R.; Sangiliyandi, G. Optimization of media composition for Nattokinase production by Bacillus subtilis using response surface methodology. Bioresour. Technol. 2008, 99, 8170–8174. [Google Scholar] [CrossRef]
- Latifian, M.; Hamidi-Esfahani, Z.; Barzegar, M. Evaluation of culture conditions for cellulase production by two Trichoderma reesei mutants under solid-state fermentation conditions. Bioresour. Technol. 2007, 98, 3634–3637. [Google Scholar] [CrossRef]
- Khusro, A.; Kaliyan, B.K.; Al-Dhabi, N.A.; Arasu, M.V.; Agastian, P. Statistical optimization of thermo-alkali stable xylanase production from Bacillus tequilensis strain ARMATI. Electron. J. Biotechnol. 2016, 22, 16–25. [Google Scholar] [CrossRef]
- Yang, V.W.; Zhuang, Z.; Elegir, G.; Jeffries, T.W. Alkaline-active xylanase produced by an alkaliphilic Bacillus sp. isolated from kraft pulp. J. Ind. Microbiol. 1995, 15, 434–441. [Google Scholar] [CrossRef]
- Ling, H.H.; Heng, K.L. Xylanase Production by Bacillus subtilis in Cost-Effective Medium Using Soybean Hull as Part of Medium Compostion under Submerged Fermentation (Smf) and Solid State Fermentation (SsF). J. Biodivers. Bioprospect. Dev. 2014, 2, 143. [Google Scholar] [CrossRef]
- Ayishal Begam, M.; Annu, A.; Shameera Banu, S.; Vishnu Priya, D. Comparison and Optimization of Thermostable Xylanase Production by Bacillus pumilus and Bacillus cereus using Corn Husk. IARJSET Int. Adv. Res. J. Sci. Eng. Technol. 2015, 9, 139–147. [Google Scholar] [CrossRef]
- Verma, D.; Satyanarayana, T. Production of cellulase-free xylanase by the recombinant Bacillus subtilis and its applicability in paper pulp bleaching. Biotechnol. Prog. 2013, 29, 1441–1447. [Google Scholar] [CrossRef]
- Tandom, D.; Sharma, N.; Vyas, G. Optimization of Process Parameters for Xylanase Production by Bacillus atropheaus [KJ 590121] SD9 Isolated from Sludge using Response Surface Methodology. Austin J. Proteom. Bioinform. Genom. 2016, 3, 1016. [Google Scholar]
- Haddar, A.; Driss, D.; Frikha, F.; Ellouz-Chaabouni, S.; Nasri, M. Alkaline xylanases from Bacillus mojavensis A21: Production and generation of xylooligosaccharides. Int. J. Biol. Macromol. 2012, 51, 647–656. [Google Scholar] [CrossRef]
- Mandal, A.; Kar, S.; Dutta, T.; Pati, B.R.; Mondal, K.C.; Das Mohapatra, P.K. Parametric optimization of submerged fermentation conditions for xylanase production by Bacillus cereus BSA1 through Taguchi Methodology. Acta Biol. Szeged. 2015, 59, 189–195. [Google Scholar]
- Mullai, P.; Fathima, N.S.A.; Rene, E.R. Statistical Analysis of Main and Interaction Effects to Optimize Xylanase Production under Submerged Cultivation Conditions. J. Agric. Sci. 2010, 2, 144. [Google Scholar] [CrossRef][Green Version]
- Patel, K.; Dudhagara, P. Optimization of xylanase production by Bacillus tequilensis strain UD-3 using economical agricultural substrate and its application in rice straw pulp bleaching. Biocatal. Agric. Biotechnol. 2020, 30, 101846. [Google Scholar] [CrossRef]
- Alokika; Singh, B. Enhanced production of bacterial xylanase and its utility in saccharification of sugarcane bagasse. Bioprocess Biosyst. Eng. 2020, 43, 1081–1091. [Google Scholar] [CrossRef]
- Uzuner, S.; Cekmecelioglu, D. Enhanced pectinase production by optimizing fermentation conditions of Bacillus subtilis growing on hazelnut shell hydrolyzate. J. Mol. Catal. B Enzym. 2015, 113, 62–67. [Google Scholar] [CrossRef]
- Kohli, P.; Sharma, N.; Gupta, R. Statistical optimization of production conditions of alkaline pectin lyase from Bacillus cereus using response surface methodology. Biocatal. Biotransform. 2017, 35, 417–426. [Google Scholar] [CrossRef]
- Guo, F.; Li, X.; Zhao, J.; Li, G.; Gao, P.; Han, X. Optimizing Culture Conditions by Statistical Approach to Enhance Production of Pectinase from Bacillus sp. Y1. BioMed Res. Int. 2019, 2019, 8146948. [Google Scholar] [CrossRef] [PubMed]
- Kuvvet, C.; Uzuner, S.; Cekmecelioglu, D. Improvement of Pectinase Production by Co-culture of Bacillus spp. Using Apple Pomace as a Carbon Source. Waste Biomass Valorization 2019, 10, 1241–1249. [Google Scholar] [CrossRef]
- Namasivayam, E.; John Ravindar, D.; Mariappan, K.; Jiji, A.; Kumar, M.; Jayaraj, R.L. Production of extracellular pectinase by Bacillus cereus isolated from market solid waste. J. Bioanal. Biomed. 2011, 3, 70–75. [Google Scholar] [CrossRef]
- Martín, M.C.; Morata de Ambrosini, V.I. Cold-active acid pectinolytic system from psychrotolerant Bacillus: Color extraction from red grape skin. Am. J. Enol. Vitic. 2013, 64, 495–504. [Google Scholar] [CrossRef]
- Sharma, D.; Sharma, G.; Mahajan, R. Development of strategy for simultaneous enhanced production of alkaline xylanase-pectinase enzymes by a bacterial isolate in short submerged fermentation cycle. Enzyme Microb. Technol. 2019, 122, 90–100. [Google Scholar] [CrossRef]
- Oumer, O.J.; Abate, D. Comparative Studies of Pectinase Production by Bacillus subtilis strain Btk 27 in Submerged and Solid-State Fermentations. BioMed Res. Int. 2018, 2018, 1514795. [Google Scholar] [CrossRef]
- Kashyap, D.R.; Chandra, S.; Kaul, A.; Tewari, R. Production, purification and characterization of pectinase from a Bacillus sp. DT7. World J. Microbiol. Biotechnol. 2000, 16, 277–282. [Google Scholar] [CrossRef]
- Prajapati, J.; Dudhagara, P.; Patel, K. Production of thermal and acid-stable pectinase from Bacillus subtilis strain BK-3: Optimization, characterization, and application for fruit juice clarification. Biocatal. Agric. Biotechnol. 2021, 35, 102063. [Google Scholar] [CrossRef]
| Variables | Values of CCD Variables | ||||
|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | |
| A. Initial pH of media | 5.5 | 7 | 8.5 | 10 | 11.5 |
| B. Temperature (°C) | 11.5 | 20 | 28.5 | 37 | 45.5 |
| C. Inoculum concentration (%) | 2 | 3 | 4 | 5 | 6 |
| D. Agitation speed (rpm) | 0 | 50 | 100 | 150 | 200 |
| Run Order | A | B | C | D | Xylanase (U/mL) | Pectinase (U/mL) | ||
|---|---|---|---|---|---|---|---|---|
| Experimental | Predicted | Experimental | Predicted | |||||
| 1 | 0 | 0 | 0 | 0 | 180.36 | 155.11 | 208.75 | 220.73 |
| 2 | −1 | +1 | +1 | +1 | 11.98 | 11.70 | 61.70 | 47.89 |
| 3 | −1 | −1 | +1 | −1 | 67.94 | 62.71 | 58.69 | 50.35 |
| 4 | +α | 0 | 0 | 0 | 24.13 | 28.32 | 42.97 | 30.62 |
| 5 | 0 | 0 | 0 | 0 | 168.46 | 155.11 | 229.34 | 220.73 |
| 6 | −1 | +1 | −1 | +1 | 4.72 | 6.65 | 0.00 | −6.54 |
| 7 | −1 | +1 | +1 | −1 | 53.05 | 50.57 | 38.16 | 36.33 |
| 8 | 0 | +α | 0 | 0 | 15.25 | 8.08 | 0.00 | −7.21 |
| 9 | −1 | +1 | −1 | −1 | 40.23 | 46.90 | 18.57 | 36.56 |
| 10 | 0 | 0 | 0 | +α | 24.37 | 24.47 | 2.86 | 12.88 |
| 11 | 0 | 0 | −α | 0 | 26.87 | 23.83 | 38.90 | 25.15 |
| 12 | +1 | −1 | +1 | +1 | 36.68 | 27.33 | 74.49 | 54.01 |
| 13 | 0 | 0 | 0 | 0 | 166.64 | 155.11 | 216.40 | 220.73 |
| 14 | +1 | −1 | +1 | −1 | 48.63 | 43.85 | 0.00 | 9.99 |
| 15 | −α | 0 | 0 | 0 | 39.72 | 41.05 | 47.21 | 58.60 |
| 16 | −1 | −1 | −1 | +1 | 27.22 | 26.04 | 0.00 | −0.87 |
| 17 | +1 | +1 | −1 | −1 | 30.78 | 29.83 | 47.35 | 51.01 |
| 18 | −1 | −1 | +1 | +1 | 24.88 | 22.98 | 96.66 | 96.45 |
| 19 | +1 | −1 | −1 | −1 | 43.59 | 41.19 | 4.60 | 15.91 |
| 20 | 0 | 0 | 0 | 0 | 157.31 | 155.11 | 222.13 | 220.73 |
| 21 | 0 | 0 | +α | 0 | 22.97 | 31.53 | 60.86 | 73.65 |
| 22 | 0 | 0 | 0 | 0 | 125.14 | 155.11 | 215.88 | 220.73 |
| 23 | +1 | +1 | −1 | +1 | 10.24 | 12.79 | 0.00 | 5.84 |
| 24 | +1 | +1 | +1 | +1 | 22.05 | 24.92 | 0.00 | 11.68 |
| 25 | +1 | +1 | +1 | −1 | 42.09 | 40.59 | 3.82 | 2.20 |
| 26 | +1 | −1 | −1 | +1 | 23.66 | 23.30 | 0.00 | 5.28 |
| 27 | 0 | −α | 0 | 0 | 18.05 | 30.73 | 0.00 | 6.25 |
| 28 | 0 | 0 | 0 | −α | 75.81 | 81.24 | 22.94 | 11.96 |
| 29 | −1 | −1 | −1 | −1 | 72.86 | 67.14 | 15.93 | 7.70 |
| 30 | 0 | 0 | 0 | 0 | 132.73 | 155.11 | 231.91 | 220.73 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value Prob > F | |
|---|---|---|---|---|---|---|
| Model | 78,687.82 | 14 | 5620.58 | 28.58 | <0.0001 | Significant |
| A | 242.82 | 1 | 242.82 | 1.23 | 0.28 | |
| B | 769.41 | 1 | 769.41 | 3.91 | 0.07 | |
| C | 88.98 | 1 | 88.98 | 0.45 | 0.51 | |
| D | 4834.05 | 1 | 4834.05 | 24.58 | 0.00 | |
| AB | 78.83 | 1 | 78.83 | 0.40 | 0.54 | |
| AC | 50.29 | 1 | 50.29 | 0.25 | 0.62 | |
| AD | 538.51 | 1 | 538.51 | 2.74 | 0.12 | |
| BC | 65.59 | 1 | 65.59 | 0.33 | 0.57 | |
| BD | 0.73 | 1 | 0.73 | 0.00 | 0.95 | |
| CD | 1.89 | 1 | 1.89 | 0.01 | 0.92 | |
| A2 | 24,859.25 | 1 | 24,859.25 | 126.41 | <0.0001 | |
| B2 | 31,566.79 | 1 | 31,566.79 | 160.52 | <0.0001 | |
| C2 | 27,834.88 | 1 | 27,834.88 | 141.55 | <0.0001 | |
| D2 | 17,925.08 | 1 | 17,925.08 | 91.15 | <0.0001 | |
| Residual | 2949.75 | 15 | 196.65 | |||
| Lack of Fit | 597.12 | 10 | 59.71 | 0.13 | 0.99 | Not significant |
| Pure Error | 2352.63 | 5 | 470.52 | |||
| Corr. Total | 81,637.57 | 29 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value Prob > F | |
|---|---|---|---|---|---|---|
| Model | 198,070.57 | 14 | 14,147.90 | 73.31 | 3.27 | Significant |
| A | 1174.60 | 1 | 1174.60 | 6.09 | 0.03 | |
| B | 271.72 | 1 | 271.72 | 1.41 | 0.25 | |
| C | 3528.71 | 1 | 3528.71 | 18.29 | 0.00 | |
| D | 1.29 | 1 | 1.29 | 0.01 | 0.94 | |
| AB | 38.86 | 1 | 38.86 | 0.20 | 0.66 | |
| AC | 2360.65 | 1 | 2360.65 | 12.23 | 0.00 | |
| AD | 4.30 | 1 | 4.30 | 0.02 | 0.88 | |
| BC | 1839.38 | 1 | 1839.38 | 9.53 | 0.01 | |
| BD | 1192.77 | 1 | 1192.77 | 6.18 | 0.025 | |
| CD | 2986.84 | 1 | 2986.84 | 15.48 | 0.001 | |
| A2 | 53,177.47 | 1 | 53,177.47 | 275.57 | <0.0001 | |
| B2 | 83,890.04 | 1 | 83,890.04 | 434.72 | <0.0001 | |
| C2 | 50,323.44 | 1 | 50,323.44 | 260.78 | <0.0001 | |
| D2 | 74,392.74 | 1 | 74,392.74 | 385.51 | <0.0001 | |
| Residual | 2894.62 | 15 | 192.97 | |||
| Lack of Fit | 2507.70 | 10 | 250.77 | 3.24 | 0.1032 | Not significant |
| Pure Error | 386.92 | 5 | 77.38 | |||
| Corr. Total | 2.01 × 105 | 29 |
| Organisms | Methods | Carbon Sources | Xylanase Activity (U/mL) | References |
|---|---|---|---|---|
| B. amyloliquefaciens ADI2 | CCD, RSM | Banana peel | 159.08 | Present investigation |
| B. tequilensis ARMATI | CCD, RSM | Birchwood xylan | 86.82 | [61] |
| Bacillus sp. | Non-statistical | Birchwood xylan | 49.00 | [62] |
| B. subtilis | Non-statistical | Pineapple peel | 18.87 | [63] |
| Bacillus sp. | CCD, RSM | Corn husk | 2.50 | [64] |
| B. subtilis | CCD, RSM | LB-Xylose | 119.00 | [65] |
| B. atropheaus | CCD, RSM | Xylan | 85.16 | [66] |
| B. subtilis SD8 | CCD, RSM | Xylan | 8.18 | [42] |
| B. mojavensis A21 | CCD, RSM | Barley bran | 7.45 | [67] |
| B. cereus BSA1 | Taguchi OA | Xylan | 7.40 | [68] |
| Bacillus sp. 2129 | CCD, RSM | Oat | 2.39 | [69] |
| B. tequilensis UD-3 | Non-statistical | Rice straw | 8.54 | [70] |
| B. substilis JJBS250 | CCD, RSM | Sugarcane bagasse | 98.16 | [71] |
| Organisms | Methods | Carbon Sources | Pectinase Activity (U/mL) | References |
|---|---|---|---|---|
| B. amyloliquefaciens ADI2 | CCD, RSM | Banana peel | 204.86 | Present investigation |
| B. subtilis | CCD, RSM | Hazelnut shell | 5.60 | [72] |
| B. pumilus | CCD, RSM | Sugar beet pulp | 33.43 | [58] |
| B. cereus | CCD, RSM | Pectin | 3.37 | [73] |
| B. mojavensis I4 | CCD, RSM | Carrot peel | 64.80 | [51] |
| Bacillus sp. Y1 | CCD, RSM | Starch, sucrose, wheat bran | 40.00 | [74] |
| Bacillus spp. | Non-statistical | Apple pomace | 11.25 | [75] |
| B. cereus | Non-statistical | Pectin | 44.00 | [76] |
| Bacillus sp. CH15 | Non-statistical | Pectin | 0.31 | [77] |
| B. pumilus AJK | CCD, RSM | Pectin, wheat bran | 109.19 | [78] |
| B. subtilis BTK27 | Non-statistical | Pectin | 66.30 | [79] |
| Bacillus sp. DT7 | Non-statistical | Pectin | 53.00 | [80] |
| B. subtilis BK-3 | CCD, RSM | Citrus peel | 31.8 | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawawi, M.H.; Ismail, K.I.; Sa’ad, N.; Mohamad, R.; Tahir, P.M.; Asa’ari, A.Z.; Saad, W.Z. Optimisation of Xylanase–Pectinase Cocktail Production with Bacillus amyloliquefaciens ADI2 Using a Low-Cost Substrate via Statistical Strategy. Fermentation 2022, 8, 119. https://doi.org/10.3390/fermentation8030119
Nawawi MH, Ismail KI, Sa’ad N, Mohamad R, Tahir PM, Asa’ari AZ, Saad WZ. Optimisation of Xylanase–Pectinase Cocktail Production with Bacillus amyloliquefaciens ADI2 Using a Low-Cost Substrate via Statistical Strategy. Fermentation. 2022; 8(3):119. https://doi.org/10.3390/fermentation8030119
Chicago/Turabian StyleNawawi, Muhammad Hariadi, Khairul Izdihar Ismail, Norazliza Sa’ad, Rosfarizan Mohamad, Paridah Md Tahir, Ainun Zuriyati Asa’ari, and Wan Zuhainis Saad. 2022. "Optimisation of Xylanase–Pectinase Cocktail Production with Bacillus amyloliquefaciens ADI2 Using a Low-Cost Substrate via Statistical Strategy" Fermentation 8, no. 3: 119. https://doi.org/10.3390/fermentation8030119
APA StyleNawawi, M. H., Ismail, K. I., Sa’ad, N., Mohamad, R., Tahir, P. M., Asa’ari, A. Z., & Saad, W. Z. (2022). Optimisation of Xylanase–Pectinase Cocktail Production with Bacillus amyloliquefaciens ADI2 Using a Low-Cost Substrate via Statistical Strategy. Fermentation, 8(3), 119. https://doi.org/10.3390/fermentation8030119






