Abstract
The inoculation of grape juice with Saccharomyces cerevisiae strains selected from indigenous yeast populations can be a suitable tool to control alcoholic fermentation, contributing to producing wines with typical flavor and aroma and, hence, the demand for native starter cultures is increasing. However, since low amounts of indigenous yeast biomasses are usually required for local winemaking, the industrial production of these yeasts can be expensive. Therefore, in this study, after selecting an indigenous S. cerevisiae strain based on relevant oenological and technological features, a pilot-plant for easy and rapid production of fresh yeast biomass directly in a winery located in Tuscany, was exploited. The selected yeast strain was used as a starter to carry out 25 and 100 hL fermentations and its enological performance was compared with that of the commercial starter normally used in the winery. Chemical and sensory analysis of the resulting wines showed that they differentiated according to the used yeast strain, with the wines produced by the indigenous S. cerevisiae strain being characterized by a distinctive aromatic and sensory profile. In conclusion, the pilot-plant effectively resulted in producing fresh yeast starter cultures in the winery to be successfully used to carry out alcoholic fermentations.
1. Introduction
In winemaking, the use of selected Saccharomyces cerevisiae strains as Active Dry Yeast (ADY) starter cultures instead of traditional spontaneous fermentations is the most diffused practice to attain a more effective management of the fermentative process [1,2,3]. However, one of the main disadvantages of using commercial starters is the wine taste evenness and the loss of the wine distinctive aroma [4,5,6]. Recently, with the aim to overcome such drawbacks, the inoculation of grape juice with S. cerevisiae strains selected from indigenous populations from restricted oenological areas has been suggested [7,8], since some evidences that grape and wine microbiota exhibit regional patterns correlating with wine chemical composition were provided [9,10,11]. Indigenous selected S. cerevisiae strains might be better adapted for the specific conditions of some alcoholic fermentations than commercial starters, contributing also to the production of wines characterized by more typical flavor and aroma [2,4]. Therefore, over the last years, an increasing number of surveys have focused on microbial diversity associated with spontaneous grape must fermentations. These studies were aimed to select indigenous yeast strains, characterized by relevant technological properties, suitable to be used as starters in the industrial wine-making process and, at the same time, able to enhance the typicity of wine by fully exploiting the characteristics of the terroir [12,13,14,15,16,17,18,19,20,21]. According to the International Organization of Vine and Wine (OIV), terroir can be defined as “an area in which collective knowledge of the interactions between the identifiable physical and biological environment and applied viticulture and oenological practices develops, providing distinctive characteristics for the products originating from this area” [22]. Several authors underlined that the yeast microbiota associated with the terroir in which grapes are cultivated (vineyard) and fermented (winery), contribute to confer typical organoleptic properties to the wine [9,11,20,21]. In particular, the occurrence of specific S. cerevisiae strains in each winery supports the potential role of these microorganisms in determining specific wine characteristics and their selection could effectively represent a resource to contribute in preserving the typicity of wines, even at the winery level [13,23,24,25,26].
Once the indigenous S. cerevisiae strains have been selected, biomass should be produced according to the requirements of the winery to inoculate grape musts, in addition to having the characteristics of vitality, stability, resistance to storage conditions and safety, as stated by standards OIV for yeast starter cultures [27]. Usually, industrial processes for the production of microbial starters as ADY involve fermenters of 50.000–350.000 L or greater and molasses as a substrate [12]. The fermentation volume is gradually increased to optimize the biomass yield. The first two phases are characterized by a relatively low fermentation volume in aseptic conditions, while a later phase is characterized by a large fermentation volume but under non-aseptic condition. Then, the biomass is washed, concentrated, dried or lyophilized and finally packed.
This productive process is very expensive; however, the production in large quantities makes it profitable. On the contrary, it is not very advantageous when low amounts of indigenous yeast biomasses are produced for a single winery. These biomasses could be produced as a fresh liquid concentrate culture on demand just before or during the harvest season, if the production plant was located inside the winery. This could be a cheaper way to obtain active yeast cultures immediately ready for grape must inoculation, without needing the previous reactivation steps.
Therefore, the aim of the present study was the selection of indigenous S. cerevisiae strains to be used as a starter in a winery and the exploitation of a pilot-plant for easy and rapid production directly in the winery of fresh yeast biomass to use in winemaking. The yeast strains, chosen to be cultured in the pilot-plant, were selected on the bases of their oenological, sensorial and technological characteristics from the indigenous yeast microbiota of a winery located in Tuscany.
2. Materials and Methods
2.1. Spontaneous Alcoholic Fermentations
Eight spontaneous alcoholic fermentations were carried out in stainless steel tanks (25 or 50 hL capacity, Pandolfini s.r.l., Tavarnelle Val di Pesa, Firenze, Italy) not previously used and located close to the winery, but separated from the cellar where fermentations by commercial yeast starters occurred. Grape juice obtained from different grape varieties (Merlot, Sangiovese, Canaiolo) were used. Yeasts were isolated in the middle and at the end of the alcoholic fermentations by plating the wine samples on a WL Nutrient Agar medium (Oxoid Ltd., Basingstoke, Hampshire, UK) containing sodium propionate (VWR International Srl, Milan, Italy) (2 g/L) and streptomycin (VWR International Srl, Milan, Italy) (30 mg/L) to inhibit mould and bacteria growth, respectively. The plates were incubated for 48 h at 30 °C, under aerobic conditions. After purification of the colonies, the yeasts were grown on a YPD medium (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose), and maintained in a solution containing 50% (v/v) glycerol at −80 °C until use.
2.2. Saccharomyces Cerevisiae Strains Isolation
Almost 400 yeast isolates were identified as belonging to the Saccharomyces cerevisiae species by amplification of the 5.8S rRNA gene and of the two ribosomal internal transcribed spacers, by using the primer pair ITS1/ITS4 followed by restriction with HaeIII (Life Technologies Italia, Monza, Itlay), as described by Granchi et al., 1999 [28]. All these isolates were also characterized at strain level by inter-δ PCR typing with δ12/δ21 primer (Thermo Fisher Diagnostics S.p.A., Rodano, Milan; Italy) pair as reported by Legras and Karst, 2003 [29]. In order to distinguish the indigenous S. cerevisiae strains, all the commercial starters commonly used in the winery (12 strains) were included in this investigation. The RFLP patterns were submitted to pairwise comparison using the Dice coefficient [30] and cluster analysis with the unweighted pair group method (UPGMA) by Gel Compare 4.0 software (Applied Math, Kortrijk, Belgium). The identified native strains were subjected to a preliminary screening of oenological traits.
2.3. Preliminary Screening of S. cerevisiae Strains for Oenological Traits
Technological characteristics including fermenting vigour, sugar consumption, resistance to sulfur dioxide, production of acetic acid and H2S were preliminarily investigated in order to select strains with proper oenological features for red wine production. 250 mL Erlenmeyer flasks sealed with a Müller valve filled with sulfuric acid and containing 160 mL of sterile grape must were inoculated with the indigenous strains as axenic culture, in order to achieve an initial cell concentration of approximately 2 × 106 CFU/mL. The chemical composition of the grape must was as follows: glucose, 121 g/L; fructose, 109 g/L; malic acid, 1.28 g/L; total acidity, 4.43 g/L; NH3, 56 mg/L; αN, 117 mg/L; total SO2, 22 ppm, pH 3.37. The fermentations were carried out in duplicate at 25 °C. The fermentations’ progress was monitored daily, recording the weight loss, until the end of the fermentation (constant weight for three consecutive days). The fermentative vigour, expressing the speed at which yeast starts the fermentation, was determined as grams of CO2 produced in 48 h from the beginning of the trial, while the sugar and acetic acid concentrations were detected at the end of the fermentations by High Pressure Liquid Chromatography (HPLC). Sulfur dioxide resistance, defined as the maximum SO2 concentration at which the yeast exhibited detectable growth, was determined by agar plate assays. 5 μL of fresh yeast culture were spotted onto plates of a YPD medium buffered to pH 3.5 with tartaric acid, added with 15 g/L agar and supplemented with variable amounts of a sterile solution of potassium metabisulfite in order to obtain total SO2 concentrations ranging from 100 to 300 mg/L. After incubation at 26 °C for 72 h, the yeast growth was compared with that obtained on the agar plates not integrated with the solution of potassium metabisulfite (negative control). The production of hydrogen sulfide was estimated by spotting 5 μL of a yeast fresh culture on BIGGY agar plates (Oxoid ltd, Basingstoke, UK). After incubation at 26 °C for 72 h, the colour of the colonies may range between white-cream and brown-black according to increasing amounts of hydrogen sulfide produced.
2.4. Lab-Scale Fermentations by Selected S. cerevisiae Strains
On the basis of the results obtained from the preliminary screening, the selected S. cerevisiae strains were inoculated as axenic cultures in stainless steel tanks (10 L of volume) containing 7 L of Sangiovese grape must, whose chemical composition is above reported. At the time of the inoculation, the grape must contained 1.4 × 104 CFU/mL of non-Saccharomyces yeasts, 2.0 × 103 CFU/mL of lactic acid bacteria and 1.7 × 102 CFU/mL of acetic acid bacteria. The fermentations were carried out in duplicate (12 in total) at 26 °C and monitored by HPLC in order to quantify the main microbial metabolites and by viable plate counts to enumerate yeasts and bacteria populations [31,32]. Yeasts were quantified on a WL Nutrient Agar medium (Oxoid Ltd., Basingstoke, Hampshire, UK) incubated 48 h at 30 °C in aerobic conditions, lactic acid bacteria on an MRS Agar medium (Oxoid Ltd., Basingstoke, Hampshire, UK) incubated five days at 30 °C in anaerobic conditions and acetic acid bacteria on a Lafon-Lafourcade medium (Glucose, 10 g/L; yeast extract, 5 g/L; peptone, 5 g/L; tomato juice broth, 2 g/L; Agar, 20 g/L; pH 5.00). As a yeast inhibitor, pimaricin at 50 mg/L was added to the MRS and Lafon-Lafourcade media. To assess if the inoculated strains were actually responsible to carry out the alcoholic fermentation, a significant number of colonies grown on a WL medium were randomly chosen from each sample and submitted to amplification of inter-δ region, following the protocol above reported. At the end of the alcoholic fermentation, the wines were transferred to 5-L glass containers and placed at 18 °C. In order to perform the malolactic fermentation, all wines were inoculated with a commercial Oenococcus oeni strain (Viniflora CH11 Chr Hansen, Hoersholm, Denmark) following manufacturer’s instructions. The malic acid degradation and the consequent formation of lactic acid were monitored by HPLC. At the end of the malolactic fermentation, the wines underwent a cold stabilization (4 °C) for 7 days. After the stabilization, they were bottled and stored at room temperature.
2.5. Characteristics of the Yeast Multiplication Pilot-Plant
The experimentation that allowed designing, sizing and optimising the pilot-plant for fresh yeast production was described by Guerrini et al., 2018 [33]. The pilot-plant is composed of two main parts: a 500 L stainless steel tank for the yeast multiplication and a vertical centrifuge to separate the cells from their growth medium and obtain the biomass. In order to control the temperature, the tank was equipped with a heat exchanger and two cooling jackets to attain the desired value (i.e., 27 °C in our trials). A compressor equipped with a 0.2-μm filter and combined with a sparger provided the sterile air used for the mixing of the growth medium and the oxygen supplying for the yeast multiplication. The water used to fill the tank was sterilized by a UV-C lamp. A continuous screw feeder automatically provided nitrogen nutrients (Nutrient Vit Start Lallemand 1.5 gr/L) and solid rectified concentrated grape must (SRCM, Naturalia Ingredients s.r.l., Mazara del Vallo-TP-Italy) at 1 g/L per hour. As SRCM consisted of crystallized sugar extracted from grapes, the sugar’s content was 100% (50% fructose and 50% glucose). The separation between the growth medium and the yeasts was provided by a disc stack centrifuge (Alfa Laval, Italy) with a flow rate of 360 L/h. Finally, the pilot-plant was equipped with a dedicated automatic cleaning programme. All these components of the pilot-plant were electronically handled with a Programmable Logic Controller.
In particular, the pilot-plant working cycle can be divided into 6 main steps. First of all the tank is filled with 90 L of sterilized tap water. During this step, 1.5 g/L of nitrogen nutrients and phosphoric acid to adjust the medium to pH 5 are manually added to the water. Then, 10 L of axenic 24 h-yeast culture are introduced into the tank. The length of this step is roughly 20 min. Then, RCM is automatically dosed (1 g/L per hour) and the first yeast multiplication phase starts. The mass is mixed with sterilized air and the temperature is kept between 26 °C and 28 °C. Temperature, pH and dissolved oxygen are continuously monitored. This step lasts 240 min. The third step represents the second tank filling. After the 6 h, 350 L of sterilized water are automatically introduced into the tank. At the end of the second filling, the system alerts the operator for the second manual introduction of 1.5 g/L of nitrogen nutrients and phosphoric acid. The fourth step is the second yeast multiplication. As in the second step, the RCM is automatically dosed (1 g/L per hour). The mass is homogenized with sterilized air and the temperature is controlled set at 27 °C. This phase lasts 900 min. During the fifth step, the yeasts are separated from the culture medium. A pump feeds the centrifuge at a flow rate of 6 L/min. At the end of the stage, the fresh yeasts are recovered from the centrifuge bowl and stored at 4 °C. The final step is the pilot-plant cleaning. The fermenter, the pipes and the centrifuge are sanitized with 40 L of a per-acetic acid water solution at 2% (v/v). After 20 min, the tank is emptied, and the system is rinsed 3 times with hot (40 °C) sterilized water. The centrifuge disks are manually cleaned with a per-acetic acid water solution at 2% (v/v).
2.6. Industrial Fermentations
The selected indigenous S. cerevisiae strain and a commercial starter strain usually used in the winery (LALVIN BM 4X4, Lallemand Inc. Castel d’Azzano, Verona-Italy) were inoculated as axenic cultures in 25 hL and 100 hL stainless steel tanks containing Sangiovese grape must (pH: 3.36). The chemical composition of the grape must was as follows: glucose, 105 g/L; fructose, 109 g/L; malic acid,1.50 g/L; total acidity, 4.80 g/L; NH3, 55 mg/L; αN, 88 mg/L; total SO2, 61 mg/L. At the inoculation time, the grape must contained 4.0 × 104 CFU/mL of non-Saccharomyces yeasts and 8.6 × 102 CFU/mL of acetic acid bacteria. The fermentations were chemically and microbiologically monitored as reported for the lab-scale fermentations. At the end of the alcoholic fermentation, the wines were transferred into 10 hL tanks and inoculated with a commercial Oenococcus oeni strain (Lalvin VP41, Lallemand Inc. Castel d’Azzano, Verona-Italy) to carry out the malolactic fermentation. At the end of the malolactic fermentation, the wines were transferred into a 54 L-wine container and cold stabilized (4 °C) for 15 days. Afterwards, the wines were bottled and stored at room temperature.
2.7. Volatile Organic Compounds (VOC) Analysis
For volatile extraction (HS-SPME), a 40 mL vial was filled with 20 mL of sample. Extraction was performed by SPME using a DVB/CAR/PDMS fiber of 50/30 mm film thickness (Supelco, Bellefonte, PA, USA). Quantitative analyses by GC/MS were performed as reported by Verzera et al., 2008 [34] and Scacco et al., 2010 [35].
2.8. Sensory Analysis
The sensory analysis of the experimental wines obtained from the lab-scale fermentations was performed according to the methods reported by the Resolution OIV/CONCOURS 332A–2009 [36]. The sensory analysis of the wines obtained from the industrial fermentations was carried out by using the “Napping” procedure according to Perrin et al., 2008 [37]. Napping positioning was carried out by an untrained panel including 30 wine professionals (winemakers, wine advisors, oenologists and students) aged between 22 and 70 years. Each judge was provided with a rectangular sheet, 60 × 40 cm, where the wine samples were positioned based on the degree of the detected sensorial differences. The positioning distances on the sheet (map) were the discrimination criterion. In this test, each judge compared a set of 5 samples (4 different wines and 1 in duplicate) with the opportunity to indicate a preference for the sample considered qualitatively superior. For each map, the Cartesian coordinates of each sample were detected and measured in cm, after setting the 0 of the axes in the lower left corner of the 60 × 40 sheet. A matrix was obtained from each map, in which the rows consisted of the list of samples, and the columns contained the x and y coordinates determined by each judge. All the matrices obtained were processed through the Generalized Procastic Analysis (Senstools v. 1.2x OP&P Product Research BV, Utrecht, The Netherlands).
2.9. Statistical Analysis
Chemical compounds determinations, performed in duplicate, were elaborated according to nonparametric ANOVA followed by Tukey’s Test. Differences were reported at a significance level of p < 0.05. A Principal Component Analysis (PCA) was used to classify samples. All of the statistical analysis was performed by Statistica 7.0 software package (Stasoft GmbH, Hamburg, Germany).
3. Results
3.1. Indigenous Saccharomyces Cerevisiae Selection
Spontaneous fermentations were carried out at industrial scale at two different moments over the same vintage in a Tuscany winery. The sampling of the yeast microbiota was performed in the middle and at the end of the alcoholic fermentation process when S. cerevisiae populations occurred at concentrations higher than 106 CFU/mL. More than 400 S. cerevisiae isolates were analyzed to detect genetically different strains using the PCR amplification of the inter-δ regions. With the aim to recognize indigenous S. cerevisiae strains, the inter-δ patterns obtained were compared with the patterns of the commercial starter strains normally used in the winery to carry out alcoholic fermentations. The comparison pointed out 18 different patterns recognized as indigenous S. cerevisiae strains. Only two fermentations did not show these patterns, while the others showed an isolation frequency ranging from 10–45% (Figure 1). Most indigenous strains occurred in the presence of 56% (v/v) ethanol.
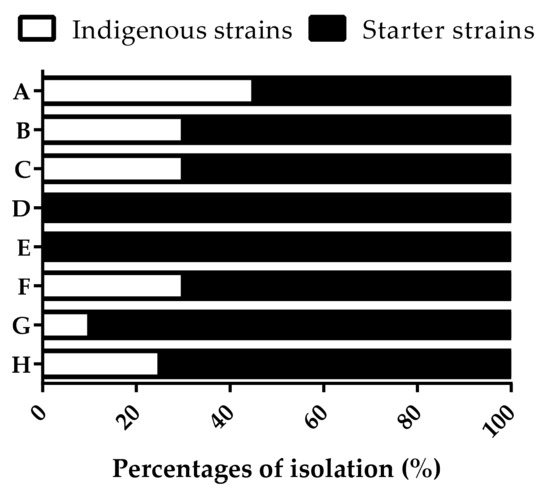
Figure 1.
Isolation frequency expressed as percentages of the indigenous and starter S. cerevisiae strains in different spontaneous fermentations carried out at industrial scale at the beginning (A, B, C, D) and the end of the same vintage (E, F, G, H).
The cluster analysis of the different inter-δ patterns highlighted the high genetic distance between 18 dominant indigenous S. cerevisiae strains and the commercial starter strains, since they grouped into two clusters at a similarity index lower than 30% (Figure 2). Only the strain VI 30 proved to be more similar to the starter strains 11 and 12.
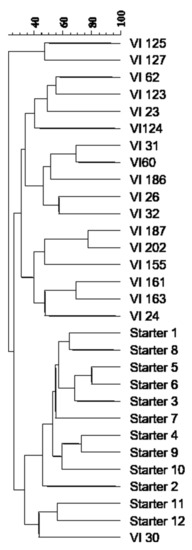
Figure 2.
Dendrogram from UPGMA clustering analysis, based on the Dice coefficient of the inter-δ patterns of the indigenous S. cerevisiae strains isolated from the spontaneous fermentations and the commercial starter strains usually used in the winery.
The 18 indigenous S. cerevisiae strains were tested through 160 mL-fermentations on sterile grape must in order to evaluate strain-specific oenological and technological properties. The parameters taken into consideration were the following: high fermentative vigour (CO2 produced in 48 h higher than 6 g/100 mL), total sugar consumption (glucose and fructose residues <2 g/L), acetic acid production lower than 0.5 g/L, SO2 production lower than 50 mg/L and lack of H2S production. The results of the screening are summarized in Table 1. The indigenous S. cerevisiae strains VI 23, VI 123, VI 127, VI 161, VI 186 and VI 202 showed the required parameters and, therefore, they were further characterized through fermentations of grape juice at laboratory scale.

Table 1.
Oenological and technological features of the 18 indigenous S. cerevisiae strains evaluated through 160 mL-fermentations with sterile grape must at 25 °C (+: presence; -: absence, underlined: S. cerevisiae strains selected for a further characterization).
3.2. Fermentation Performances of Selected Yeast Strains
The six selected S. cerevisiae strains were used as a starter in lab-scale fermentations carried out in stainless steel tanks containing 7 L of grape juice. Each strain was inoculated, as axenic culture, at the concentration of 2 × 106 CFU/mL. Table 2 shows the results of chemical and microbiological analyses performed at the end of each alcoholic fermentation and precisely after 10 days from the yeast strains inoculation. After this period, the sugar consumption was completed for the tested strains, with the exception of the strain VI 186 that showed a significant fructose residue. All the tested strains, despite strain-specific properties, confirmed the ability to produce low acetic acid and sulphur dioxide content (Table 2).

Table 2.
Chemical and microbiological analyses performed at the end of the alcoholic fermentations carried out by the six selected indigenous S. cerevisiae strains (LAB = lactic acid bacteria, AAB = acetic acid bacteria). Values with different superscript letters (a, b, c) within a row indicate significant different concentrations between strains (ANOVA, p < 0.05).
At the end of the fermentations, molecular analysis of the yeast populations confirmed the dominance of the inoculated S. cerevisiae strains. After the racking, a commercial Oenoccoccus oeni strain was inoculated in order to carry out malolactic fermentation. On the 35th day, when the malic acid was degraded, the wines were stabilized, bottled and finally analysed to quantify the volatile compounds. Considering that different S. cerevisiae strains can significantly affect the aromatic profile of wines [38,39,40,41,42], an SPE/GC-MS analysis of the various wines was performed allowing the identification and quantification of a number of volatile compounds that are by-products of yeast metabolism (Table 3). The concentration of total esters was higher in wines obtained by indigenous S. cerevisiae strains VI 161 and VI 186, while the concentration of total acetates was higher in those obtained by strains VI 123, VI 161 and VI 23. Finally, the wine produced by the strain VI 127 showed the lowest concentration of total alcohols.

Table 3.
SPE/GC-MS analysis of the wines obtained with the six indigenous strains. Values with different superscript letters (a, b, c) within a row indicate significant different concentrations between strains (ANOVA, p < 0.05).
Analysing the results in more detail, the concentration of some compounds exhibited statistically significant differences among some strains (ANOVA, p < 0.05). These compounds were 2-methyl-1-propanol, 3-methylbutyl acetate, hexanol, diethyl succinate, ethyl 9-decenoate, ethyl-dodecanoate, ethyl tetradecanoate, ethyl 9-hexadecenoate, ethyl hydrogen succinate, ethyl octadecenoate, ethyl 9-octadecenoate and dodecanoic acid. The perception thresholds and the corresponding aroma descriptor of some of these compounds are reported in literature [40,41,42,43]. Based on this information, 3-methylbutyl acetate (descriptor, banana; threshold, 30 μg/L), ethyl dodecanoate (descriptor, medicinal and grassy; threshold, 1500–2000 μg/L), ethyl tetradecanoate (descriptor, fruity and floral; threshold, 800 μg/L), ethyl 9-hexadecenoate (descriptor, pleasant sweet; aroma threshold, 1500 μg/L) and ethyl 9-octadecenoate (descriptor, coconut; threshold, 0.87 ng/mL) exceeded the perception threshold in wine. Principal component analysis (PCA) was applied to the matrix of multivariate data comprising concentrations of the volatile compounds reported in Table 3. Therefore, Figure 3A,B shows PCA scores and loadings biplots, respectively, for all the experimental wines originating from grape juice fermentation by the different strains. Examination of the data by PCA showed that PC1 and PC2 accounted for 71.06% of variation in the dataset. The S. cerevisiae strain VI 186 did not group with the others strains and was located to the left at the bottom, the strains VI 202 and VI 127 were positioned to the left at the top and the remaining strains on the right. The loadings of each compound on the principal components showed that 3-methylbutyl acetate, ethyl hexanoate, hexyl acetate, ethyl 9-decenoate and 2,4-bis(1,1-dimethylethyl)-phenol were mainly responsible for the cluster of indigenous strains VI 123, VI 161 and VI 23, while hexanol, methyl decanoate, beta-damascenone and ethyl 9-octadecenoate for the cluster of strains VI 202 and VI 127. Finally, ethyl dodecanoate, ehyl octanoate, ethyl decanoate, trans-nerolidol, ethyl tetradecanoate, octanoic acid and 4-ethylphenol were mainly responsible for the differentiation of the VI 186 strain.
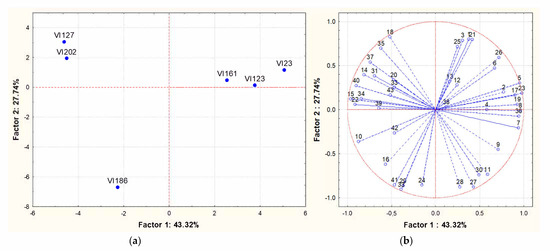
Figure 3.
Principal Component Analysis carried out on volatile compounds content in wines obtained by different S. cerevisiae indigenous strains (VI 23 -VI 123 - VI 127 -VI 161 - VI 186 and VI 202). (a): similarity map determined by Principal Component (Factor) 1 and 2; (b): projection of the variables on the factor plane. Variables: (1) Ethyl acetate; (2) Ethyl butanoate; (3) 1-propanol; (4) 2-methyl-1-propanol; (5) 3-methylbutyl acetate; (6) 3-methyl-1-butanol; (7) Ethyl hexanoate; (8) Hexyl acetate; (9) Ethyl lactate; (10) Hexanol; (11) Ethyl octanoate; (12) Ethyl nonanoate; (13) 2,3-butanediol; (14) Linalool; (15) Methyl decanoate; (16) Ethyl decanoate; (17) 3-methylbutyl octanoate; (18) Diethyl succinate; (19) Ethyl 9-decenoate; (20) alpha-terpineol; (21) Ethylphenyl acetate; (22) Beta-damascenone; (23) 2-Phenylethyl acetate; (24) Ethyl dodecanoate; (25) Hexanoic acid; (26) 2-phenylethanol; (27) 4-ethylguaiacol; (28) trans-nerolidol; (29) Ethyl tetradecanoate; (30) Octanoic acid; (31) 3-methylphenol; (32) Nonanoic Acid; (33) 4-ethylphenol; (34) Ethyl hexadecanoate; (35) Ethyl 9-hexadecenoate; (36) 2,4-bis(1,1-dimethylethyl)-phenol; (37) Ethyl hydrogen succinate; (38) Benzoic acid; (39) Ethyl octadecenoate; (40) Ethyl 9-octadecenoate; (41) Dodecanoic acid; (42) Tetradecanoic acid; (43) Hexadecanoic acid.
To assess whether the different concentrations of volatile compounds observed among the wines obtained by fermentations of different S. cerevisiae indigenous strains corresponded to different sensory profiles, a sensorial characterization of the wines was performed using the experimental approach proposed by the OIV [36]. The panel was composed of 12 experts and the chosen descriptors were: clarity, appearance, frankness, aromatic intensity and general impression. The results were expressed as a 5-point scale and analyzed with a statistical software. The perceptive map obtained from PCA applied to the mean intensity data of significant attributes is shown in Figure 4. The total variance explained for the sensory attributes based on the first two significant dimensions was 96%, with PC1 and PC2 accounting, respectively, for 78% and 18%. As shown in the PCA of the volatile compounds, the VI 186 strain did not group with the others strains, while the strains VI 23, VI 123 and VI 202 grouped in the same cluster characterized by a positive correlation with all the chosen descriptors, in particular, with the so-called “general impression”. Within this group, the S. cerevisiae strain VI 123 was chosen as a starter to induce an industrial-scale fermentation and, hence, it was multiplied into the pilot-plant.
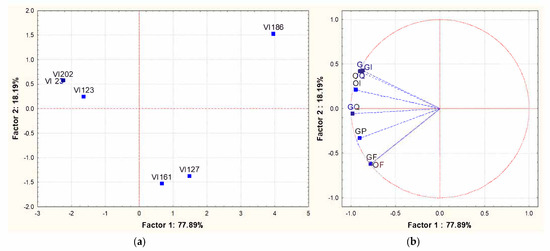
Figure 4.
Principal Component Analysis carried out on sensory analysis of the wines obtained with different S. cerevisiae indigenous strains (VI 23-VI 123-VI 127-VI 161-VI 186 and VI 202). The experimental approach is the one proposed by the OIV [36]. (a): similarity map determined by Principal Component (Factor) 1 and 2; (b): projection of the variables on the factor plane. Variables: OF: olfactory frankness; OI: olfactory intensity; OQ: olfactory quality; GF: gustatory frankness; GI: gustatory intensity; GQ: gustatory quality; GP: gustatory persistence; G: Global.
3.3. Production of Fresh Yeast Starter Culture by Pilot-Plant in the Winery
The S. cerevisiae strain VI 123 was produced as a fresh yeast starter culture by using the developed pilot-plant [33], and it was utilised to induce alcoholic fermentations at industrial scale. At the time of inoculation into the grape juice, the fresh yeast culture contained (1.15 ± 0.21) × 1010 CFU/g viable yeasts, a microbial contamination (total microbial count) of (2.90 ± 0.14) × 104 CFU/g and Enterobacteriaceae below the limit of detection (<10 CFU/g). The mortality rate of yeasts when the fresh biomass was stored for 40 days at 4 °C was 5%, while the microbial contamination remained unchanged.
3.4. Use of the Produced Yeast Culture at Industrial Scale
The enological performance of the indigenous strain VI 123 was compared with that of a commercial S. cerevisiae starter strain (LALVIN BM 4 × 4) normally used by the winery where the experimentation was accomplished. The strain VI 123 and the commercial strain BM 4 × 4 were inoculated separately to carry out industrial fermentations of two different volumes of grape must (25 hL and 100 hL). In Figure 5 and Figure 6 the chemical and microbiological data during the wine fermentations monitoring are reported. Both strains finished the fermentation after seven days
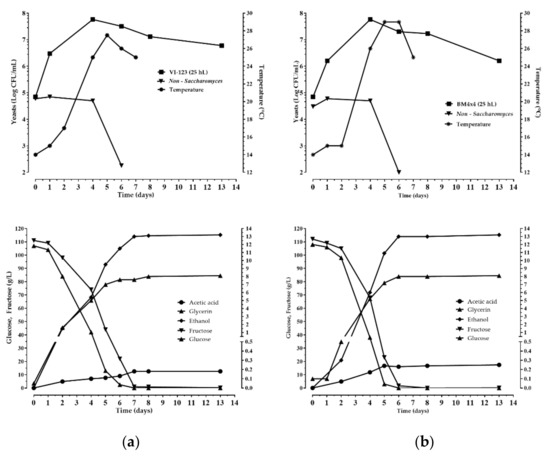
Figure 5.
Chemical and microbiological monitoring of the wine fermentations obtained inoculating the indigenous S. cerevisiae strain VI-123 (a), produced using the plant, and the commercial starter BM4 × 4 (b) in 25 hL of grape must.
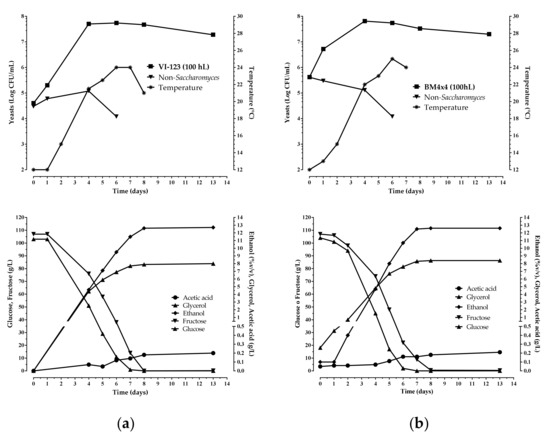
Figure 6.
Chemical and microbiological monitoring of the wine fermentations obtained inoculating the indigenous S. cerevisiae strain VI-123 (a), produced using the plant, and the commercial starter BM4 × 4 (b) in 100 hL of grape must.
Molecular analysis (PCR of the interdelta region), carried out on the yeast isolates collected at the seventh day from the inoculation in the different fermentations demonstrated that the inoculated yeast strains dominated the alcoholic fermentation (data not shown). After racking, a commercial O. oeni strain was inoculated in each wine to conduct malolactic fermentation. On the 30th day, when the malic acid was degraded, the wines were transferred into filled containers and kept at 4 °C for fifteen days. After cold stabilization, the wines were bottled and analyzed to quantify the volatile compounds (Table 4). The analysis of the variance (ANOVA, p < 0.05) showed that statistically significant differences among the various aromatic compounds in some cases were due to the yeast strain used (Ethyl lactate), whereas in other cases, to the tank size (ethyl acetate, ethyl hexanoate, methylethyl dodecanoate, decanoic acid), or otherwise to both (ethyl octanoate, diethyl succinate, 3-methylbutyl acetate, 3-methyl-1-butanol, benzaldehyde).

Table 4.
SPE/GC-MS analysis of the wines obtained from 25 and 100 hL fermentations inoculated by the indigenous S. cerevisiae strain VI-123 (produced using the plant) and the commercial starter BM4 × 4. Different letters indicate significant different concentrations between strains (ANOVA, p < 0.05).
Principal component analysis (PCA) was applied to the matrix of multivariate data comprising concentrations of the volatile compounds reported in Table 4, and Figure 7 shows PCA scores (a) and loadings biplots (b), respectively, for the industrial wines. Examination of the data by PCA showed that PC1 and PC2 accounted for 87.37% of variation in the dataset. The strains VI 123 and BM4 × 4 produced wines with a very similar volatile composition when inoculated in 100 hL of grape must. When the same strains were inoculated in 25 hL of grape must, the resulting wines were both characterized by a higher volatile complexity, even if they were very different from each other. In particular, they were principally characterized by a different combination of esters and alcohols. In other words, the difference between the volatile profiles of wines obtained from two yeast strains was more evident when they were inoculated in 25 hL of must than in 100 hL.
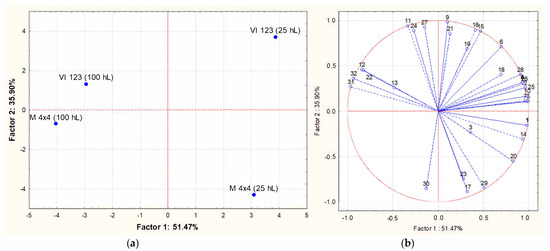
Figure 7.
Principal Component Analysis carried out on volatile compounds content in wines obtained inoculating the indigenous strain VI 123 and the commercial strain BM4 × 4 in 25 or 100 hL of grape must. (a): similarity map determined by Principal Component (Factor) 1 and 2; (b): projection of the variables on the factor plane. Variables: (1) Ethyl acetate; (2) Ethyl butanoate; (3) 1-propanol; (4) Ethyl 2-methylbutyrate; (5) Ethyl 3-methylbutyrate; (6) 2-methyl-1-propanol; (7) 3-methylbutyl acetate; (8) Limonene; (9) 3-methyl-1-butano; (10) Ethyl hexanoate; (11) Ethyl lactate; (12); Hexanol; (13) Methyl octanoate; (14) Ethyl octanoate; (15) Benzaldehyde; (16) Ethyl nonanoate; (17) 2.3-butanediol; (18) 1-octanol; (19) Methyl decanoate; (29) Ethyl decanoate; (21) 3-methylbutyl octanoate; (22) Diethyl succinate; (23) Ethylphenyl acetate; (24) 2-Phenylethyl acetate; (25) Methylethyl dodecanoate; (26) Hexanoic acid; (27) 2-phenylethanol; (28) Octanoic acid; (29) 4-methylphenol; (30) 3-methylphenol; (31) Ethyl hydrogen succinate; (32) Decanoic acid.
Once again, the sensorial characterization of the wines was carried out using the experimental approach and the related statistical elaboration previously described. PCA analysis showed that the total variance explained for the sensory attributes based on the first two significant dimensions was 99%, with PC1 and PC2 accounting, respectively, for 75% and 24% (Figure 8). The wines did not group either based on the inoculated strain or on the volume of fermented must. The strain VI 123 inoculated in 25 hL and the strain BM4 × 4 inoculated in 100 hL resulted characterized by a positive correlation with all the chosen descriptors, with the exception of “frankness”.
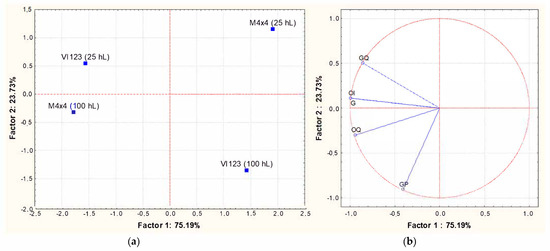
Figure 8.
Principal Component Analysis carried out on sensorial characterizations of wines obtained inoculating the indigenous strain VI 123 and the commercial strain BM4 × 4 in 25 or 100 hL of grape must. The experimental approach is the one proposed by the OIV [36]. (a): similarity map determined by Principal Component (Factor) 1 and 2; (b): projection of the variables on the factor plane. Variables: OF: olfactory frankness; OI: olfactory intensity; OQ: olfactory quality; GF: gustatory frankness; GI: gustatory intensity; GQ: gustatory quality; GP: gustatory persistence; G: Global.
Finally, to verify if an untrained panel was actually able to perceive the differences between wines obtained with the indigenous S. cerevisiae strain and those obtained with the commercial starter, the so-called “Napping method” was used. This approach is based on the method known as the “projective map” developed by Risvik, et al., 1994 [44] that allows subjects to express perceived similarities through the positioning of the samples along the two dimensions of a plane. In such a way the distant samples are those perceived as different, and those near are perceived as similar. To evaluate the effective judgment of the subjects involved in the test, a sample is usually provided in duplicate. The method provides that the sensorial characteristics of the products are evaluated according to a criterion chosen individually by each subject and does not provide for an analytical approach through defined descriptors. For these reasons, the Napping method is considered a “holistic” approach to the sensory evaluation of a product. In addition, this method offers to the involved subjects the opportunity to express their preference for a sample. Therefore, a map relating to the positioning of the various wines tasted was drawn for each subject so that 30 maps were obtained. The statistical treatment of these 30 individual maps through multivariate analysis (Generalized Procastic Analysis—GPA), allowed the building of a single consensus map that is reported in Figure 9. The two components explained the 56.11% of the total variance (44.76% and 11.35%, respectively). The proximity of two replicated samples indicated a good discriminatory performance of the panel. Both yeast strains demonstrated different sensory performances based on the size of the fermentation tank, confirming the results obtained previously. Regarding the preferences expressed, the most appreciated wine was obtained with the indigenous S. cerevisiae strain VI123 inoculated in 100 hL of grape must (67% of preferences). The wine obtained with the commercial strain inoculated in 100 hL of grape must was the second choice with 19% of the preferences. The remaining preferences (14%) were expressed for the wine obtained with the indigenous strain inoculated in 25 hL of grape must. No preference was expressed for the commercial strain when it was inoculated in 25 hL.
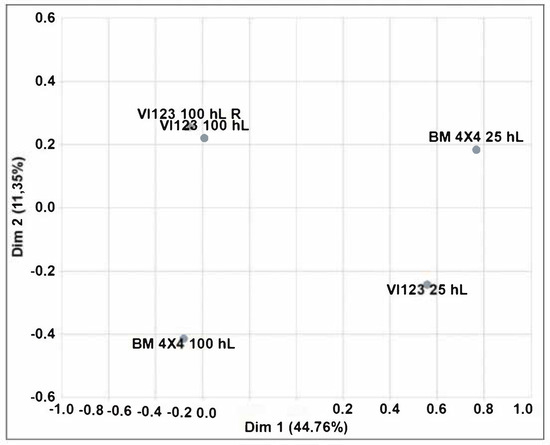
Figure 9.
Generalized Procastic Analysis carried out on 30 individual maps of the Napping test performed on wines obtained inoculating the indigenous strain VI 123 and the commercial strain BM4 × 4 in 25 or 100 hL of grape must. (R = replicate).
4. Discussion
In recent years, evidences that specific indigenous S. cerevisiae strains conferring distinctive chemical and sensory wine properties may be associated with a terroir caused an increasing interest among winemakers in selecting local strains with the aim to obtain tailored starter cultures potentially well adapted to specific ecosystems [4,5,6,7,8,9,10,11,23,45,46,47,48]. In this context, firstly, we performed a selection of indigenous S. cerevisiae strains in a winery located in Tuscany, where commercial ADY starters were usually inoculated into grape juice. Despite the repeated use of this practice in the alcoholic fermentation management in this winery, different indigenous S. cerevisiae strains possessing technological properties comparable to commercial starters were isolated. Other studies carried out on the implantation of starter cultures, as well as on the competition between indigenous and commercial inoculated strains, highlighted that the inoculation of tanks with dried yeasts does not always assure their implantation during the fermentation process and, hence, some indigenous S. cerevisiae strains are capable to participate in or conduct the alcoholic fermentation [46,48,49,50,51,52,53]. It is underlined that in our work, in order to have more chances in isolating indigenous S. cerevisiae strains, spontaneous wine fermentations were carried out by using musts from different grape varieties, new steel stainless tanks and new pumps located inside an area close to the cellar, but different from the place where ADY starters were usually used. Therefore, this procedure limiting the possible presence of the winery-resident yeasts might be a suitable strategy to perform a successful selection indigenous yeast program in wineries which extensively used commercial yeast starters. However, in the winery here taken into consideration, up to 12 different commercial yeast strains were used, thus, making the implantation/persistence of commercial strains in musts or in winery facilities more difficult.
In total, 18 indigenous S. cerevisiae strains were isolated from eight spontaneous fermentations, demonstrating that natural yeast populations showed a high genetic diversity as stated by different studies [23,25,46,49,53]. Probably, the vineyard in addition to winery becomes a very important reservoir of interesting genetic resources as, also in the present work, some indigenous S. cerevisiae strains possessing suitable oenological characteristics were found. Moreover, comparison of interdelta profiles between indigenous and commercial yeast strains revealed that the latter grouped in the same cluster at a low level of similarity of approximately 25%. Only the indigenous strain VI30 exhibited a higher similarity level to two commercial S. cerevisiae strains and might represent yeast population descended from commercial strains, as suggested by some authors [46,48,53].
After selecting the most performant indigenous S. cerevisiae strain based on its oenological properties, we exploited a pilot-plant previously developed [33] in order to produce a concentrate fresh biomass directly available for using in the winery as a starter culture. Indeed, the chance to produce local wine yeasts on demand in a production plant close to wineries just before or during the harvest season might be an advantage because industrial production of low amounts of a single yeast strain as active dry yeast (ADY) would be very expensive [12,33]. In addition, the fresh yeast cultures were ready for must inoculation, since the reactivation (i.e., rehydration) stage was not required, and, thus, vats were more quickly inoculated. By using the pilot-plant, in a 24 h-working-cycle, the fresh biomass to inoculate 100 hL of grape must was obtained. As a matter of fact, the amount of viable cells per gram of fresh yeast after the centrifugation step was (1.15 ± 0.21) × 1010 CFU/g and no contamination by wild yeasts, acetic acid bacteria, coliforms or filamentous fungi was observed, in agreement with the standards OIV for yeast starter cultures [27]. Moreover, evidence that fresh biomass can be stored at 4 °C for 40 days without significant vitality loss, made this yeast production procedure very convenient for the single winery needs.
During the vintage, biomass of the indigenous S. cerevisiae strain produced locally in the winery was used to inoculate 25 and 100 hL of Sangiovese must in comparison with a commercial yeast strain usually used to carry out red wine fermentations. The alcoholic fermentation performed at industrial scale by both S. cerevisiae strains showed regular time-course, attaining complete sugar degradation in a week. During the fermentation process the implantation of the indigenous, as well as the commercial, strain was demonstrated by an interdelta profiles technique. These results confirmed the promising technological features exhibited by the indigenous yeast strain in laboratory-scale fermentations and, thus, the effectiveness of the selection process adopted to select indigenous yeasts to be used as a starter culture in winemaking. The wines obtained with the indigenous S. cerevisiae strain and the wine obtained with the commercial starter differed in the concentration of some aromatic compounds. These differences were sensory perceptible not only from a trained panel, but also from an untrained panel comprising professionals operating in the wine sector, such as winemakers, wine advisors, oenologists, university students, etc. Furthermore, a predilection of the tasters toward the wine produced with the indigenous yeast strain was pointed out, suggesting that the use of selected indigenous starter cultures seems a suitable tool for differentiation and diversification of wines produced in a specific oenological area as supported by other surveys [50,51,52,53].
In conclusion, this study demonstrated the possibility for the wineries to self-produce their own starter thanks to a low cost pilot-plant and an effective selection process of the winery indigenous yeasts. The selection of indigenous S. cerevisiae strains adapted to a specific winery, limiting the use of commercial yeasts, has great importance in the biodiversity preservation and exploitation and may contribute to the maintenance of typical sensory properties of the wines of each specific region, and to the development of a more competitive regional wine industry.
Author Contributions
Conceptualization, S.G., L.G. (Lisa Granchi), A.P.; methodology, G.B., D.B., L.G. (Lorenzo Guerrini); validation, D.B., L.G. (Lorenzo Guerrini), G.B., Y.R.; laboratory analysis, E.M., S.M., D.B.; writing—original draft preparation, S.G., L.G. (Lorenzo Guerrini); writing—review and editing, L.G. (Lisa Granchi), V.G.; funding acquisition, S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the FEASR (Piano di Sviluppo Rurale 2014–2020 della Regione Toscana sottomisura 16.2) contribution. Project title “Produzione in cantina di lieviti starter autoctoni—VICAStart”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors would like to thanks Cantina Sociale Vicas soc. coop., Cantina Tuscania srl and Parsec srl for their technical support during the trials and the plant development.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fleet, G.H. Wine yeasts for the future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef] [Green Version]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Suárez-Lepe, J.; Morata, A. New trends in yeast selection for winemaking. Trends Food Sci. Technol. 2012, 23, 39–50. [Google Scholar] [CrossRef]
- Francesca, N.; Chiurazzi, M.; Romano, R.; Aponte, M.; Settanni, L.; Moschetti, G. Indigenous yeast communities in the environment of “Rovello bianco” grape variety and their use in commercial white wine fermentation. World J. Microbiol. Biotechnol. 2010, 26, 337–351. [Google Scholar] [CrossRef] [Green Version]
- Di Maio, S.; Polizzotto, G.; Di Gangi, E.; Foresta, G.; Genna, G.; Verzera, A.; Scacco, A.; Amore, G.; Oliva, D. Biodiversity of Indigenous Saccharomyces Populations from Old Wineries of South-Eastern Sicily (Italy): Preservation and Economic Potential. PLoS ONE 2012, 7, e30428. [Google Scholar] [CrossRef]
- Csoma, H.; Zakany, N.; Capece, A.; Romano, P.; Sipiczki, M. Biological diversity of Saccharomyces yeasts of spontaneously fermenting wines in four wine regions: Comparative genotypic and phenotypic analysis. Int. J. Food Microbiol. 2010, 140, 239–248. [Google Scholar] [CrossRef]
- Esteve-Zarzoso, B.; Gostíncar, A.; Bobet, R.; Uruburu, F.; Querol, A. Selection and molecular characterization of wine yeasts isolated from the ‘El Penedès’ area (Spain). Food Microbiol. 2000, 17, 553–562. [Google Scholar] [CrossRef]
- Orlic, S.; Redžepović, S.; Jeromel, A.; Herjavec, S.; Iacumin, L. Influence of indigenous Saccharomyces paradoxus strains on Chardonnay wine fermentation aroma. Int. J. Food Sci. Technol. 2007, 42, 95–101. [Google Scholar] [CrossRef]
- Knight, S.; Klaere, S.; Fedrizzi, B.; Goddard, M.R. Regional microbial signatures positively correlate with differential wine phenotypes: Evidence for a microbial aspect to terroir. Sci. Rep. 2015, 5, 14233. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among Wine Grape Microbiome, Metabolome, and Fermentation Behavior Suggest Microbial Contribution to Regional Wine Characteristics. mBio 2016, 7, e00631-16. [Google Scholar] [CrossRef] [Green Version]
- Anagnostopoulos, D.A.; Kamilari, E.; Tsaltas, D. Contribution of the Microbiome as a Tool for Estimating Wine’s Fermentation Output and Authentication. In Advances in Grape and Wine Biotechnology; IntechOpen: London, UK, 2019; pp. 1–21. [Google Scholar]
- Maqueda, M.; Nevado, F.P.; Regodón, J.A.; Zamora, E.; Álvarez, M.L.; Rebollo, J.E.; Ramírez, M. A low-cost procedure for production of fresh autochthonous wine yeast. J. Ind. Microbiol. Biotechnol. 2011, 38, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Ganucci, D.; Guerrini, S.; Mangani, S.; Vincenzini, M.; Granchi, L. Quantifying the Effects of Ethanol and Temperature on the Fitness Advantage of Predominant Saccharomyces cerevisiae Strains Occurring in Spontaneous Wine Fermentations. Front. Microbiol. 2018, 9, 1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tristezza, M.; Vetrano, C.; Bleve, G.; Spano, G.; Capozzi, V.; Logrieco, A.; Mita, G.; Grieco, F. Biodiversity and safety aspects of yeast strains characterized from vineyards and spontaneous fermentations in the Apulia Region, Italy. Food Microbiol. 2013, 36, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Tristezza, M.; Fantastico, L.; Vetrano, C.; Bleve, G.; Corallo, D.; Grieco, F.; Mita, G. Molecular and Technological Characterization ofSaccharomyces cerevisiaeStrains Isolated from Natural Fermentation of Susumaniello Grape Must in Apulia, Southern Italy. Int. J. Microbiol. 2014, 2014, 897428. [Google Scholar] [CrossRef]
- Tristezza, M.; Vetrano, C.; Bleve, G.; Grieco, F.; Tufariello, M.; Quarta, A.; Mita, G.; Spano, G. Autochthonous fermentation starters for the industrial production of Negroamaro wines. J. Ind. Microbiol. Biotechnol. 2012, 39, 81–92. [Google Scholar] [CrossRef]
- Capozzi, V.; Russo, P.; Beneduce, L.; Weidmann, S.; Grieco, F.; Guzzo, J.; Spano, G. Technological properties ofOenococcus oenistrains isolated from typical southern Italian wines. Lett. Appl. Microbiol. 2010, 50, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Garofalo, C.; Chiriatti, M.A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2015, 181, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Cappello, M.S.; Stefani, D.; Grieco, F.; Logrieco, A.F.; Zapparoli, G. Genotyping by Amplified Fragment Length Polymorphism and malate metabolism performances of indigenous Oenococcus oeni strains isolated from Primitivo wine. Int. J. Food Microbiol. 2008, 127, 241–245. [Google Scholar] [CrossRef]
- Garofalo, C.; El Khoury, M.; Lucas, P.; Bely, M.; Russo, P.; Spano, G.; Capozzi, V. Autochthonous starter cultures and indigenous grape variety for regional wine production. J. Appl. Microbiol. 2015, 118, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Grieco, F.; Tristezza, M.; Vetrano, C.; Bleve, G.; Panico, E.; Mita, G.; Logrieco, A. Exploitation of autochthonous microorganism potential to enhance the quality of Apulian wines. Ann. Microbiol. 2011, 61, 67–73. [Google Scholar] [CrossRef]
- Anonymous. Definition of Vitinicultural Terroir; RESOLUTION OIV/VITI 333/2010; International Organization of Vine and Wine General Assembly (OIV): Tbilisi, Georgia, 2010. [Google Scholar]
- Granchi, L.; Ganucci, D.; Buscioni, G.; Mangani, S.; Guerrini, S. The Biodiversity of Saccharomyces cerevisiae in Spontaneous Wine Fermentation: The Occurrence and Persistence of Winery-Strains. Fermentation 2019, 5, 86. [Google Scholar] [CrossRef] [Green Version]
- Vezinhet, F.; Hallet, J.N.; Valade, M.; Poulard, A. Ecological survey of wine yeast strains by molecular methods of identi-fication. Am. J. Enol. Vitic. 1992, 43, 83–86. [Google Scholar]
- Augruso, S.; Ganucci, D.; Buscioni, G.; Granchi, L.; Vincenzini, M. A ogni cantina il suo lievito? VQ 2008, 5, 58–65. [Google Scholar]
- Aponte, M.; Blaiotta, G. Potential Role of Yeast Strains Isolated from Grapes in the Production of Taurasi DOCG. Front. Microbiol. 2016, 7, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anonymous. Monograph of Saccharomyces Yeasts; RESOLUTION OIV/576A-2017; International Organization of Vine and Wine General Assembly (OIV): Sofia, Bulgaria, 2017. [Google Scholar]
- Granchi, L.; Bosco, M.; Messini, A.; Vincenzini, M. Rapid detection and quantification of yeast species during spontaneous wine fermentation by PCR-RFLP analysis of the rDNA ITS region. J. Appl. Microbiol. 1999, 87, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Legras, J.-L.; Karst, F. Optimisation of interdelta analysis forSaccharomyces cerevisiaestrain characterisation. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy. In The Principles and Practice of Numerical Classification; W.H. Freeman and Company: San Francisco, CA, USA, 1973; p. 573. [Google Scholar]
- Schneider, A.; Gerbi, V.; Redoglia, M. A rapid HPLC method for separation and determination of major organic acids in grape musts and wines. Am. J. Enol. Vitic. 1987, 38, 151–155. [Google Scholar]
- Granchi, L.; Ganucci, D.; Messini, A.; Rossellini, D.; Vincenzini, M. Dynamics of Yeast Populations during the Early Stages of Natural Fermentations for the Production of Brunello di Montalcino Wines. Food Technol. Biotech. 1998, 36, 313–318. [Google Scholar]
- Guerrini, L.; Parenti, A.; Angeloni, G.; Masella, P.; Barbato, D.; Mari, E.; Romboli, Y.; Buscioni, G.; Mangani, S.; Guerrini, S.; et al. Development of a Machine for the Production of Liquid Wine Yeast to Induce Grape Juice Fermentations. EurAgEng Conference, Wageningen, The Netherlands, 8–12 July 2018; pp. 1–6. [Google Scholar]
- Verzera, A.; Ziino, M.; Scacco, A.; Lanza, C.M.; Mazzaglia, A.; Romeo, V.; Condurso, C. Volatile Compound and Sensory Analysis for the Characterization of an Italian White Wine from “Inzolia” Grapes. Food Anal. Methods 2008, 1, 144–151. [Google Scholar] [CrossRef]
- Scacco, A.; Verzera, A.; Lanza, C.M.; Sparacio, A.; Genna, G.; Raimondi, S.; Tripodi, G.; Dima, G. Influence of Soil Salinity on Sensory Characteristics and Volatile Aroma Compounds of Nero d’Avola Wine. Am. J. Enol. Vitic. 2010, 61, 498–505. [Google Scholar] [CrossRef]
- Anonymous. OIV Standard for International Wine and Spirituous Beverages of Vitivinicultural Origin Competitions Resolution; OIV/CONCOURS 332A/2009; International Organization of Vine and Wine General Assembly (OIV): Zagreb, Croatia, 2009. [Google Scholar]
- Perrin, L.; Symoneaux, R.; Maître, I.; Asselin, C.; Jourjon, F.; Pagès, J. Comparison of three sensory methods for use with the Napping® procedure: Case of ten wines from Loire valley. Food Qual. Prefer. 2008, 19, 1–11. [Google Scholar] [CrossRef]
- Ugliano, M.; Henschke, P.A. Yeasts and Wine Flavour. In Wine Chemistry and Biochemistry; Moreno-Arribas, M.V., Polo, M.C., Eds.; Springer Science Business Media, LLC: New York, NY, USA, 2009; pp. 313–392. [Google Scholar]
- Romano, P. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef]
- Guerrini, L.; Masella, P.; Spugnoli, P.; Spinelli, S.; Calamai, L.; Parenti, A. A Condenser to Recover Organic Volatile Compounds during Vinification. Am. J. Enol. Vitic. 2016, 67, 163–168. [Google Scholar] [CrossRef]
- Bartowsky, E.J.; Pretorius, I.S. Microbial Formation and Modification of Flavor and Off-Flavor Compounds in Wine. In Biology of Microorganisms on Grapes, in Must and in Wine; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2009; pp. 209–231. [Google Scholar]
- Englezos, V.; Rantsiou, K.; Cravero, F.; Torchio, F.; Pollon, M.; Fracassetti, D.; Ortiz-Julien, A.; Gerbi, V.; Rolle, L.; Cocolin, L. Volatile profile of white wines fermented with sequential inoculation of Starmerella bacillaris and Saccharomyces cerevisiae. Food Chem. 2018, 257, 350–360. [Google Scholar] [CrossRef] [PubMed]
- McKay, M.; Buica, A. Factors Influencing Olfactory Perception of Selected Off-flavourcausing Compounds in Red Wine—A Review. South Afr. J. Enol. Vitic. 2020, 41, 56–71. [Google Scholar] [CrossRef]
- Risvik, E.; McEwan, J.A.; Colwill, J.S.; Rogers, R.; Lyon, D.H. Projective mapping: A tool for sensory analysis and consumer research. Food Qual. Prefer. 1994, 5, 263–269. [Google Scholar] [CrossRef]
- Agarbati, A.; Canonico, L.; Ciani, M.; Comitini, F. Fitness of Selected Indigenous Saccharomyces cerevisiae Strains for White Piceno DOC Wines Production. Fermentation 2018, 4, 37. [Google Scholar] [CrossRef] [Green Version]
- Aponte, M.; Romano, R.; Villano, C.; Blaiotta, G. Dominance of S. cerevisiae Commercial Starter Strains during Greco di Tufo and Aglianico Wine Fermentations and Evaluation of Oenological Performances of Some Indigenous/Residential Strains. Foods 2020, 9, 1549. [Google Scholar] [CrossRef]
- Feghali, N.; Bianco, A.; Zara, G.; Tabet, E.; Ghanem, C.; Budroni, M. Selection of Saccharomyces cerevisiae Starter Strain for Merwah Wine. Fermentation 2020, 6, 43. [Google Scholar] [CrossRef]
- Capece, A.; Pietrafesa, R.; Siesto, G.; Romaniello, R.; Condelli, N.; Romano, P. Selected Indigenous Saccharomyces cerevisiae Strains as Profitable Strategy to Preserve Typical Traits of Primitivo Wine. Fermentation 2019, 5, 87. [Google Scholar] [CrossRef] [Green Version]
- Tello, J.; Cordero-Bueso, G.; Aporta, I.; Cabellos, J.; Arroyo, T. Genetic diversity in commercial wineries: Effects of the farming system and vinification management on wine yeasts. J. Appl. Microbiol. 2012, 112, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Barrajón, N.; Arévalo-Villena, M.; Úbeda, J.; Briones, A. Competition between Spontaneous and Commercial Yeasts in Winemaking: Sudy of Possible Factors Involved. In Communicating Current Research and Ed-Ucational Topics and Trends in Applied Microbiology; Méndez-Vilas, I.A., Ed.; Microbiology Book Series; Elsevier: Amsterdam, The Netherlands, 2011; Volume 2, ISBN 978-84-614-6195-0. [Google Scholar]
- Lange, J.N.; Faasse, E.; Tantikachornkiat, M.; Gustafsson, F.S.; Halvorsen, L.C.; Kluftinger, A.; Ledderhof, D.; Durall, D.M. Implantation and persistence of yeast inoculum in Pinot noir fermentations at three Canadian wineries. Int. J. Food Microbiol. 2014, 180, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Martiniuk, J.T.; Pacheco, B.; Russell, G.; Tong, S.; Backstrom, I.; Measday, V. Impact of Commercial Strain Use on Saccharomyces cerevisiae Population Structure and Dynamics in Pinot Noir Vineyards and Spontaneous Fermentations of a Canadian Winery. PLoS ONE 2016, 11, e0160259. [Google Scholar] [CrossRef] [Green Version]
- Capece, A.; Granchi, L.; Guerrini, S.; Mangani, S.; Romaniello, R.; Vincenzini, M.; Romano, P. Diversity of Saccharomyces cerevisiae Strains Isolated from Two Italian Wine-Producing Regions. Front. Microbiol. 2016, 7, 1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).