Effect of Lactobacillus plantarum Inoculation on Chemical Composition, Fermentation, and Bacterial Community Composition of Ensiled Sweet Corn Whole Plant or Stover
Abstract
:1. Introduction
2. Materials and Methods
2.1. Forage Harvest, Ensiling Conditions, and Sampling
2.2. pH Determination
2.3. Determination of Ammonia Nitrogen, Lactic Acid, Volatile Fatty Acids, and Ethanol
2.4. Proximate Composition
2.5. DNA Extraction, qPCR, and Pyrosequencing
2.6. Statistical Analyses
3. Results
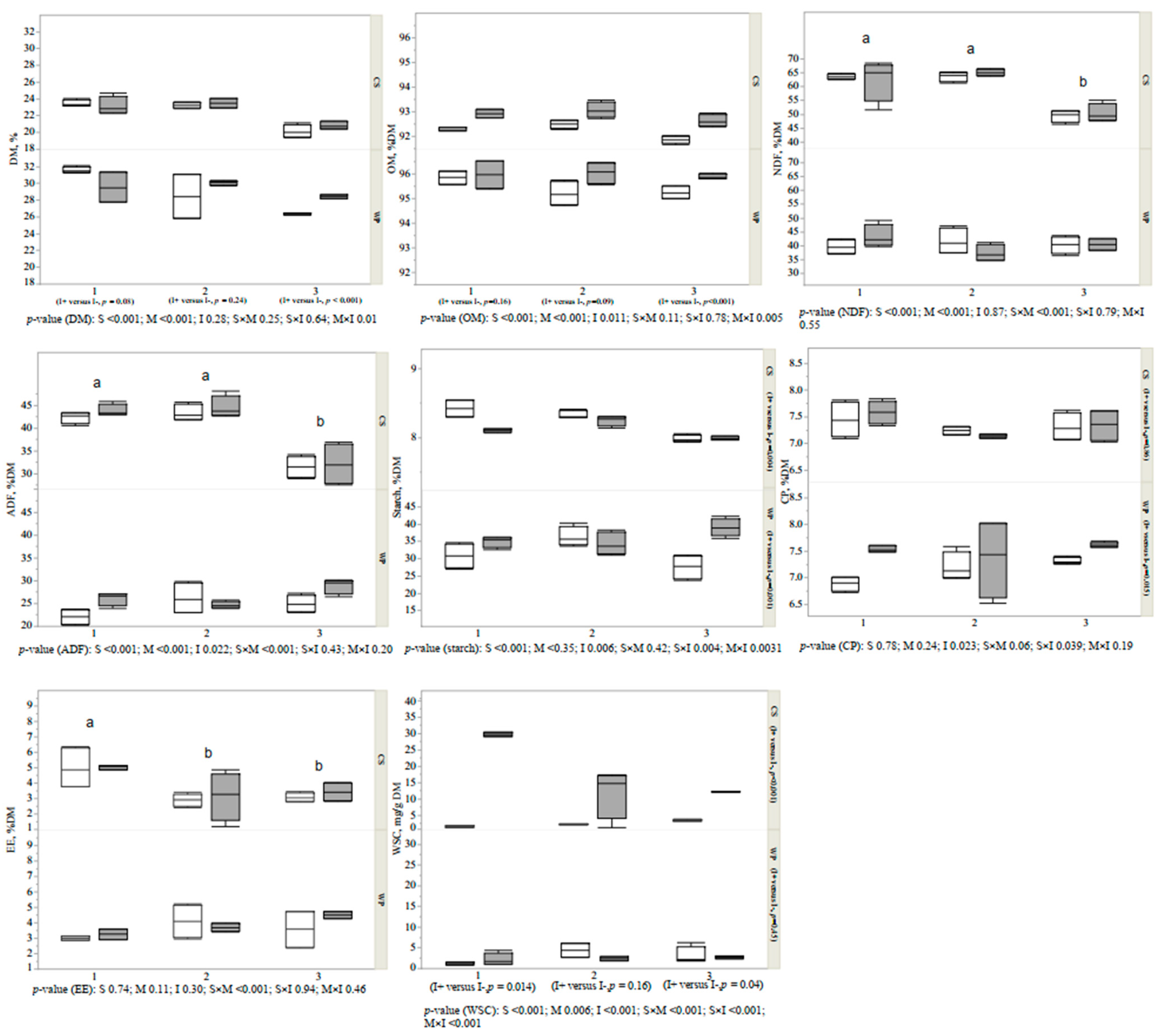
3.1. Proximate Composition of Forages and Silages
3.2. Fermentation Variables
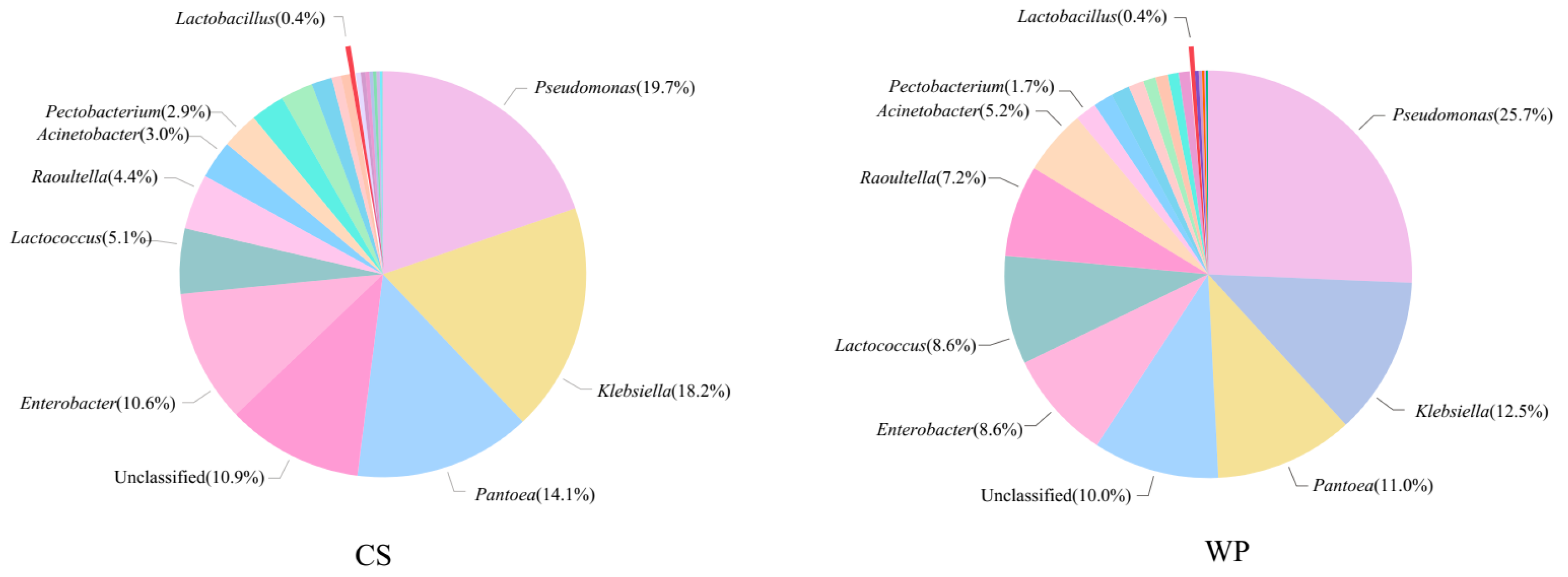
3.3. Microbial Community Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coşkun, M.B.; Yalçın, İ.; Özarslan, C. Physical properties of sweet corn seed (Zea mays saccharata Sturt.). J. Food Eng. 2006, 74, 523–528. [Google Scholar] [CrossRef]
- Azanza, F.; Bar-Zur, A.; Juvik, J.A. Variation in sweet corn kernel characteristics associated with stand establishment and eating quality. Euphytica 1996, 87, 7–18. [Google Scholar] [CrossRef]
- Lazcano, C.; Revilla, P.; Malvar, R.A.; Dominguez, J. Yield and fruit quality of four sweet corn hybrids (Zea mays) under conventional and integrated fertilization with vermicompost. J. Sci. Food Agric. 2011, 91, 1244–1253. [Google Scholar] [CrossRef] [PubMed]
- FAO. FAOStat: Food and Agriculture Data. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 12 January 2021).
- Li, K.; Huang, C. Current production status, problem and countermeasure on sweet corn industry in China. Sugar Crops China 2021, 43, 67–71. (In Chinese) [Google Scholar] [CrossRef]
- Olsen, J.K.; McMahon, C.R.; Hammer, G.L. Prediction of sweet corn phenology in subtropical environments. Agron. J. 1993, 85, 410–415. [Google Scholar] [CrossRef]
- Zhang, J.G.; Liu, Q.H.; Yang, F.Y. The chemical composition and ensiling characteristics of sweet corn processing by-products. Adv. Mater. Res. 2011, 236–238, 305–308. [Google Scholar] [CrossRef]
- Barros-Ríos, J.; Romaní, A.; Garrote, G.; Ordas, B. Biomass, sugar, and bioethanol potential of sweet corn. Glob. Chang. Biol. Bioenergy 2015, 7, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Shinners, K.J.; Binversie, B.N. Whole-Plant Corn Harvesting for Biomass: Comparision of Single-Pass and Ultiple-Pass Harvest Systems; Paper number 036089; ASAE Annual Meeting, 2003; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2003. [Google Scholar]
- Cui, T. Finishing effects comparison trials between whole corn silage and corn silage without ear. J. Yellow Cattle Sci. 2002, 3. (In Chinese) [Google Scholar]
- Muck, R.E. Factors Influencing Silage Quality and Their Implications for Management. J. Dairy Sci. 1988, 71, 2992–3002. [Google Scholar] [CrossRef]
- Yahaya, M.S.; Kawai, M.; Takahashi, J.; Matsuoka, S. The effects of different moisture content and ensiling time on silo degradation of structural carbohydrate of orchardgrass. Asian Austral. J. Anim. 2002, 15, 213–217. [Google Scholar] [CrossRef]
- Kurle, J.E.; Sheaffer, C.C.; Crookston, R.K.; Peterson, R.H.; Chester-Jones, H.; Lueschen, W.E. Popcorn, Sweet Corn, and Sorghum as Alternative Silage Crops. J. Prod. Agric. 2013, 4, 432–436. [Google Scholar] [CrossRef]
- Zhou, X.; Ouyang, Z.; Zhang, X.; Wei, Y.; Tang, S.; Ma, Z.; Tan, Z.; Zhu, N.; Teklebrhan, T.; Han, X. Sweet corn stalk treated with saccharomyces cerevisiae alone or in combination with Lactobacillus plantarum: Nutritional composition, fermentation traits and aerobic stability. Animals 2019, 9, 598. [Google Scholar] [CrossRef] [Green Version]
- Parvin, S.; Wang, C.; Li, Y.; Nishino, N. Effects of inoculation with lactic acid bacteria on the bacterial communities of Italian ryegrass, whole crop maize, guinea grass and rhodes grass silages. Anim. Feed Sci. Technol. 2010, 160, 160–166. [Google Scholar] [CrossRef]
- Fijałkowska, M.; Przemieniecki, S.W.; Purwin, C.; Lipiński, K.; Kurowski, T.P.; Karwowska, A. The effect of an additive containing three Lactobacillus species on the fermentation pattern and microbiological status of silage. J. Sci. Food Agric. 2020, 100, 1174–1184. [Google Scholar] [CrossRef]
- Bernardes, T.F.; Gervasio, J.; De Morais, G.; Casagrande, D.R. Technical note: A comparison of methods to determine pH in silages. J. Dairy Sci. 2019, 102, 9039–9042. [Google Scholar] [CrossRef]
- Playne, M.J. Determination of ethanol, volatile fatty acids, lactic and succinic acids in fermentation liquids by gas chromatography. J. Sci. Food Agric. 1985, 36, 638–644. [Google Scholar] [CrossRef]
- AOAC. Official Method of Analysis, 15th ed.; AOAC: Washington, DC, USA, 1990. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Karthner, R.J.; Theurer, B. Comparison of hydrolysis methods used in feed, digesta, and fecal starch analysis. J. Agric. Food Chem. 1981, 29, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Zahiroddini, H.; Baah, J.; Absalom, W.; Mcallister, T.A. Effect of an inoculant and hydrolytic enzymes on fermentation and nutritive value of whole crop barley silage. Anim. Feed Sci. Technol. 2004, 117, 317–330. [Google Scholar] [CrossRef]
- Denman, S.E.; McSweeney, C.S. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 2006, 58, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Wang, P.; He, Z.; Tang, S.; Zhou, C.; Han, X.; Wang, M.; Wu, D.; Kang, J.; Tan, Z. In vitro evaluation on neutral detergent fiber and cellulose digestion by post-ruminal microorganisms in goats. J. Sci. Food Agric. 2014, 94, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Inceoglu, O.; Al-Soud, W.A.; Salles, J.F.; Semenov, A.V.; van Elsas, J.D. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS ONE 2011, 6, e23321. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Halim, R.A.; Aimera, F.S.; Noraniza, M.; Abdullah, N.; Rahman, N.A. Changes in Water Soluble Carbohydrate for Corn Silage Treated with Bacterial Cultures at Different Fermentation Durations; Malaysian Society of Animal Production: Sarawak, Malaysia, 2014. [Google Scholar]
- Xu, Z.; Zhang, S.; Zhang, R.; Li, S.; Kong, J. The changes in dominant lactic acid bacteria and their metabolites during corn stover ensiling. J. Appl. Microbiol. 2018, 125, 675–685. [Google Scholar] [CrossRef]
- Drouin, P.; Tremblay, J.; Chaucheyras-Durand, F. Dynamic succession of microbiota during ensiling of whole plant corn following inoculation with Lactobacillus buchneri and Lactobacillus hilgardii alone or in combination. Microorganisms 2019, 7, 595. [Google Scholar] [CrossRef] [Green Version]
- Dunière, L.; Sindou, J.; Chaucheyras-Durand, F.; Chevallier, I.; Thévenot-Sergentet, D. Silage processing and strategies to prevent persistence of undesirable microorganisms. Anim. Feed Sci. Technol. 2013, 182, 1–15. [Google Scholar] [CrossRef]
- Blajman, J.E.; Paez, R.B.; Vinderola, C.G.; Lingua, M.S.; Signorini, M.L. A meta-analysis on the effectiveness of homofermentative and heterofermentative lactic acid bacteria for corn silage. J. Appl. Microbiol. 2018, 125, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
- Gharechahi, J.; Kharazian, Z.A.; Sarikhan, S.; Jouzani, G.S.; Aghdasi, M.; Hosseini Salekdeh, G. The dynamics of the bacterial communities developed in maize silage. Microb. Biotechnol. 2017, 10, 1663–1676. [Google Scholar] [CrossRef] [PubMed]
- Keshri, J.; Chen, Y.; Pinto, R.; Kroupitski, Y.; Weinberg, Z.G.; Sela, S. Microbiome dynamics during ensiling of corn with and without Lactobacillus plantarum inoculant. Appl. Microbiol. Biotechnol. 2018, 102, 4025–4037. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Bolsen, K.K.; Brent, B.E.; Hart, R.A.; Dickerson, J.T.; Feyerherm, A.M.; Aimutis, W.R. Epiphytic microflora on alfalfa and whole-plant corn. J. Dairy Sci. 1992, 75, 2484–2493. [Google Scholar] [CrossRef]
- Cai, Y.; Benno, Y.; Ogawa, M.; Ohmomo, S.; Kumai, S.; Nakase, T. Influence of Lactobacillus spp. from An inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 1998, 64, 2982–2987. [Google Scholar] [CrossRef] [Green Version]
- Kung, L.J.; Robinson, J.R.; Ranjit, N.K.; Chen, J.H.; Golt, C.M.; Pesek, J.D. Microbial populations, fermentation end-products, and aerobic stability of corn silage treated with ammonia or a propionic acid-based preservative. J. Dairy Sci. 2000, 83, 1479–1486. [Google Scholar] [CrossRef]
- Der Bedrosian, M.C.; Nestor, K.J.; Kung, L.J. The effects of hybrid, maturity, and length of storage on the composition and nutritive value of corn silage. J. Dairy Sci. 2012, 95, 5115–5126. [Google Scholar] [CrossRef] [Green Version]
- Ranjit, N.K.; Kung, L.J. The effect of Lactobacillus buchneri, Lactobacillus plantarum, or a chemical preservative on the fermentation and aerobic stability of corn silage. J. Dairy Sci. 2000, 83, 526–535. [Google Scholar] [CrossRef]
- Addah, W.; Baah, J.; Groenewegen, P.; Okine, E.K.; McAllister, T.A. Comparison of the fermentation characteristics, aerobic stability and nutritive value of barley and corn silages ensiled with or without a mixed bacterial inoculant. Can. J. Anim. Sci. 2011, 91, 133–146. [Google Scholar] [CrossRef]
- Driehuis, F.; Oude Elferink, S.J.W.H.; Van Wikselaar, P.G. Fermentation characteristics and aerobic stability of grass silage inoculated with Lactobacillus buchneri, with or without homofermentative lactic acid bacteria. Grass Forage Sci. 2001, 56, 330–343. [Google Scholar] [CrossRef]
- Arriola, K.G.; Kim, S.C.; Adesogan, A.T. Effect of applying inoculants with heterolactic or homolactic and heterolactic bacteria on the fermentation and quality of corn silage. J. Dairy Sci. 2011, 94, 1511–1516. [Google Scholar] [CrossRef]
- Aksu, T.; Baytok, E.; Karslı, M.A.; Muruz, H. Effects of formic acid, molasses and inoculant additives on corn silage composition, organic matter digestibility and microbial protein synthesis in sheep. Small Rumin. Res. 2006, 61, 29–33. [Google Scholar] [CrossRef]
- Filya, I.; Sucu, E.; Karabulut, A. The effects of Propionibacterium acidipropionici and Lactobacillus plantarum, applied at ensiling, on the fermentation and aerobic stability of low dry matter corn and sorghum silages. J. Ind. Microbiol. Biotechnol. 2006, 33, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Cheng, H.; Liu, D.; Wei, C.; An, W.; Wang, Y.; Sun, H.; Song, E. Treatment of Whole-Plant Corn Silage With Lactic Acid Bacteria and Organic Acid Enhances Quality by Elevating Acid Content, Reducing pH, and Inhibiting Undesirable Microorganisms. Front. Microbiol. 2020, 11, 593088. [Google Scholar] [CrossRef] [PubMed]
- Mogodiniyai Kasmaei, K.; Rustas, B.O.; Spörndly, R.; Udén, P. Prediction models of silage fermentation products on crop composition under strict anaerobic conditions: A meta-analysis. J. Dairy Sci. 2013, 96, 6644–6649. [Google Scholar] [CrossRef]
- Lindgren, S.E.; Dobrogosz, W.J. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol. Rev. 1990, 7, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Y.; Wang, X.F.; Liu, J.B.; Gao, L.J.; Ishii, M.; Igarashi, Y.; Cui, Z.J. Effects of water-soluble carbohydrate content on silage fermentation of wheat straw. J. Biosci. Bioeng. 2006, 101, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Amer, S.; Hassanat, F.; Berthiaume, R.; Seguin, P.; Mustafa, A.F. Effects of water soluble carbohydrate content on ensiling characteristics, chemical composition and in vitro gas production of forage millet and forage sorghum silages. Anim. Feed Sci. Technol. 2012, 177, 23–29. [Google Scholar] [CrossRef]
- Przemieniecki, S.W.; Purwin, C.; Mastalerz, J.; Borsuk, M.; Lipiński, K.; Kurowski, T. Biostimulating effect of L-tryptophan on the yield and chemical and microbiological quality of perennial ryegrass (Lolium perenne) herbage and silage for ruminant. J. Sci. Food Agric. 2020, 101, 3969–3974. [Google Scholar] [CrossRef]
- Schaefer, D.M.; Brotz, P.G.; Arp, S.C.; Cook, D.K. Inoculation of corn silage and high-moisture corn with lactic acid bacteria and its effects on the subsequent fermentations and on feedlot performance of beef steers. Anim. Feed Sci. Technol. 1989, 25, 23–38. [Google Scholar] [CrossRef]
- Lynch, J.P.; O’Kiely, P.; Waters, S.M.; Doyle, E.M. Conservation characteristics of corn ears and stover ensiled with the addition of Lactobacillus plantarum MTD-1, Lactobacillus plantarum 30114, or Lactobacillus buchneri 11A44. J. Dairy Sci. 2012, 95, 2070–2080. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Nishino, N. Monitoring the bacterial community of maize silage stored in a bunker silo inoculated with Enterococcus faecium, Lactobacillus plantarum and Lactobacillus buchneri. J. Appl. Microbiol. 2011, 110, 1561–1570. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.H.; Shao, T.; Zhang, J.G. Determination of aerobic deterioration of corn stalk silage caused by aerobic bacteria. Anim Feed Sci. Technol. 2013, 183, 124–131. [Google Scholar] [CrossRef]
- Lo, Y.C.; Lu, W.C.; Chen, C.Y.; Chen, W.M.; Chang, J.S. Characterization and high-level production of xylanase from an indigenous cellulolytic bacterium Acinetobacter junii F6-02 from southern Taiwan soil. Biochem. Eng. J. 2010, 53, 77–84. [Google Scholar] [CrossRef]
- Pourramezan, Z.; Ghezelbash, G.R.; Romani, B.; Ziaei, S.; Hedayatkhah, A. Screening and identification of newly isolated cellulose-degrading bacteria from the gut of xylophagous termite Microcerotermes diversus (Silvestri). Microbiology 2012, 81, 736–742. [Google Scholar] [CrossRef]
- Kornberg, H.L.; Krebs, H.A. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 1957, 179, 988–991. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Z.-Y.; Ungerfeld, E.; Ouyang, Z.; Zhou, X.-L.; Han, X.-F.; Zeng, Y.-Q.; Tan, Z.-L. Effect of Lactobacillus plantarum Inoculation on Chemical Composition, Fermentation, and Bacterial Community Composition of Ensiled Sweet Corn Whole Plant or Stover. Fermentation 2022, 8, 24. https://doi.org/10.3390/fermentation8010024
Ma Z-Y, Ungerfeld E, Ouyang Z, Zhou X-L, Han X-F, Zeng Y-Q, Tan Z-L. Effect of Lactobacillus plantarum Inoculation on Chemical Composition, Fermentation, and Bacterial Community Composition of Ensiled Sweet Corn Whole Plant or Stover. Fermentation. 2022; 8(1):24. https://doi.org/10.3390/fermentation8010024
Chicago/Turabian StyleMa, Zhi-Yuan, Emilio Ungerfeld, Zhu Ouyang, Xiao-Ling Zhou, Xue-Feng Han, Yan-Qin Zeng, and Zhi-Liang Tan. 2022. "Effect of Lactobacillus plantarum Inoculation on Chemical Composition, Fermentation, and Bacterial Community Composition of Ensiled Sweet Corn Whole Plant or Stover" Fermentation 8, no. 1: 24. https://doi.org/10.3390/fermentation8010024
APA StyleMa, Z.-Y., Ungerfeld, E., Ouyang, Z., Zhou, X.-L., Han, X.-F., Zeng, Y.-Q., & Tan, Z.-L. (2022). Effect of Lactobacillus plantarum Inoculation on Chemical Composition, Fermentation, and Bacterial Community Composition of Ensiled Sweet Corn Whole Plant or Stover. Fermentation, 8(1), 24. https://doi.org/10.3390/fermentation8010024





