The Effect of Incubation Temperature, Substrate and Initial pH Value on Plantaricin Activity and the Relative Transcription of pln Genes of Six Sourdough Derived Lactiplantibacillus plantarum Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Effect of Parameters Related to Sourdough Preparation on Growth and Plantaricin Activity Kinetics of Lp. plantarum Strains
2.3. Effect of Parameters Related to Sourdough Preparation on the Transcription of Plantaricin Genes
2.4. Statistical Analysis
3. Results
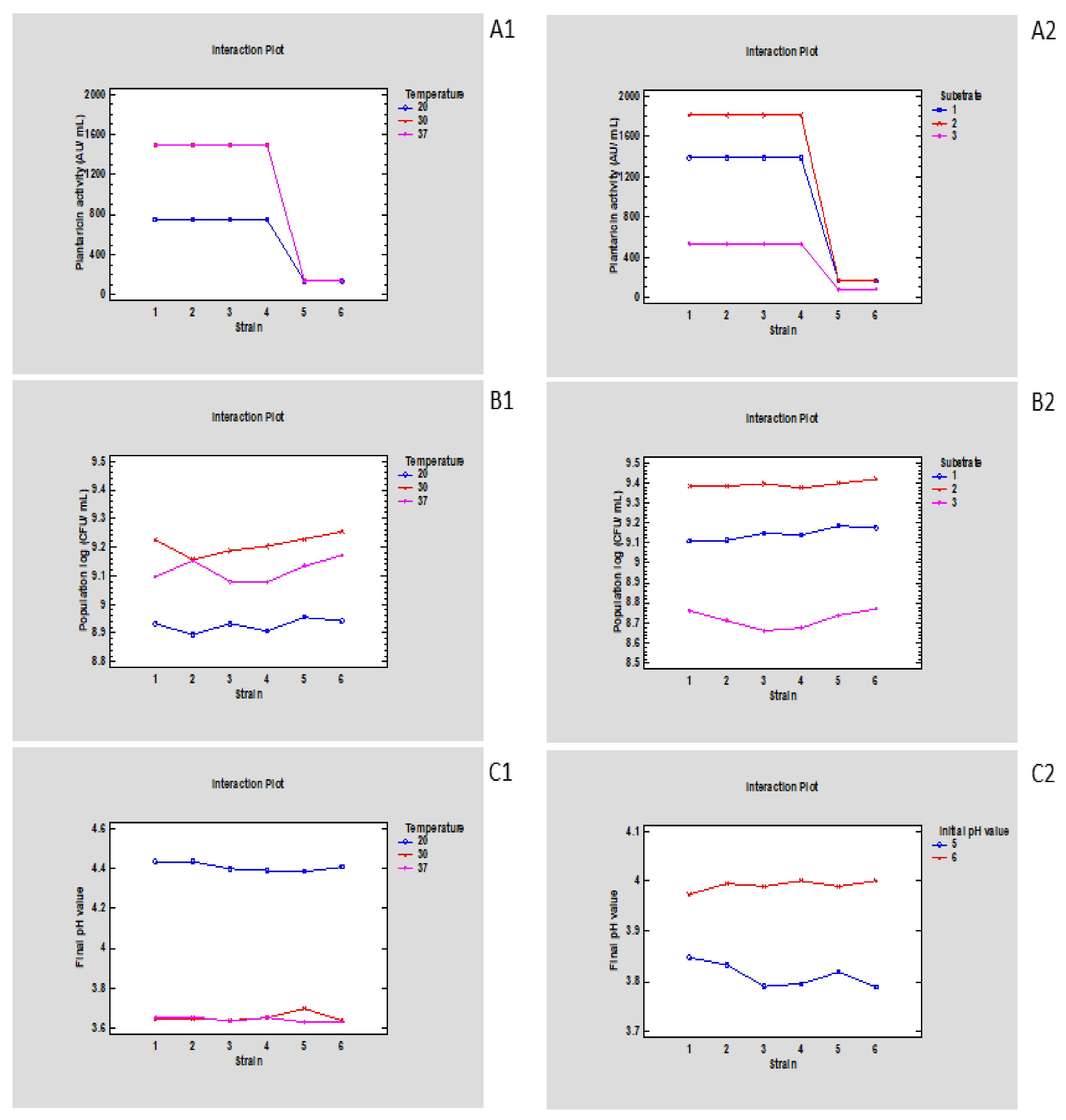
3.1. The Effect of Parameters Related to Sourdough Preparation on Growth and Plantaricin Activity Kinetics of Lp. plantarum Strains
3.2. The Effect of Parameters Related to Sourdough Preparation on the Transcription of Plantaricin Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barbosa, M.S.; Todorov, S.D.; Ivanova, I.V.; Belguesmia, Y.; Choiset, Y.; Rabesona, H.; Chobert, J.M.; Haertle, T.; Franco, B.D.G.M. Characterization of a two-peptide plantaricin produced by Lactobacillus plantarum MBSa4 isolated from Brazilian salami. Food Control 2016, 60, 103–112. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, N.; Battista, N.; Prete, R.; Corsetti, A. Health-promoting role of Lactiplantibacillus plantarum isolated from fermented foods. Microorganisms 2021, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Sabo, S.; Vitolo, M.; González, J.M.D.; Oliveira, R.P.S. Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Res. Int. 2014, 64, 527–536. [Google Scholar] [CrossRef]
- Paramithiotis, S.; Tsiasiotou, S.; Drosinos, E.H. Comparative study of spontaneously fermented sourdoughs originating from two regions of Greece: Peloponnesus and Thessaly. Eur. Food Res. Technol. 2010, 231, 883–890. [Google Scholar] [CrossRef]
- Corsetti, A. Technology of sourdough fermentation and sourdough applications. In Handbook on Sourdough Biotechnology; Gobbetti, M., Gänzle, M., Eds.; Springer: Boston, MA, USA, 2013; pp. 85–103. [Google Scholar]
- Syrokou, M.K.; Themeli, C.; Paramithiotis, S.; Mataragas, M.; Bosnea, L.; Argyri, A.; Chorianopoulos, N.G.; Skandamis, P.N.; Drosinos, E.H. Microbial ecology of Greek wheat sourdoughs identified by culture-dependent and culture-independent approach. Foods 2020, 9, 1603. [Google Scholar] [CrossRef]
- Diep, D.B.; Håvarstein, L.S.; Nes, I.F. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 1996, 178, 4472–4483. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Boekhorst, J.; van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.E.J.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, A.; Ruiz-Barba, J.L.; Jimenez-Diaz, R. Purification and genetic characterization of plantaricin NC8, a novel coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl. Environ. Microbiol. 2003, 69, 383–389. [Google Scholar] [CrossRef]
- Navarro, L.; Rojo-Bezares, B.; Sáenz, Y.; Diez, L.; Zarazaga, M.; Ruiz-Larrea, F.; Torres, C. Comparative study of the pln locus of the quorum-sensing regulated bacteriocin- producing L. plantarum J51 strain. Int. J. Food Microbiol. 2008, 128, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Bezares, B.; Sáenz, Y.; Navarro, L.; Jimιnez-Dνaz, R.; Zarazaga, M.; Ruiz-Larrea, F.; Torres, C. Characterisation of a new organisation of the plantaricin locus in the inducible bacteriocin- producing Lactobacillus plantarum J23 of grape must origin. Arch. Microbiol. 2008, 189, 491–499. [Google Scholar] [CrossRef]
- Barbosa, J.; Albano, H.; Silva, B.; Almeida, M.H.; Nogueira, T.; Teixeira, P. Characterization of a Lactiplantibacillus plantarum R23 isolated from Arugula by whole-genome sequencing and its bacteriocin production ability. Int. J. Environ. Res. Public Health 2021, 18, 5515. [Google Scholar] [CrossRef] [PubMed]
- Tenea, G.N.; Ortega, C. Genome characterization of Lactiplantibacillus plantarum strain UTNGt2 originated from Theobroma grandiflorum (White Cacao) of Ecuadorian Amazon: Antimicrobial peptides from safety to potential applications. Antibiotics 2021, 10, 383. [Google Scholar] [CrossRef]
- Diep, D.B.; Straume, D.; Kjos, M.; Torres, C.; Nes, I.F. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides 2009, 30, 1562–1574. [Google Scholar] [CrossRef]
- Tai, H.F.; Foo, H.L.; Rahim, R.A.; Loh, T.C.; Abdullah, M.P.; Yoshinobu, K. Molecular characterisation of new organisation of plnEF and plw loci of bacteriocin genes harbour concomitantly in Lactobacillus plantarum I-UL4. Microb. Cell Fact. 2015, 14, 89. [Google Scholar] [CrossRef]
- Todorov, S.D. Bacteriocins from Lactobacillus plantarum—Production, genetic organization and mode of action. Braz. J. Microbiol. 2009, 40, 209–221. [Google Scholar]
- Straume, D.; Johansen, R.F.; Bjørås, M.; Nes, I.F.; Diep, D.B. DNA binding kinetics of tworesponse regulators, PlnC and PlnD, from the bacteriocin regulon of Lactobacillus plantarum C11. BMC Biochem. 2009, 10, 17. [Google Scholar] [CrossRef]
- Syrokou, M.K.; Paramithiotis, S.; Drosinos, E.H.; Bosnea, L.; Mataragas, M. Comparative genomics and safety assessment of six Lactiplantibacillus plantarum subsp. argentoratensis strains isolated from spontaneously fermented Greek wheat sourdoughs for potential biotechnological application. Food Res. Int. under review.
- Paramithiotis, S.; Papadelli, M.; Pardali, E.; Mataragas, M.; Drosinos, E.H. Evaluation of plantaricin genes expression during fermentation of Raphanus sativus roots with a plantaricin-producing Lactobacillus plantarum starter. Curr. Microbiol. 2019, 76, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Syrokou, M.K.; Tziompra, S.; Psychogiou, E.-E.; Mpisti, S.-D.; Paramithiotis, S.; Bosnea, L.; Mataragas, M.; Skandamis, P.N.; Drosinos, E.H. Technological and safety attributes of lactic acid bacteria and yeasts isolated from spontaneously fermented Greek wheat sourdoughs. Microorganisms 2021, 9, 671. [Google Scholar] [CrossRef]
- Andritsos, N.; Mataragas, M.; Paramithiotis, S.; Drosinos, E.H. Quantifying Listeria monocytogenes prevalence and concentration in minced pork meat and estimating performance of three culture media from presence/absence microbiological testing using a deterministic and stochastic approach. Food Microbiol. 2013, 36, 395–405. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Paramithiotis, S.; ·Drosinos, E.H. Genetic analysis of the Listeria Pathogenicity Island 1 of Listeria monocytogenes 1/2a and 4b isolates. Current Microbiol. 2018, 75, 857–865. [Google Scholar] [CrossRef]
- Paramithiotis, S. Study on the Symbiosis of Wild Yeasts and Lactic Acid Bacteria in Sourdough. Ph.D. Thesis, Agricultural University of Athens, Athens, Greece, 2001. [Google Scholar]
- Gobbetti, M. The sourdough microflora: Interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 1998, 9, 267–274. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Coda, R.; De Angelis, M.; Di Cagno, R.; Carnevali, P.; Gobbetti, M. Long-term fungal inhibitory activity of water-soluble extract from Amaranthus spp. seeds during storage of gluten-free and wheat flour breads. Int. J. Food Microbiol. 2009, 131, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Syrokou, M.K.; Paramithiotis, S.; Skandamis, P.N.; Drosinos, E.H.; Bosnea, L.; Mataragas, Μ. High-quality draft genome sequence data of six Lactiplantibacillus plantarum subsp. argentoratensis strains isolated from various Greek sourdoughs. Data Brief 2021, 37, 107172. [Google Scholar] [CrossRef] [PubMed]
- Van der Veen, S.; Abee, T. Importance of SigB for Listeria monocytogenes static and continuous-flow bioflm formation and disinfectant resistance. Appl. Environ. Microbiol. 2010, 76, 7854–7860. [Google Scholar] [CrossRef] [PubMed]
- Hadjilouka, A.; Andritsos, N.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Listeria monocytogenes serotype prevalence and biodiversity in diverse food products. J. Food Prot. 2014, 77, 2115–2120. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Mataragas, M.; Alessandria, V.; Cocolin, L. Expression of virulence genes of Listeria monocytogenes in food. J. Food Saf. 2012, 32, 161–168. [Google Scholar] [CrossRef]
- Olesen, I.; Jorgensen, K.-F.; Jespersen, L. Gene transcription and virulence potential of Listeria monocytogenes strains after exposure to acidic and NaCl stress. Foodborne Pathog. Dis. 2009, 6, 669–680. [Google Scholar] [CrossRef]
- Ben Omar, N.; Abriouel, H.; Lucas, R.; Martínez-Cañamero, M.; Guyot, J.-P.; Gálvez, A. Isolation of bacteriocinogenic Lactobacillus plantarum strains from ben saalga, a traditional fermented gruel from Burkina Faso. Int. J. Food Microbiol. 2006, 112, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Hadjilouka, A.; Gkolfakis, P.; Patlaka, A.; Grounta, A.; Vourli, G.; Paramithiotis, S.; Touloumi, G.; Triantafyllou, K.; Drosinos, E.H. In vitro gene transcription of Listeria monocytogenes after exposure to human gastric and duodenal aspirates. J. Food Prot. 2019, 83, 89–100. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization—Applied to bladder- and colon-cancer data-sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef]
- Messens, W.; Neysens, P.; Vansieleghem, W.; Vanderhoeven, J.; De Vuyst, L. Modeling growth and bacteriocin production by Lactobacillus amylovorus DCE 471 in response to temperature and pH values used for sourdough fermentations. Appl. Environ. Microbiol. 2002, 68, 1431–1435. [Google Scholar] [CrossRef] [PubMed]
- Neysens, P.; Messens, W.; De Vuyst, L. Effect of sodium chloride on growth and bacteriocin production by Lactobacillus amylovorus DCE 471. Int. J. Food Microbiol. 2003, 15, 29–39. [Google Scholar] [CrossRef]
- Leroy, F.; De Winter, T.; Adriany, T.; Neysens, P.; De Vuyst, L. Sugars relevant for sourdough fermentation stimulate growth of and bacteriocin production by Lactobacillus amylovorus DCE 471. Int. J. Food Microbiol. 2006, 112, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Prévost, H.; Lebois, M.; Dousset, X.; LeBlanc, J.G.; Franco, B.D.G.M. Bacteriocinogenic Lactobacillus plantarum ST16Pa isolated from papaya (Carica papaya)—From isolation to application: Characterization of a bacteriocin. Food Res. Int. 2011, 44, 1351–1363. [Google Scholar] [CrossRef]
- Parlindungan, E.; Dekiwadia, C.; Jones, O.A.H. Factors that influence growth and bacteriocin production in Lactiplantibacillus plantarum B21. Process Biochem. 2021, 107, 18–26. [Google Scholar] [CrossRef]
- Van Reenen, C.A.; Van Zyl, W.H.; Dicks, L.M.T. Expression of the immunity protein of plantaricin 423, produced by Lactobacillus plantarum 423, and analysis of the plasmid encoding the bacteriocin. Appl. Environ. Microbiol. 2006, 72, 7644–7651. [Google Scholar] [CrossRef]
- Mataragas, M.; Metaxopoulos, J.; Galiotou, M.; Drosinos, E.H. Influence of pH and temperature on growth and bacteriocin production by Leuconostoc mesenteroides L124 and Lactobacillus curvatus L442. Meat Sci. 2003, 64, 265–271. [Google Scholar] [CrossRef]
- Sidooski, T.; Brandelli, A.; Bertoli, S.L.; de Souza, C.K.; de Carvalho, L.F. Physical and nutritional conditions for optimized production of bacteriocins by lactic acid bacteria—A review. Crit. Rev. Food Sci. Nutr. 2019, 59, 2839–2849. [Google Scholar] [CrossRef]
- Drosinos, E.H.; Mataragas, M.; Paramithiotis, S. Antimicrobial activity of bacteriocins and their applications. In Meat Biotechnology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 375–397. [Google Scholar]
- Śliżewska, K.; Chlebicz-Wójcik, A. Growth kinetics of probiotic Lactobacillus strains in the alternative, cost-efficient semi-solid fermentation medium. Biology 2020, 9, 423. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Lactobacillus plantarum isolated from molasses produces bacteriocins active against Gram-negative bacteria. Enzym. Microb. Technol. 2005, 36, 318–326. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Medium components effecting bacteriocin production by two strains of Lactobacillus plantarum ST414BZ and ST664BZ isolated from boza. Biologia 2006, 61, 269–274. [Google Scholar] [CrossRef][Green Version]
- Todorov, S.D. Bacteriocin production by Lactobacillus plantarum AMA-K isolated from Amasi, a Zimbabwean fermented milk product and study of the adsorption of bacteriocin AMA-K to Listeria sp. Braz. J. Microbiol. 2008, 39, 178–187. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Callewaert, R.; Crabbe, K. Primary metabolite kinetics of bacteriocin biosynthesis by Lactobacillus amylovorus and evidence for stimulation of bacteriocin production under unfavourable growth conditions. Microbiology 1996, 142, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Uguen, P.; Hamelin, J.; Le Pennec, J.-P.; Blanco, C. Influence of osmolarity and the presence of an osmoprotectant on Lactococcus lactis growth and bacteriocin production. Appl. Environ. Microbiol. 1999, 65, 291–293. [Google Scholar] [CrossRef]
- Leal-Sanchez, M.V.; Jimenez-Diaz, R.; Maldonado-Barragan, A.; Garrido-Fernandez, A.; Ruiz-Barba, J.L. Optimization of bacteriocin production by batch fermentation of Lactobacillus plantarum LPCO10. Appl. Environ. Microbiol. 2002, 68, 4465–4471. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; De Vuyst, L. The presence of salt and a curing agent reduces bacteriocin production by Lactobacillus sakei CTC 494, a potential starter culture for sausage fermentation. Appl. Environ. Microbiol. 1999, 65, 5350–5356. [Google Scholar] [CrossRef]
- Himelbloom, B.; Nilsson, L.; Gram, L. Factors affecting production of an antilisterial bacteriocin by Carnobacterium piscicola strain A9b in laboratory media and model fish systems. J. Appl. Microbiol. 2001, 91, 506–513. [Google Scholar] [CrossRef]
- Verluyten, J.; Messens, W.; De Vuyst, L. Sodium chloride reduces production of curvacin A, a bacteriocin produced by Lactobacillus curvatus strain LTH 1174, originating from fermented sausage. Appl. Environ. Microbiol. 2004, 70, 2271–2278. [Google Scholar] [CrossRef]
- Drosinos, E.H.; Mataragas, M.; Nasis, P.; Galiotou, M.; Metaxopoulos, J. Growth and bacteriocin production kinetics of Leuconostoc mesenteroides E131. J. Appl. Microbiol. 2005, 99, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Messens, W.; Verluyten, J.; Leroy, F.; De Vuyst, L. Modelling growth and bacteriocin production by Lactobacillus curvatus LTH 1174 in response to temperature and pH values used for European sausage fermentation processes. Int. J. Food Microbiol. 2002, 81, 41–52. [Google Scholar] [CrossRef]
- Nilsen, T.; Nes, I.F.; Holo, H. An exported inducer peptide regulates bacteriocin production in Enterococcus faecium CTC 492. J. Bacteriol. 1998, 180, 1848–1854. [Google Scholar] [CrossRef]
- Nilsson, L.; Nielsen, M.K.; Ng, Y.; Gram, L. Role of acetate in production of an autoinducible class IIa bacteriocin in Carnobacterium piscicola A9b. Appl. Environ. Microbiol. 2002, 68, 2251–2260. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Todorov, S.D.; Holzapfel, W.; Nero, L.A. Characterization of a novel bacteriocin produced by Lactobacillus plantarum ST8SH and some aspects of its mode of action. Ann. Microbiol. 2016, 66, 949–962. [Google Scholar] [CrossRef]
- Yang, E.; Fan, L.; Yan, J.; Jiang, Y.; Doucette, C.; Fillmore, S.; Walker, B. Influence of culture media, pH and temperature on growth and bacteriocin production of bacteriocinogenic lactic acid bacteria. AMB Express 2018, 8, 10. [Google Scholar] [CrossRef]
- Hurtado, A.; Ben Othman, N.; Chammem, N.; Hamdi, M.; Ferrer, S.; Reguant, C.; Bordons, A.; Rozès, N. Characterization of Lactobacillus isolates from fermented olives and their bacteriocin gene profiles. Food Microbiol. 2011, 28, 1514–1518. [Google Scholar] [CrossRef]
- Doulgeraki, A.I.; Paraskevopoulos, N.; Nychas, G.J.E.; Panagou, E.Z. An in vitro study of Lactobacillus plantarum strains for the presence of plantaricin genes and their potential control of the table olive microbiota. Antonie Van Leeuwenhoek 2013, 103, 821–832. [Google Scholar] [CrossRef]
- Picard, F.; Dressaire, C.; Girbal, L.; Cocaign-Bousquet, M. Examination of post-transcriptional regulations in prokaryotes by integrative biology. C. R. Biol. 2009, 332, 958–973. [Google Scholar] [CrossRef]
| Genes | Primer | Sequence | Amplicon Size (bp) | PCR Efficiency | References |
|---|---|---|---|---|---|
| Reference genes | |||||
| 16S | 16SF | GATGCATAGCCGACCTGAGA | 114 | 2.05 | [27] |
| 16SR | CTCCGTCAGACTTTCGTCCA | [28] | |||
| IGS | IGSF | GGCCTATAGCTCAGCTGGTTA | 135 | 2.03 | [29] |
| IGSR | GCTGAGCTAAGGCCCCGTAAA | ||||
| rpob | rpobF | CCGCGATGCGAAAACAAT | 69 | 2.04 | [30] |
| rpobR | CCWACAGAGATACGGTTATCRAATGC | ||||
| Plantaricin genes | |||||
| plNC8a | plNC8aF | GGCGGTGATTTAACAACCAAG | 70 | 2.05 | this study |
| plNC8aR | AATTCCAACGTGCTTTCTTGC | ||||
| plNC8b | plNC8bF | CGGATCAGTCCCAACTTCAGTA | 80 | 2.01 | this study |
| plNC8bR | TTTCAATCGTTTTGCGATGCT | ||||
| plNC8c | plNC8cF | AGCGTAAAAGCAGCAGTGAATA | 98 | 2.01 | this study |
| plNC8cR | AGTACGTGGCAAATGCCTAAAA | ||||
| pln423 (plaA) | 423F | TGTGGTAAACATTCCTGCTCTG | 86 | 2.06 | this study |
| 423R | CACTTTCCATGACCGAAGTTAGC | ||||
| plaB | plaBF | CGGTGAAAAACCCTGAGGCA | 151 | 1.98 | this study |
| plaBR | TAGCTACCGTTCCAACCTGC | ||||
| plaD | plaDF | GCCAAAACAACTGCTGACGG | 95 | 1.96 | this study |
| plaDR | TCCATATCAGCACGCACAGC | ||||
| plNC8-HK | plNC8-HKF | GGTGAAAAACCCTGAGGCAT | 148 | 2.00 | this study |
| plNC8-HKR | GCTACCGTTCCAACCTGCT | ||||
| plNC8-IF | plNC8-IFF | ATAAGCTTGATGTCGGGGTTG | 70 | 2.00 | this study |
| plNC8-IFR | GATGGCCTCCAAGTGCTTTT | ||||
| plnD | plnDF | GTGGTTTTGTTGAGTACATCGAAAT | 126 | 1.98 | this study |
| plnDR | GCATCGGAAAAATTGCGGATAC | [31] | |||
| plnE | plnEF | TGGTTTTAATCGGGGCGGT | 87 | 2.03 | this study |
| plnER | ATACCACGAATGCCTGCAAC | ||||
| plnF | plnFF | TGCTATTTCAGGTGGCGTTT | 94 | 2.08 | this study |
| plnFR | GCTAATGACCCAATCGGCAG | ||||
| plnG | plnGF | TGCGGTTATCAGTATGTCAAAG | 453 | 1.96 | [31] |
| plnGR | CCTCGAAACAATTTCCCCC | ||||
| plnH | plnHF | AACTGTTCAACCGACCGGAA | 90 | 2.07 | this study |
| plnHR | ACTCGCGCACCTTCAACTAA | ||||
| plnI | plnIF | CTGGCTGCCATTAGTGTCCA | 100 | 2.09 | this study |
| plnIR | GAGCTTCCATTGGCCCGTTA | ||||
| plnJ | plnJF | TTGAACGGGGTTGTTGGGG | 81 | 2.03 | this study |
| plnJR | GCCAGCTTCGCCATCATAAA | ||||
| plnK | plnKF | GGCCGTCGGAGTCGTAAAAA | 90 | 2.05 | this study |
| plnKR | ATCCCTTGAACCACCAAGCA | ||||
| plnL | plnLF | GGGTGCATCGTATTTGCGTG | 113 | 2.01 | this study |
| plnLR | TTTGCAGATCGCCATGAAGC | ||||
| plnM | plnMF | AGCAGTGGGAAGATGCTTGA | 109 | 2.02 | this study |
| plnMR | TGCCAACCTGCTTTACCTGT | ||||
| plnR | plnRF | GCGCTTATTGTCGTTTTCGC | 88 | 2.01 | this study |
| plnRR | CAGCAGCCCCATCACTAAGC | ||||
| plnS | plnSF | TATGGCACCGGCGTATCTTT | 121 | 2.02 | this study |
| plnSR | AACTCGTGCTGTATGCCGAT | ||||
| plnY | plnYF | GATTGGGGTACCCACGTCAC | 91 | 2.07 | this study |
| plnYR | AAAGAATCGTCCTAGCCGCA | ||||
| LQC 2320 | LQC 2520 | |||||||
|---|---|---|---|---|---|---|---|---|
| log2(FC) | Effect of Temperature 1 | Effect of Substrate 2 | Effect of pH 3 | Effect of Temperature | Effect of Substrate | Effect of pH | Effect of Strain 4 | |
| plNC8α | <−1 | 2 (11.1) | 2 (11.1) | 2 (22.2) | 0 (0.0) | 4 (22.2) | 0 (0.0) | 3 (16.7) |
| −1 to 1 | 12 (66.7) | 12 (66.7) | 7 (77.8) | 9 (50.0) | 14 (77.8) | 8 (88.9) | 12 (66.7) | |
| >1 | 4 (22.2) | 4 (22.2) | 0 (0.0) | 9 (50.0) | 0 (0.0) | 1 (11.1) | 3 (16.7) | |
| plNC8β | <−1 | 2 (11.1) | 3 (16.7) | 1 (11.1) | 1 (5.6) | 5 (27.8) | 0 (0.0) | 1 (5.6) |
| −1 to 1 | 9 (50.0) | 10 (55.6) | 8 (88.9) | 8 (44.4) | 13 (72.2) | 7 (77.8) | 15 (83.3) | |
| >1 | 7 (38.9) | 5 (27.8) | 0 (0.0) | 9 (50.0) | 0 (0.0) | 2 (22.2) | 2 (11.1) | |
| plnNC8c | <−1 | 2 (11.1) | 2 (11.1) | 2 (22.2) | 1 (5.6) | 5 (27.8) | 0 (0.0) | 5 (27.8) |
| −1 to 1 | 11 (61.1) | 12 (66.7) | 7 (77.8) | 8 (44.4) | 13 (72.2) | 8 (88.9) | 10 (55.6) | |
| >1 | 5 (27.8) | 4 (22.2) | 0 (0.0) | 9 (50.0) | 0 (0.0) | 1 (11.1) | 3 (16.7) | |
| plnR | <−1 | 4 (22.2) | 7 (38.9) | 2 (22.2) | 2 (11.1) | 7 (38.9) | 0 (0.0) | 3 (16.7) |
| −1 to 1 | 7 (38.9) | 8 (44.4) | 7 (77.8) | 8 (44.4) | 11 (61.1) | 7 (77.8) | 12 (66.7) | |
| >1 | 7 (38.9) | 3 (16.7) | 0 (0.0) | 8 (44.4) | 0 (0.0) | 2 (22.2) | 3 (16.7) | |
| plnL | <−1 | 4 (22.2) | 7 (38.9) | 2 (22.2) | 2 (11.1) | 7 (38.9) | 0 (0.0) | 3 (16.7) |
| −1 to 1 | 7 (38.9) | 8 (44.4) | 7 (77.8) | 8 (44.4) | 11 (61.1) | 7 (77.8) | 12 (66.7) | |
| >1 | 7 (38.9) | 3 (16.7) | 0 (0.0) | 2 (11.1) | 0 (0.0) | 2 (22.2) | 3 (16.7) | |
| plnK | <−1 | 5 (27.8) | 7 (38.9) | 2 (22.2) | 2 (11.1) | 7 (38.9) | 0 (0.0) | 4 (22.2) |
| −1 to 1 | 6 (33.3) | 8 (44.4) | 7 (77.8) | 8 (44.4) | 11 (61.1) | 7 (77.8) | 11 (61.1) | |
| >1 | 7 (38.9) | 3 (16.7) | 0 (0.0) | 8 (44.4) | 0 (0.0) | 2 (22.2) | 3 (16.7) | |
| plnJ | <−1 | 1 (5.6) | 7 (38.9) | 0 (0.0) | 2 (11.1) | 7 (38.9) | 0 (0.0) | 3 (16.7) |
| −1 to 1 | 10 (55.6) | 8 (44.4) | 8 (88.9) | 7 (38.9) | 11 (61.1) | 7 (77.8) | 12 (66.7) | |
| >1 | 7 (38.9) | 3 (16.7) | 1 (11.1) | 9 (50.0) | 0 (0.0) | 2 (22.2) | 3 (16.7) | |
| plnE | <−1 | 3 (16.7) | 3 (16.7) | 0 (0.0) | 3 (16.7) | 4 (22.2) | 0 (0.0) | 1 (5.6) |
| −1 to 1 | 14 (77.8) | 11 (61.1) | 9 (100.0) | 12 (66.7) | 13 (72.2) | 7 (77.8) | 14 (77.8) | |
| >1 | 1 (5.6) | 4 (22.2) | 0 (0.0) | 3 (16.7) | 1 (5.6) | 2 (22.2) | 3 (16.7) | |
| plnF | <−1 | 3 (16.7) | 6 (33.3) | 0 (0.0) | 2 (11.1) | 6 (33.3) | 0 (0.0) | 1 (5.6) |
| −1 to 1 | 8 (44.4) | 6 (33.3) | 9 (100.0) | 9 (50.0) | 12 (66.7) | 7 (77.8) | 14 (77.8) | |
| >1 | 7 (38.9) | 6 (33.3) | 0 (0.0) | 7 (38.9) | 0 (0.0) | 2 (22.2) | 3 (16.7) | |
| plnH | <−1 | 2 (11.1) | 8 (44.4) | 2 (22.2) | 2 (11.1) | 6 (33.3) | 0 (0.0) | 5 (27.8) |
| −1 to 1 | 9 (50.0) | 5 (27.8) | 7 (77.8) | 7 (38.9) | 10 (55.6) | 7 (77.8) | 10 (55.6) | |
| >1 | 7 (38.9) | 5 (27.8) | 0 (0.0) | 9 (50.0) | 2 (11.1) | 2 (22.2) | 3 (16.7) | |
| plnS | <−1 | 3 (16.7) | 6 (33.3) | 1 (11.1) | 2 (11.1) | 6 (33.3) | 0 (0.0) | 3 (16.7) |
| −1 to 1 | 9 (50.0) | 7 (38.9) | 8 (88.9) | 7 (38.9) | 12 (66.7) | 7 (77.8) | 12 (66.7) | |
| >1 | 6 (33.3) | 5 (27.8) | 0 (0.0) | 9 (50.0) | 0 (0.0) | 2 (22.2) | 3 (16.7) | |
| plnY | <−1 | 1 (5.6) | 6 (33.3) | 1 (11.1) | 1 (5.6) | 6 (33.3) | 0 (0.0) | 1 (5.6) |
| −1 to 1 | 8 (44.4) | 9 (50.0) | 8 (88.9) | 7 (38.9) | 12 (66.7) | 7 (77.8) | 14 (77.8) | |
| >1 | 9 (50.0) | 3 (16.7) | 0 (0.0) | 10 (55.6) | 0 (0.0) | 2 (22.2) | 3 (16.7) | |
| plNC8-IF | <−1 | 4 (22.2) | 6 (33.3) | 0 (0.0) | 1 (5.6) | 6 (33.3) | 0 (0.0) | 1 (5.6) |
| −1 to 1 | 13 (72.2) | 10 (55.6) | 9 (100.0) | 12 (66.7) | 12 (66.7) | 7 (77.8) | 13 (72.2) | |
| >1 | 1 (5.6) | 2 (11.1) | 0 (0.0) | 5 (27.8) | 0 (0.0) | 2 (22.2) | 4 (22.2) | |
| plNC8-HK | <−1 | 4 (22.2) | 6 (33.3) | 0 (0.0) | 2 (11.1) | 6 (33.3) | 0 (0.0) | 1 (5.6) |
| −1 to 1 | 13 (72.2) | 10 (55.6) | 9 (100.0) | 11 (61.1) | 12 (66.7) | 7 (77.8) | 13 (72.2) | |
| >1 | 1 (5.6) | 2 (11.1) | 0 (0.0) | 5 (27.8) | 0 (0.0) | 2 (22.2) | 4 (22.2) | |
| plnD | <−1 | 4 (22.2) | 6 (33.3) | 0 (0.0) | 1 (5.6) | 6 (33.3) | 0 (0.0) | 1 (5.6) |
| −1 to 1 | 13 (72.2) | 10 (55.6) | 8 (88.9) | 12 (66.7) | 12 (66.7) | 7 (77.8) | 13 (72.2) | |
| >1 | 1 (5.6) | 2 (11.1) | 1 (11.1) | 5 (27.8) | 0 (0.0) | 2 (22.2) | 4 (22.2) | |
| plnM | <−1 | 5 (27.8) | 7 (38.9) | 1 (11.1) | 3 (16.7) | 6 (33.3) | 0 (0.0) | 4 (22.2) |
| −1 to 1 | 7 (38.9) | 6 (33.3) | 7 (77.8) | 5 (27.8) | 11 (61.1) | 6 (66.7) | 10 (55.6) | |
| >1 | 6 (33.3) | 5 (27.8) | 1 (11.1) | 10 (55.6) | 1 (5.6) | 3 (33.3) | 4 (22.2) | |
| plnI | <−1 | 3 (16.7) | 7 (38.9) | 2 (22.2) | 2 (11.1) | 6 (33.3) | 1 (11.1) | 3 (16.7) |
| −1 to 1 | 9 (50.0) | 6 (33.3) | 7 (77.8) | 6 (33.3) | 12 (66.7) | 6 (66.7) | 12 (66.7) | |
| >1 | 6 (33.3) | 5 (27.8) | 0 (0.0) | 10 (55.6) | 0 (0.0) | 2 (22.2) | 3 (16.7) | |
| plnG | <−1 | 3 (16.7) | 5 (27.8) | 1 (11.1) | 1 (5.6) | 7 (38.9) | 1 (11.1) | 3 (16.7) |
| −1 to 1 | 7 (38.9) | 8 (44.4) | 7 (77.8) | 6 (33.3) | 9 (50.0) | 5 (55.6) | 12 (66.7) | |
| >1 | 8 (44.4) | 5 (27.8) | 1 (11.1) | 11 (61.1) | 2 (11.1) | 3 (33.3) | 3 (16.7) | |
| Total | <−1 | 55 (17.0) | 101 (31.2) | 19 (11.7) | 30 (9.3) | 107 (33.0) | 2 (1.2) | 46 (14.2) |
| −1 to 1 | 172 (53.1) | 154 (47.5) | 139 (85.8) | 150 (46.3) | 211 (65.1) | 124 (76.5) | 221 (68.2) | |
| >1 | 97 (29.9) | 69 (21.3) | 4 (2.5) | 144 (44.4) | 6 (1.9) | 36 (22.2) | 57 (17.6) | |
| log2(FC) | LQC 2441 | LQC 2422 | LQC 2485 | LQC 2516 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect of Temperature 1 | Effect of Substrate 2 | Effect of pH 3 | Effect of Temperature | Effect of Substrate | Effect of pH | Effect of Temperature | Effect of Substrate | Effect of pH | Effect of Temperature | Effect of Substrate | Effect of pH | Effect of Strain 4 | ||
| pln423 | <−1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (11.1) | 0 (0.0) | 1 (11.1) | 3 (16.7) | 0 (0.0) | 1 (11.1) | 2 (11.1) | 1 (5.6) | 1 (11.1) | 5 (4.6) |
| −1 to 1 | 17 (94.4) | 10 (55.6) | 7 (77.8) | 15 (83.3) | 7 (38.9) | 7 (77.8) | 13 (72.2) | 8 (44.4) | 7 (77.8) | 14 (77.8) | 6 (33.3) | 7 (77.8) | 46 (42.6) | |
| >1 | 1 (5.6) | 8 (44.4) | 2 (22.2) | 1 (5.6) | 11 (61.1) | 1 (11.1) | 2 (11.1) | 10 (55.6) | 1 (11.1) | 2 (11.1) | 11 (61.1) | 1 (11.1) | 57 (52.8) | |
| plaB | <−1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (11.1) | 0 (0.0) | 1 (11.1) | 3 (16.7) | 0 (0.0) | 1 (11.1) | 2 (11.1) | 1 (5.6) | 1 (11.1) | 7 (6.5) |
| −1 to 1 | 17 (94.4) | 10 (55.6) | 7 (77.8) | 15 (83.3) | 7 (38.9) | 7 (77.8) | 13 (72.2) | 8 (44.4) | 7 (77.8) | 14 (77.8) | 5 (27.8) | 7 (77.8) | 47 (43.5) | |
| >1 | 1 (5.6) | 8 (44.4) | 2 (22.2) | 1 (5.6) | 11 (61.1) | 1 (11.1) | 2 (11.1) | 10 (55.6) | 1 (11.1) | 2 (11.1) | 12 (66.7) | 1 (11.1) | 54 (50.0) | |
| plaD | <−1 | 5 (27.8) | 8 (44.4) | 2 (22.2) | 2 (11.1) | 6 (33.3) | 7 (77.8) | 4 (22.2) | 7 (38.9) | 2 (22.2) | 5 (27.8) | 3 (16.7) | 1 (11.1) | 41 (38.0) |
| −1 to 1 | 9 (50.0) | 5 (27.8) | 3 (33.3) | 11 (61.1) | 10 (55.6) | 2 (22.2) | 9 (50.0) | 9 (50.0) | 4 (44.4) | 9 (50.0) | 12 (66.7) | 5 (55.6) | 52 (48.1) | |
| >1 | 4 (22.2) | 5 (27.8) | 4 (44.4) | 5 (27.8) | 2 (11.1) | 0 (0.0) | 5 (27.8) | 2 (11.1) | 3 (33.3) | 4 (22.2) | 3 (16.7) | 3 (33.3) | 15 (13.9) | |
| Total | <−1 | 5 (9.3) | 8 (14.8) | 2 (7.4) | 6 (11.1) | 6 (11.1) | 9 (33.3) | 10 (18.5) | 7 (13.0) | 4 (14.8) | 9 (16.7) | 5 (9.3) | 3 (11.1) | 53 (16.4) |
| −1 to 1 | 43 (79.6) | 25 (46.3) | 17 (63.0) | 41 (75.9) | 24 (44.4) | 16 (59.3) | 35 (64.8) | 25 (46.3) | 18 (66.7) | 37 (68.5) | 23 (42.6) | 19 (70.4) | 145 (44.8) | |
| >1 | 6 (11.1) | 21 (38.9) | 8 (29.6) | 7 (13.0) | 24 (44.4) | 2 (7.4) | 9 (16.7) | 22 (40.7) | 5 (18.5) | 8 (14.8) | 26 (48.1) | 5 (18.5) | 126 (38.9) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syrokou, M.K.; Stasinopoulou, P.; Paramithiotis, S.; Bosnea, L.; Mataragas, M.; Papadopoulos, G.K.; Skandamis, P.N.; Drosinos, E.H. The Effect of Incubation Temperature, Substrate and Initial pH Value on Plantaricin Activity and the Relative Transcription of pln Genes of Six Sourdough Derived Lactiplantibacillus plantarum Strains. Fermentation 2021, 7, 320. https://doi.org/10.3390/fermentation7040320
Syrokou MK, Stasinopoulou P, Paramithiotis S, Bosnea L, Mataragas M, Papadopoulos GK, Skandamis PN, Drosinos EH. The Effect of Incubation Temperature, Substrate and Initial pH Value on Plantaricin Activity and the Relative Transcription of pln Genes of Six Sourdough Derived Lactiplantibacillus plantarum Strains. Fermentation. 2021; 7(4):320. https://doi.org/10.3390/fermentation7040320
Chicago/Turabian StyleSyrokou, Maria K., Panagiota Stasinopoulou, Spiros Paramithiotis, Loulouda Bosnea, Marios Mataragas, Georgios K. Papadopoulos, Panagiotis N. Skandamis, and Eleftherios H. Drosinos. 2021. "The Effect of Incubation Temperature, Substrate and Initial pH Value on Plantaricin Activity and the Relative Transcription of pln Genes of Six Sourdough Derived Lactiplantibacillus plantarum Strains" Fermentation 7, no. 4: 320. https://doi.org/10.3390/fermentation7040320
APA StyleSyrokou, M. K., Stasinopoulou, P., Paramithiotis, S., Bosnea, L., Mataragas, M., Papadopoulos, G. K., Skandamis, P. N., & Drosinos, E. H. (2021). The Effect of Incubation Temperature, Substrate and Initial pH Value on Plantaricin Activity and the Relative Transcription of pln Genes of Six Sourdough Derived Lactiplantibacillus plantarum Strains. Fermentation, 7(4), 320. https://doi.org/10.3390/fermentation7040320










