Abstract
Oxidative stress, which can cause imbalance in the body by damaging cells and tissues, arises from the immoderate production of reactive oxygen species (ROS)/reactive nitrogen species (RNS). Therefore, external supplements having antioxidant activity are required for reducing oxidative stress. In our study, we investigated DPPH and ABTS radical scavenging ability, and the inhibition effect on the nitric oxide (NO) production of 15 food-derived bacterial strains in LPS-activated RAW264.7 cells. Among these LAB strains, eight strains with an excellent inhibition effect on NO production were selected through comparisons within the same genera. Moreover, the selected strains, including Leuconostoc mesenteroides MG860, Leu. citreum MG210, Pediococcus acidilactici MG5001, P. pentosaceus MG5078, Weissella cibaria MG5090, Levilactobacillus brevis MG5306, Latilactobacillus curvatus MG5020, and Latilactobacillus sakei MG5048 diminished the inducible nitric oxide synthase (iNOS)/cyclooxygenase-2 (COX-2) expression. In addition, the stability and adhesion ability of the eight LAB strains in the gastrointestinal tract were determined. In conclusion, the selected strains have potential as new probiotics with antioxidant effects.
1. Introduction
Reactive oxygen species (ROS) and reactive nitrogen species (RNS) are usually generated during normal metabolic processes and are unavoidable products for aerobic organisms [1]. Appropriate ROS/RNS levels play a critical role in immunoreaction against invading microbes and regulating intercellular communication [2]. However, the excessive production of ROS/RNS causes oxidative stress, which leads to protein oxidation, lipid peroxidation, and DNA damage [3,4].
Bacterial endotoxins, such as lipopolysaccharide (LPS), are known to promote ROS production in macrophages as well as the production of nitric oxide (NO), a type of RNS, via the expression of inducible nitric oxide synthase (iNOS) [5]. NO interacts with superoxide anion (∙O2−), an anionic radical, to form peroxynitrite (ONOO−), which is a powerful oxidizing agent and cytotoxic RNS, causing septic shock, brain damage, and local or systemic inflammatory disorders [5,6,7,8]. Additionally, RNS is known to induce iNOS and cyclooxygenase-2 (COX-2) by activating nuclear factor-κB (NF-κB), a pro-inflammatory transcription factor [9,10]. Therefore, it is necessary to maintain homeostasis by regulating oxidative stress through ROS/RNS-related enzymes and transcription factors [2,11].
Probiotics are living microorganisms—mainly lactic acid bacteria (LAB) such as Lactobacillus, Bifidobacterium, Streptococcus, and Enterococcus—and they produce beneficial effects on the host when ingested at an appropriate level [12,13]. LAB have been known to improve intestinal transit, balance intestinal flora, and maintain the acid–base equilibrium in the colon, resulting in immune system regulation and the lowering of the serum cholesterol level [14]. Research on LAB is being actively conducted due to increasing interest in their antioxidant ability. For example, bacteria belonging to genera Bifidobacterium and Lactobacillus, and Enterococcus durans LAB 18 have been shown to decrease the levels of 2,2-diphenyl-1-picrylhydrazyl (DPPH) and 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) free radicals [15,16,17,18]. In a previous study, heat-killed L. salivarius MG242, L. plantarum MG989, B. bifidum MG731 and B. lactis MG741 were confirmed to reduce oxidative stress by inhibiting NO production through a decrease in iNOS/COX-2 gene expression in LPS-activated RAW264.7 [19]. Although functional activities of various LAB strains have been reported, studies on some genera are insufficient.
Therefore, we evaluated DPPH and ABTS radical scavenging activity and NO inhibition of LAB to identify various LAB strains with antioxidant ability. Moreover, we investigated the gene expression of iNOS/COX-2, which affects NO production, in LPS-activated RAW264.7 treated with LAB. In addition, to assess the practical application of LAB as probiotics, the survival rate and adhesion ability of LAB in the gastrointestinal tract were investigated.
2. Materials and Methods
2.1. Preparation of Sample
All LAB strains (Leuconostoc mesenteroides MG860, Leu. citreum MG210, Pediococcus acidilactici MG5001, P. pentosaceus MG5078, Weissella cibaria MG5223, MG5090, MG5215, MG5285, Levilactobacillus brevis MG5250, MG5280, MG5306, MG5311, Latilactobacillus curvatus MG5020, and Latilactobacillus sakei MG5048, MG5031) were collected from MEDIOGEN (Jecheon, Korea) and LAB used in our study were isolated from fermented foods. The LAB were cultured in MRS medium (Difco, MI, USA), at 37 °C for 18 h. After stationary culture, the absorbance of the culture medium was adjusted to an OD600 of 1.0 (108–109 CFU/mL), centrifuged at 4000 rpm for 10 min, and filtered using a 0.20 μm filter (Advantec, Tokyo, Japan). This sample (cell free supernatant, CFS) was used for further cell culture experiments.
2.2. DPPH and ABTS Radical Scavenging Activity
The effect of bacterial strains on DPPH and ABTS radical scavenging activity was performed with slight modifications to the method of Kim et al. and Lee et al. [20,21]. Samples for antioxidant evaluation were live cells and were prepared as follows. After culturing the strains in MRS medium at 37 °C for 18 h, it was rinsed twice with phosphate-buffered saline (PBS). After centrifugation at 4000 rpm for 5 min, the pellet was resuspended in PBS to OD600 1.0.
In the DPPH method, 0.2 mM DPPH (Sigma-Aldrich, St. Louis, MO, USA), and sample were mixed at a ratio of 1:2 (v/v) in the amber tube and reacted for 30 min at RT. In the ABTS method, first, ABTS+ solution was prepared by mixing 7 mM ABTS (Sigma-Aldrich) and 2.45 mM potassium persulfate (Daejung Chemical and Metals, Siheung, Korea) in 1:1 (v/v) ratio. The ABTS+ solution was adjusted by adding distilled water, so that the absorbance at 734 nm became 0.7 ± 0.01. Next, 300 µL of the sample and 600 µL of ABTS+ solution were mixed in the amber tube and reacted for 30 min at RT. Absorbance was measured at 517 nm for DPPH method and at 734 nm for ABTS method. PBS and L-ascorbic acid were used as the control and positive control groups, respectively. The radical scavenging activity was computed using the following equation:
where Asample = absorbance of sample group; ACTL = absorbance of control group.
Radical scavenging activity (%) = (1 − Asample/ACTL) × 100
2.3. Cell Culture
RAW264.7 (mouse macrophage) was purchased from ATCC (Manassas, VA, USA). Cells were maintained in DMEM (Gibco, Grand Island, NY, USA) containing 10% FBS and 100 units/mL of penicillin/streptomycin (P/S) at 37 °C in a 5% CO2 atmosphere.
2.4. NO Production and Cell Viability
NO production was analyzed by some modification to the method of Kang et al. [22]. RAW264.7 were seeded in 96-well plate at 2 × 106 cells/mL and incubated for 24 h. The seeded cells were treated with FBS-free DMEM diluted with 10% CFS and 500 ng/mL of E. coli LPS (Sigma-Aldrich) and further incubated for 24 h. After transferring the cell culture supernatant (100 µL) to a new plate, the equal amount of fresh Griess solution was added and reacted at RT for 10 min. Absorbance was then determined at 550 nm using a microplate spectrophotometer (Biotek, Winooski, VT, USA). NO was quantified using a standard curve of NaNO2 (Sigma-Aldrich). Cytotoxicity was measured by the MTT (Sigma-Aldrich) method [23].
2.5. mRNA Extraction and Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
The gene expression of iNOS and COX-2 was confirmed using qRT-PCR. After treating RAW264.7 with CFS and LPS for 24 h, NucleoZOL (Macherey-Nagel, Duren, Germany) was added to extract the total RNA, and the same amount of RNA (1 µg) was synthesized as cDNA using a reverse transcription premix (ELPIS Biotech, Daejeon, Korea). The synthesized cDNA was subjected to PCR using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the housekeeping gene, and the sequence of the primers (Macrogen, Seoul, Korea) were as follows. GAPDH, forward 5′- TCTCCCTCACAATTTCCATCC-3′, reverse 5′-GGGTGCAGCGAACTTTATTG-3′; iNOS, forward 5′-CCTCACGCTTGGGTCTTGTT-3′, reverse 5′-GCACAAGGGGTTTTCTTCACG-3′; COX-2, forward 5′-TGCTGTACAAGCAGTGGCA-3′, reverse 5′-GCAGCCATTTCCTTCTCTCC-3′. The relative target mRNA expression was normalized to GAPDH gene and calculated using the 2−ΔΔCT method.
2.6. Simulated Gastrointestinal Tract Stability
LAB survival in the stomach and intestine tenue is crucial to reach the colon and to be used as probiotics. Therefore, in this study, viability of LAB was evaluated under conditions similar to those of the intestinal environment [24]. The LAB strain was cultured for 18 h, rinsed twice with PBS (pH 7.4), and resuspended at OD600 of 1.0. The suspension solution of 1 mL was collected, mixed with 9 mL of PBS (pH 2.5) containing 0.3% pepsin (simulated gastric fluid, SGF; Sigma-Aldrich), and incubated at 37 °C for 2 h, with stirring at 200 rpm. Then, 10 mL of PBS adjusted to pH 7.4, containing 1% pancreatin and 1% bile salt (stimulated intestinal fluid, SIF; Sigma-Aldrich) was added to the pellet obtained by centrifugation at 4000 rpm for 5 min and incubated at 37 °C for 2.5 h, with stirring at 200 rpm. The viable cell count was analyzed by the plate count method using MRS agar, and the survival rate was represented as the percentage of viable cells after 4.5 h compared to the initial number.
2.7. Adhesion to HT-29 Cells
The adhesion ability of the selected LAB strain was assessed in HT-29 cells (human colorectal adenocarcinoma cell line), as described previously [25]. HT-29 were seeded in a 12-well plate at 1 × 106 cells/mL and incubated in 5% CO2 at 37 °C for 24 h. The strain was cultured in MRS medium at 37 °C for 24 h. Then, LAB strain was resuspended at an OD600 of 1.0 in FBS and P/S-free DMEM, and inoculated into cells. After 2 h, the cells were rinsed with PBS (pH 7.4) and then scrapped. The number of LAB attached to the cells was counted by the plate count method using MRS agar and expressed as log CFU/mL.
2.8. Statistical Analysis
All experimentations were measured in triplicate and represented as the mean ± standard deviation (SD). For statistical analysis, one-way ANOVA was carried out using the SPSS statistical software (version 25; IBM Inc., Armonk, NY, USA), and the Tukey’s multiple comparison test was used to settle the significance of differences (p < 0.05) among treatments.
3. Results
3.1. Antioxidant Activity of the LAB Strains
DPPH and ABTS radical scavenging activity were analyzed to estimate antioxidant activity of 15 food-derived bacterial strains, and the results are shown in Table 1. DPPH radical scavenging activity of LAB was in the range 2.55–6.88%, and ABTS radical scavenging activity was 19.69 to 86.26%.

Table 1.
Antioxidant activity of isolated lactic acid bacteria.
3.2. LAB Strains Suppressed NO Production in LPS-Activated RAW264.7 Cells
The suppression of NO production was investigated in LPS-activated RAW264.7 treated with LAB CFS. LPS and MRS as a vehicle treatment increased NO production to 62.50 and 52.20 µM, respectively, but treatment with all bacterial strains significantly reduced it by 19.38–38.74 µM (inhibition rate, 38.01–69.00%) (Figure 1a). In particular, the NO inhibitory effect of L. sakei MG5048 (69.00 ± 0.02%) was the highest in RAW264.7 compared with the LPS-treated group. No cytotoxicity (cell viability > 90%) was observed upon LAB CFS treatment (Figure 1b).
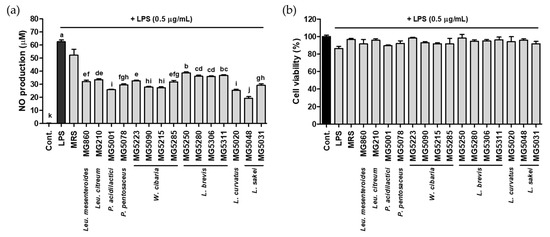
Figure 1.
Effect of CFS from 15 bacterial strains on (a) NO production and (b) cytotoxicity in LPS-activated RAW264.7. Data are represented as mean ± SD and bar with different letters indicating significance at p < 0.05.
Based on the results of antioxidant activity and NO inhibition, Leu. mesenteroides MG860, Leu. citreum MG210, P. acidilactici MG5001, P. pentosaceus MG5078, W. cibaria MG5090, L. brevis MG5306, L. curvatus MG5020, and L. sakei MG5048 were selected, and the gene expression of iNOS/COX-2 was confirmed using these strains.
3.3. LAB Strains Reduced iNOS/COX-2 Gene Expression in LPS-Activated RAW264.7 Cells
The effect of the selected LAB strains on the gene expression of iNOS/COX-2 was assessed using qRT-PCR. Although the iNOS/COX-2 expression was significantly increased in LPS-activated RAW264.7, LAB treatment markedly suppressed gene expression. iNOS expression level was most significantly decreased by L. sakei MG5048 to 0.23-fold; other strains also showed inhibitory effects ranging from 0.32–0.50-fold (Figure 2a). COX-2 expression level was the lowest after treatment with P. pentosaceus MG5078 and L. sakei MG5048, 0.13-fold and 0.12-fold, respectively; other strains showed COX-2 expression level from 0.41–0.70-fold (Figure 2b).
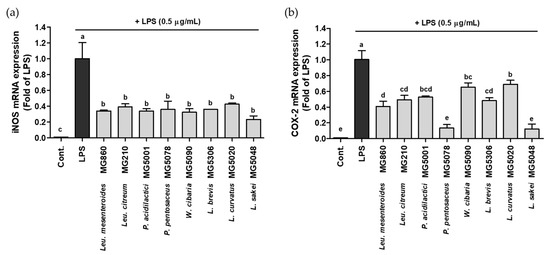
Figure 2.
Effect of CFS from LAB strains on gene expression of (a) iNOS and (b) COX-2 in LPS-activated RAW264.7. Data are represented as mean ± SD and bar with different letters indicating significance at p < 0.05.
3.4. Viability of LAB Strains in Simulated Gastrointestinal Fluid
In this study, the viability of the strains was determined by culturing the selected strains in SGF and SIF for a certain period of time (Table 2). The results confirmed that the viability of the selected strains ranged from 38.02–87.17%. Among them, P. acidilactici MG5001 (82.42%) and L. brevis MG5306 (87.17%) showed higher survival rates than those of other strains.

Table 2.
Survival rate of the LAB strains in simulated gastrointestinal fluid.
3.5. Adhesion of the LAB Strains on HT-29 Cells
The intestinal cell adhesion ability of the LAB strains was investigated using HT-29 cells (Table 3). The results revealed that the adhesion ability of the selected strains ranged from 63.02–84.91%. Among them, P. pentosaceus MG5078 (84.91%) and L. brevis MG5306 (82.35%) showed higher adhesion rates than those of other strains.

Table 3.
Adhesion ability of the LAB strains on HT-29 cells.
4. Discussion
Oxidative stress results from a disequilibrium between oxidant and antioxidant actions. Humans have an antioxidant defense system in the body, and when the accumulated ROS exceeds its intrinsic antioxidant capacity, excessive oxidation causes damage to cells or tissue. Thus, external antioxidants are required to decrease oxidative stress by improving individual antioxidant ability. Various antioxidant assays, involving stable non-biological radicals, have been conducted to determine the free-radical scavenging activity of probiotics [26]. LAB are safe bacteria with the ability to produce several compounds, such as organic acid and bacteriocin, and have the advantage of having few side effects [27]. When LAB are attached to the intestinal lumen, LAB and metabolites of LAB increase to remove ROS, thereby maintaining the intestinal oxidation–reduction balance. In addition, it is known that LAB constitute a strong natural xenogeneic antioxidant in consideration of nutritive value, safety, and functions of probiotic [28]. Therefore, it was attempted to confirm the antioxidant activity using various LAB.
Recently, much attention has been paid to CFS containing biologically active metabolites secreted by living bacteria [29]. Moreover, unlike CFS, which contain only extracellular metabolites, probiotics may be more effective in improving health through a broader range of mechanisms [30]. CFS of probiotics is used as an in vitro model for the purpose of evaluating functional effects [31]. In our study, the antioxidant activity of CFS from 15 food-derived bacterial strains was measured through the DPPH and ABTS radical scavenging method (Table 1). All LAB has DPPH and ABTS radical scavenging activity. Among them, MG5020 is the highest radical scavenging activity. The protective ability of probiotics against oxidative stress has been elucidated by ROS scavenging, chelation of metal ion, and the reduction of ascorbate autoxidation [32]. The antioxidant components in LAB are bacterial exopolysaccharides, bioactive peptides, antioxidant enzymes, and manganese ions. Moreover, the gut microflora can produce bioactive dietary antioxidants through bioconversion processes using various enzymatic reactions [33].
NO is produced by various immune cells via the gene expression of iNOS, and these cells are stimulated by substrates of the microbe and various cytokines. The major functions of NO are antimicrobial, antitumor, tissue-damaging, anti-inflammatory, and immunosuppressive effects [34]. However, NO, which plays a physiologically important role, is a kind of reactive radical, and when excessively generated reacts with an anion radical to form peroxynitrite, a more powerful oxidizing agent, causing an inflammatory reaction [5]. Therefore, reducing NO production is one of the effective treatment strategies. In our study, 15 food-derived bacterial strains showed no cytotoxicity and inhibited NO production in RAW264.7 cells (Figure 1). Based on these results, MG860, MG210, MG5001, MG5078, MG5090, MG5346, MG5020 and MG5048 were selected for evaluating iNOS/COX-2 gene expression. All LAB reduced iNOS/COX-2 gene expression: among them, MG5048 showed the highest inhibitory activity of iNOS/COX-2 (Figure 2). iNOS/COX-2 is expressed in LPS-activated macrophages and is known to be a factor related to NO production [10]. In addition, Choi et al. and Wang et al. reported that LAB treatment reduced NO production by decreasing the iNOS/COX-2 expression in LPS-activated cells [35,36]. Lactobacillus rhamnosus GG (LGG) was used as positive control primarily in probiotics study. Yang et al. reported that NO inhibition rate of LGG was 37% [29]. Our LAB strains showed higher efficacy compared with LGG with a NO inhibition rate of 38 to 69%. Therefore, the selected LAB can exhibit antioxidant potential by inhibiting NO production, by reducing the expression of iNOS/COX-2 in LPS-activated RAW264.7.
Probiotic bacteria are exposed to an acidic environment of the stomach (pH 1.5–3.5) and bile salt (pH 5.0–6.0), and should be able to survive under these conditions [37]. The results confirmed that the viability of the selected strains ranged from 38.02–87.17% (Table 2). Due to the difference in the pH of the gastric and intestines and the detergent action of bile salts, bacterial viability is limited. Bacterial strains belonging to the genera Bifidobacterium and Lactobacillus present in the intestine possess the bile salt hydrolase that converts primary bile acids into relatively less active secondary bile salts [38]. Therefore, the selected LAB showed the survival rate of 40% in the resistance of bile salt (Table 2). Overall, the strains MG5306 and MG5001 showed the highest survival rate in the gastrointestinal tract.
The adhesion ability of LAB is a strain-specific property, which indicates its probiotic potential. The health benefits of probiotics are associated with the composition of gut microbiota and their adhesion to the epithelial and mucosal surfaces. Therefore, the intestinal adhesion ability of probiotics is an important parameter that can reveal their functional property [39]. The adhesion process associates complex interactions between the host cell surfaces and the bacterial cell membrane. It has been suggested that bacteria can improve colonization in intestinal epithelial cells through the surface layer binding of proteins [40]. Generally, the Lactobacillus genus is known to have adhesion ability in the range of 2 to 10% [41]. In this study, the selected bacterial strains showed more than 60% adhesion ability; particularly, MG5078 and MG5306 strains showed excellent adhesion ability of 84.91% and 82.35%, respectively (Table 3). Therefore, these strains are expected to have excellent survival and adhesion in the gastrointestinal tract, suggesting that they will be useful for improving the intestinal environment; however, further research on the safety of these strains is needed before being used as probiotics.
5. Conclusions
Our results suggested that LAB treatment relieved oxidative stress by scavenging DPPH and ABTS radicals. In addition, LAB treatment suppressed NO production and reduced the levels of iNOS and COX-2 in LPS-activated RAW264.7 cells. Thus, LAB, including Leu. mesenteroides MG860, Leu. citreum MG210, P. acidilactici MG5001, P. pentosaceus MG5078, W. cibaria MG5090, L. brevis MG5306, L. curvatus MG5020, and L. sakei MG5048, have a strong potential for being used as probiotics and antioxidant functional foods.
Author Contributions
Conceptualization, C.-H.K.; methodology, S.K. and Y.J.; investigation, S.K. and Y.J.; resources, C.-H.K.; data curation, S.K. and J.Y.L.; writing—original draft preparation, S.K.; writing—review and editing, S.K., J.Y.L. and C.-H.K.; project administration, C.-H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available in a publicly accessible repository.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [Green Version]
- Fenster, C.P.; Weinsier, R.L.; Darley-Usmar, V.M.; Patel, R.P. Obesity, aerobic exercise, and vascular disease: The role of oxidant stress. Obes. Res. 2002, 10, 964–968. [Google Scholar] [CrossRef]
- Jeon, H.-J.; Choi, H.-S.; Lee, O.-H.; Jeon, Y.-J.; Lee, B.-Y. Inhibition of reactive oxygen species (ROS) and nitric oxide (NO) by Gelidium elegans using alternative drying and extraction conditions in 3T3-L1 and RAW264.7 cells. Prev. Nutr. Food Sci. 2012, 17, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.-S.; Hsu, W.-H.; Huang, J.-J.; Wu, S.-C. Antioxidant and anti-inflammatory effects of pigeon pea (Cajanus cajan L.) extracts on hydrogen peroxide-and lipopolysaccharide-treated RAW264. 7 macrophages. Food Funct. 2012, 3, 1294–1301. [Google Scholar] [CrossRef]
- Sekhar, S.; Sampath-Kumara, K.K.; Niranjana, S.R.; Prakash, H.S. Attenuation of reactive oxygen/nitrogen species with suppression of inducible nitric oxide synthase expression in RAW264.7 macrophages by bark extract of Buchanania lanzan. Pharmacogn. Mag. 2015, 11, 283. [Google Scholar]
- Albrecht, E.W.; Stegeman, C.A.; Tiebosch, A.T.; Tegzess, A.M.; Van Goor, H. Expression of inducible and endothelial nitric oxide synthases, formation of peroxynitrite and reactive oxygen species in human chronic renal transplant failure. Am. J. Transplant. 2002, 2, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Takumida, M.; Popa, R.; Anniko, M. Lipopolysaccharide-induced expression of reactive oxygen species and peroxynitrite in the guinea pig vestibular organ. ORL 1998, 60, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, N.D.; Liang, K.; Ding, Y. Increased nitric oxide inactivation by reactive oxygen species in lead-induced hypertension. Kidney Int. 1999, 56, 1492–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.J.; Kim, Y.A.; Yokozawa, T. Attenuation of oxidative stress and inflammation by gravinol in high glucose-exposed renal tubular epithelial cells. Toxicology 2010, 270, 106–111. [Google Scholar] [CrossRef]
- Kim, Y.J. Rhamnazin inhibits LPS-induced inflammation and ROS/RNS in raw macrophages. J. Nutr. Health 2016, 49, 288–294. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Park, J.-Y.; Kim, D.H.; Choi, S.J.; Jang, G.Y.; Seo, K.H. Chemical composition and protective effect of essential oils derived from medicinal plant on PC12 neuro-cells induced by oxidative stress. Korean J. Food Nutr. 2020, 33, 215–221. [Google Scholar]
- Kang, C.-H.; Han, S.H.; Kim, Y.; Jeong, Y.; Paek, N.-S. Antibacterial activity and probiotic properties of lactic acid bacteria isolated from traditional fermented foods. KSBB J. 2017, 32, 199–205. [Google Scholar] [CrossRef]
- AlKalbani, N.S.; Turner, M.S.; Ayyash, M.M. Isolation, identification, and potential probiotic characterization of isolated lactic acid bacteria and in vitro investigation of the cytotoxicity, antioxidant, and antidiabetic activities in fermented sausage. Microb. Cell Fact. 2019, 18, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riane, K.; Sifour, M.; Ouled-Haddar, H.; Idoui, T.; Bounar, S.; Boussebt, S. Probiotic properties and antioxidant efficiency of Lactobacillus plantarum 15 isolated from milk. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 516–520. [Google Scholar] [CrossRef]
- Cizeikiene, D.; Jagelaviciute, J. Investigation of antibacterial activity and probiotic properties of strains belonging to Lactobacillus and Bifidobacterium genera for their potential application in functional food and feed products. Probiotics Antimicrob. Proteins 2021, 13, 1387–1403. [Google Scholar] [CrossRef]
- Li, S.; Zhao, Y.; Zhang, L.; Zhang, X.; Huang, L.; Li, D.; Niu, C.; Yang, Z.; Wang, Q. Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. 2012, 135, 1914–1919. [Google Scholar] [CrossRef] [PubMed]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef] [Green Version]
- Song, M.W.; Chung, Y.; Kim, K.T.; Hong, W.S.; Chang, H.J.; Paik, H.D. Probiotic characteristics of Lactobacillus brevis B13-2 isolated from kimchi and investigation of antioxidant and immune-modulating abilities of its heat-killed cells. LWT-Food Sci. Technol. 2020, 128, 109452. [Google Scholar] [CrossRef]
- Kang, C.H.; Kim, J.S.; Park, H.M.; Kim, S.; Paek, N.S. Antioxidant activity and short-chain fatty acid production of lactic acid bacteria isolated from Korean individuals and fermented foods. 3 Biotech 2021, 11, 217. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.S.; Kim, Y.G.; Jeong, Y.; Kim, J.E.; Pack, N.S.; Kang, C.H. Antioxidant and probiotic properties of Lactobacilli and Bifidobacteria of human origins. Biotechnol. Bioprocess Eng. 2020, 25, 421–430. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, J.Y.; Park, C.G.; Kim, D.H.; Ji, Y.J.; Choi, S.J.; Oh, M.W.; Lee, Y.; Jeong, J.; Lee, J.H.; et al. Validation of a method and evaluation of antioxidant activity for the simultaneous determination of riboflavin and coixol in Coix lacryma-jobi var. ma-yuen stapf sprouts. Korean J. Crop Sci. 2019, 64, 452–458. [Google Scholar]
- Kang, C.H.; Koo, J.R.; So, J.S. Inhibitory effects of Aralia cordata thunb extracts on nitric oxide synthesis in RAW264.7 macrophage cells. Korean J. Food Sci. Technol. 2012, 44, 621–627. [Google Scholar] [CrossRef]
- Aziz, D.M. Assessment of bovine sperm viability by MTT reduction assay. Anim. Reprod. Sci. 2006, 92, 1–8. [Google Scholar] [CrossRef]
- Qi, X.; Simsek, S.; Chen, B.; Rao, J. Alginate-based double-network hydrogel improves the viability of encapsulated probiotics during simulated sequential gastrointestinal digestion: Effect of biopolymer type and concentrations. Int. J. Biol. Macromol. 2020, 165, 1675–1685. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, H.; Jeong, Y.; Kang, C.H. Lactic acid bacteria exert a hepatoprotective effect against ethanol-induced liver injury in HepG2 cells. Microorganisms 2021, 9, 1844. [Google Scholar] [CrossRef]
- Lin, M.Y.; Yen, C.L. Antioxidative ability of lactic acid bacteria. J. Agric. Food. Chem. 1999, 47, 1460–1466. [Google Scholar] [CrossRef]
- Hamed, E. Isolation, characterization and identification of lactic acid bacteria as probiotic. Ann. Agric. Sci. Moshtohor 2021, 59, 311–322. [Google Scholar] [CrossRef]
- Tang, W.; Li, C.; He, Z.; Pan, F.; Pan, S.; Wang, Y. Probiotic properties and cellular antioxidant activity of Lactobacillus plantarum MA2 isolated from Tibetan Kefir grains. Probiotics Antimicrob. Proteins 2018, 10, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Chae, S.A.; Bang, W.Y.; Ban, O.H.; Kim, S.J.; Jung, Y.H.; Yang, J. Anti-inflammatory Effect of Lactobacillus Plantarum IDCC 3501 and Its Safety Evaluation. Braz. J. Microbiol. 2021, 52, 2299–2306. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.Z.A.; Lu, Y.; Liu, S.Q. In vitro bioactivities of coffee brews fermented with the probiotics Lacticaseibacillus rhamnosus GG and Saccharomyces boulardii CNCM-I745. Food Res. Int. 2021, 149, 110693. [Google Scholar] [CrossRef] [PubMed]
- De Marco, S.; Sichetti, M.; Muradyan, D.; Piccioni, M.; Traina, G.; Pagiotti, R.; Pietrella, D. Probiotic cell-free supernatants exhibited anti-inflammatory and antioxidant activity on human gut epithelial cells and macrophages stimulated with LPS. Evid. Based Complement. Alternat. Med. 2018, 2018, 1756308. [Google Scholar] [CrossRef]
- Amaretti, A.; di Nunzio, M.; Pompei, A.; Raimondi, S.; Rossi, M.; Bordoni, A. Antioxidant properties of potentially probiotic bacteria: In vitro and in vivo activities. Appl. Microbiol. Biotechnol. 2013, 97, 809–817. [Google Scholar] [CrossRef]
- Davis, C.D.; Milner, J.A. Gastrointestinal microflora, food components and colon cancer prevention. J. Nutr. Btochem. 2009, 20, 743–752. [Google Scholar] [CrossRef] [Green Version]
- Bogdan, C. Nitric oxide and the immune response. Nat. Immunol. 2001, 2, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Lee, S.H.; Kim, M.G.; Lee, H.J.; Kim, G.B. Lactobacillus plantarum CAU1055 ameliorates inflammation in lipopolysaccharide-induced RAW264. 7 cells and a dextran sulfate sodium–induced colitis animal model. J. Dairy Sci. 2019, 102, 6718–6725. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, L.; Xu, S.; Pan, J.; Zhang, Q.; Lu, R. Surface-layer protein from Lactobacillus acidophilus NCFM inhibits lipopolysaccharide-induced inflammation through MAPK and NF-κB signaling pathways in RAW264. 7 cells. J. Agric. Food Chem. 2018, 66, 7655–7662. [Google Scholar] [CrossRef] [PubMed]
- Son, S.H.; Jeon, H.L.; Yang, S.J.; Sim, M.H.; Kim, Y.J.; Lee, N.K.; Paik, H.D. Probiotic lactic acid bacteria isolated from traditional Korean fermented foods based on β-glucosidase activity. Food Sci. Biotechnol. 2018, 27, 123–129. [Google Scholar] [CrossRef]
- Douillard, F.P.; Ribbera, A.; Kant, R.; Pietilä, T.E.; Järvinen, H.M.; Messing, M.; Randazzo, C.L.; Paulin, L.; Laine, P.; Ritari, J.; et al. Comparative Genomic and Functional Analysis of 100 Lactobacillus rhamnosus Strains and Their Comparison with Strain GG. PLoS Genet. 2013, 9, e1003683. [Google Scholar] [CrossRef]
- Kleeman, E.G.; Klaenhammer, T.R. Adherence of Lactobacillus species to human fetal intestinal cells. J. Dairy Sci. 1982, 65, 2063–2069. [Google Scholar] [CrossRef]
- Devi, S.M.; Kurrey, N.K.; Halami, P.M. In vitro anti-inflammatory activity among probiotic Lactobacillus species isolated from fermented foods. J. Funct. Foods 2018, 47, 19–27. [Google Scholar] [CrossRef]
- Lee, N.K.; Kim, S.Y.; Han, K.J.; Eom, S.J.; Paik, H.D. Probiotic potential of Lactobacillus strains with anti-allergic effects from kimchi for yogurt starters. LWT-Food Sci. Technol. 2014, 58, 130–134. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).