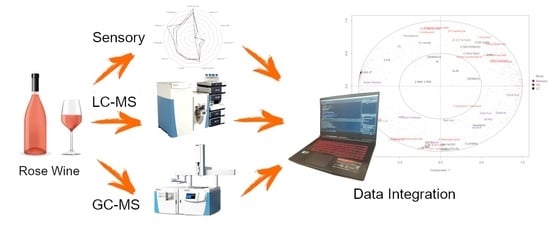
A Statistical Workflow to Evaluate the Modulation of Wine Metabolome and Its Contribution to the Sensory Attributes
Abstract
1. Introduction
2. Materials and Methods
2.1. Wine Samples
2.2. Analysis, Data Acquisition and Processing of Headspace Solid-Phase Microextraction Coupled with Gas Chromatography-Mass Spectrometry (HS-SPME-GC-MS) Data
2.3. Analysis, Data Acquisition and Processing of Using Ultra-High-Performance Liquid Chromatography High-Resolution Mass Spectrometry (UHPLC-HRMS) Data
2.4. Sensory Analysis
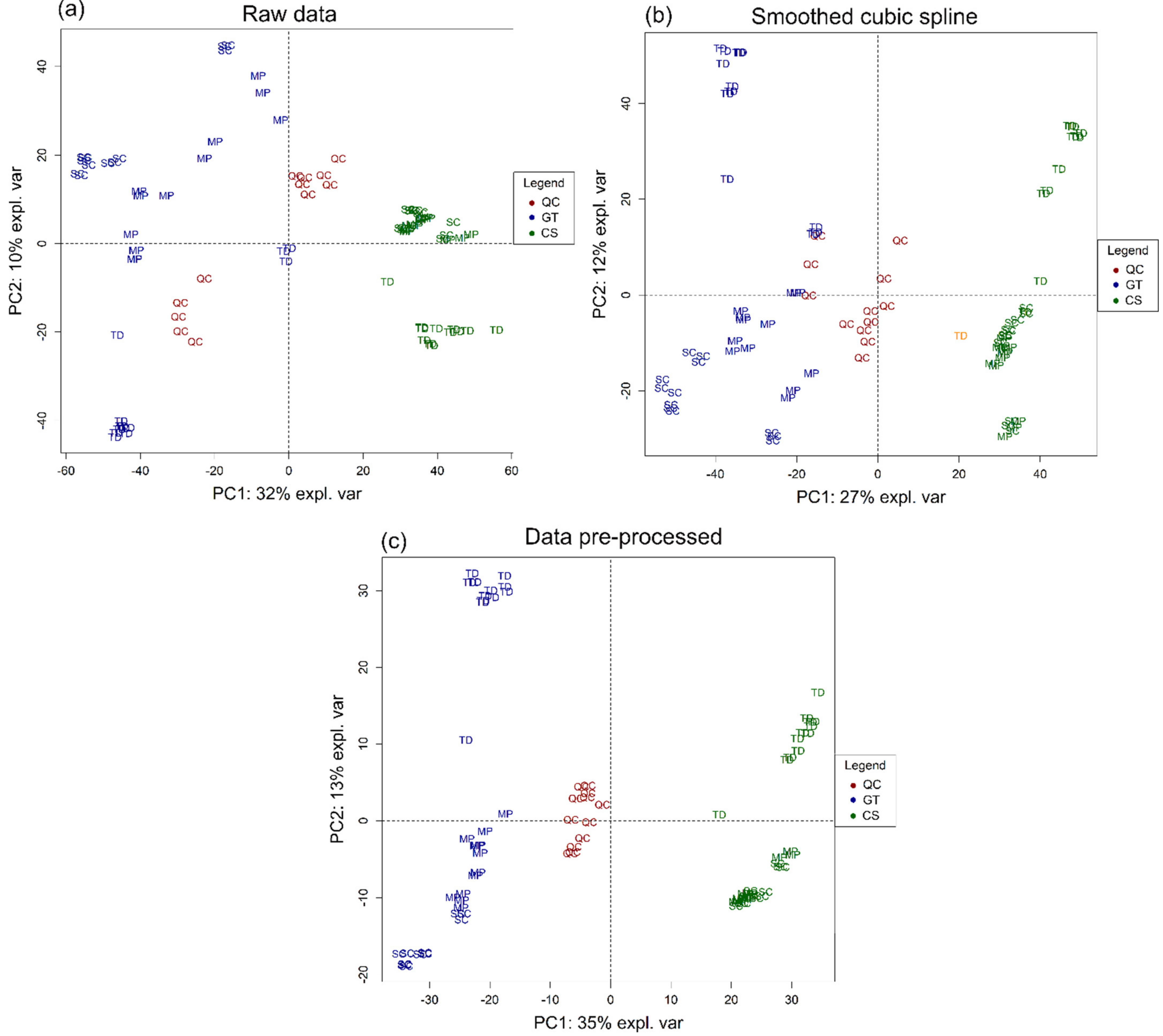
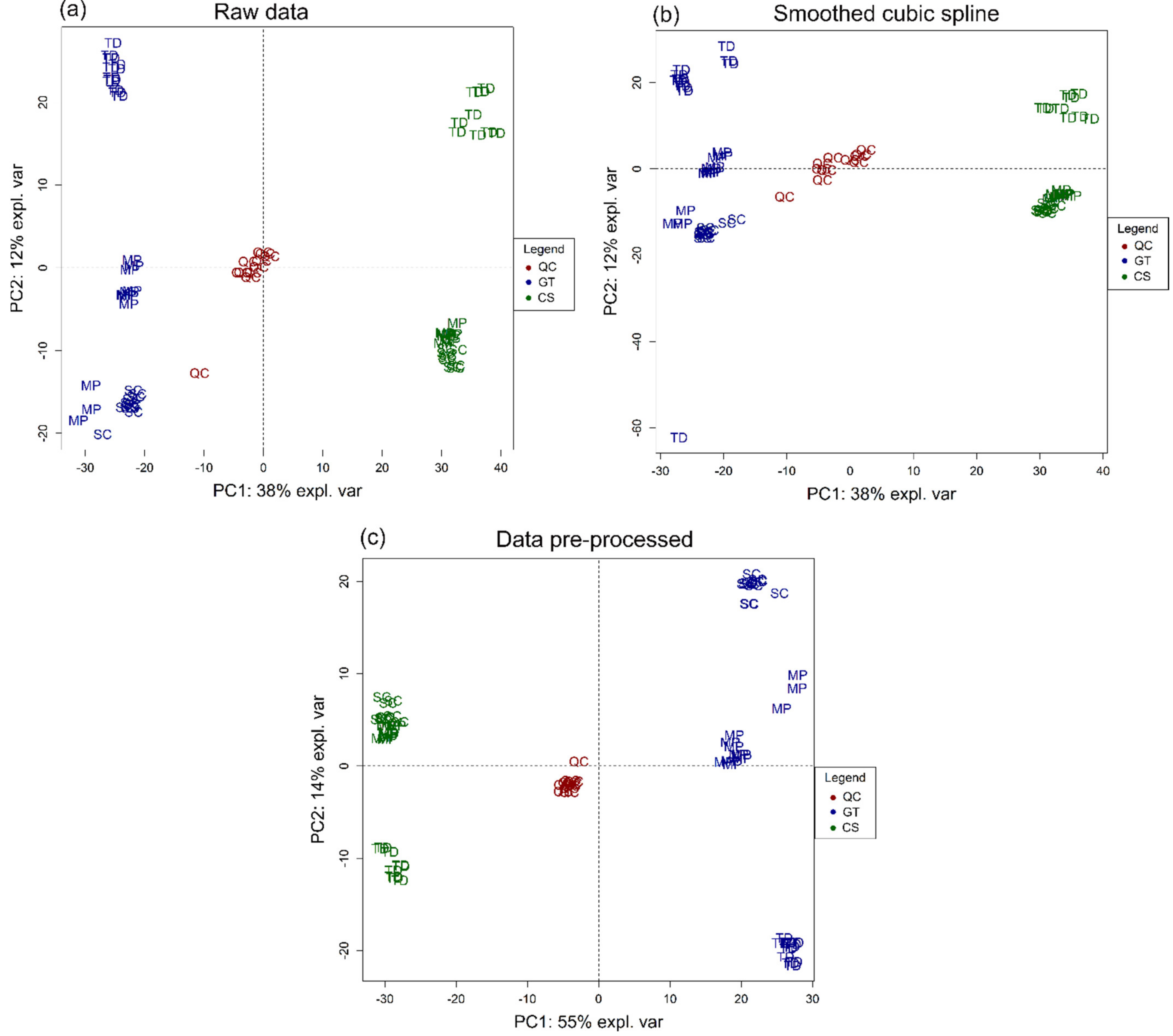
2.5. Statistical Analysis
3. Results
3.1. Volatile Profile
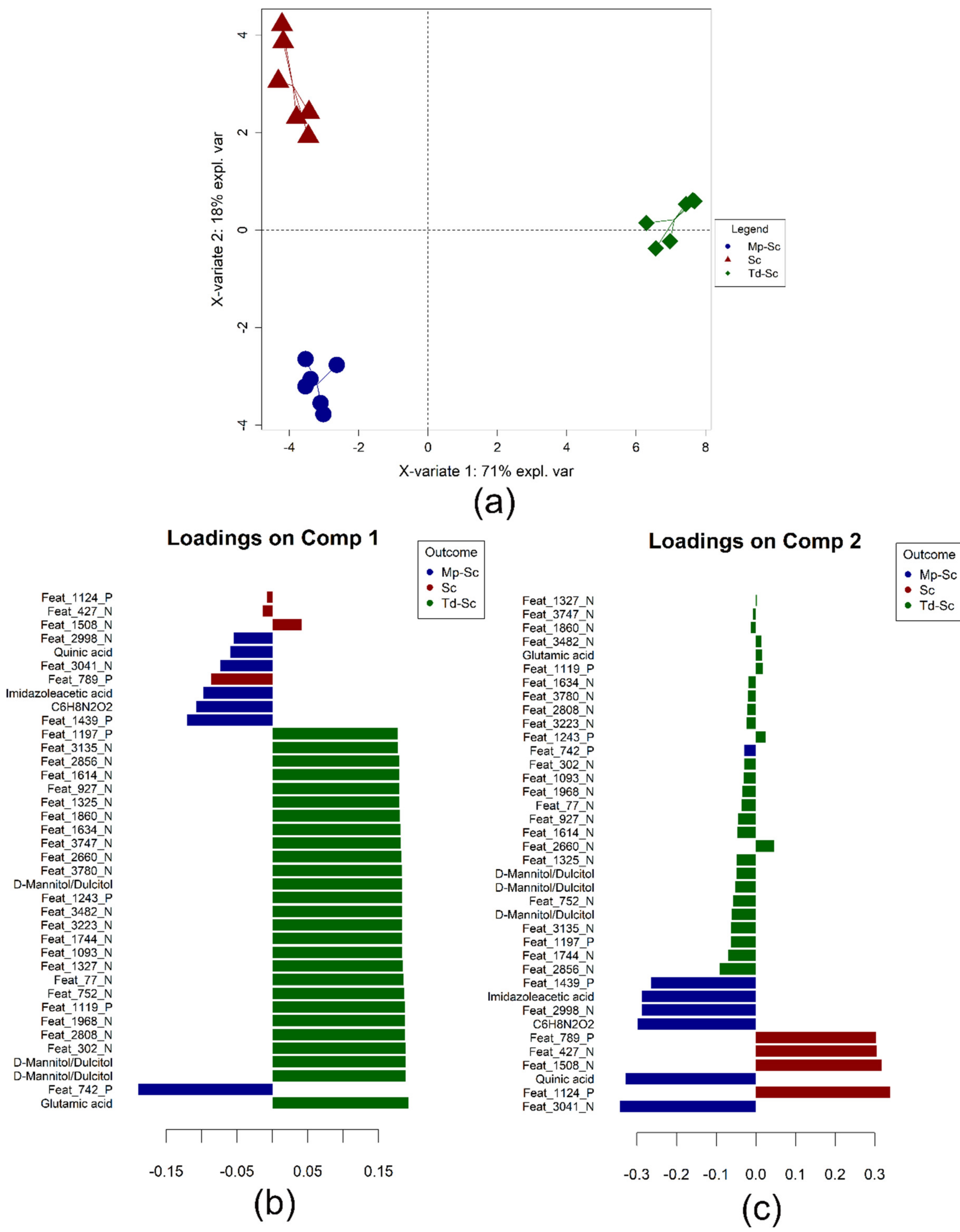
3.2. Non-Volatile Profile
3.3. Sensory Attributes
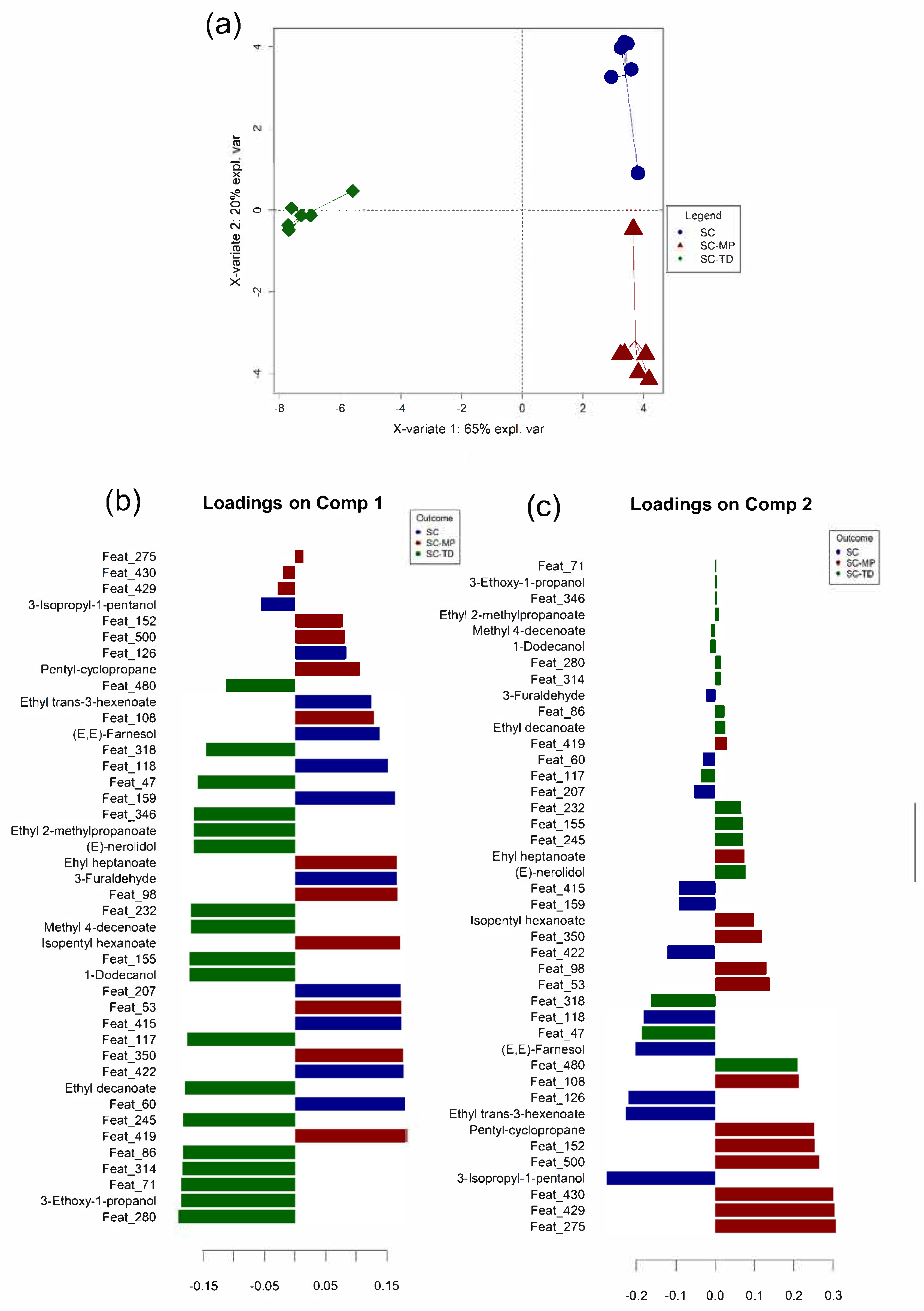
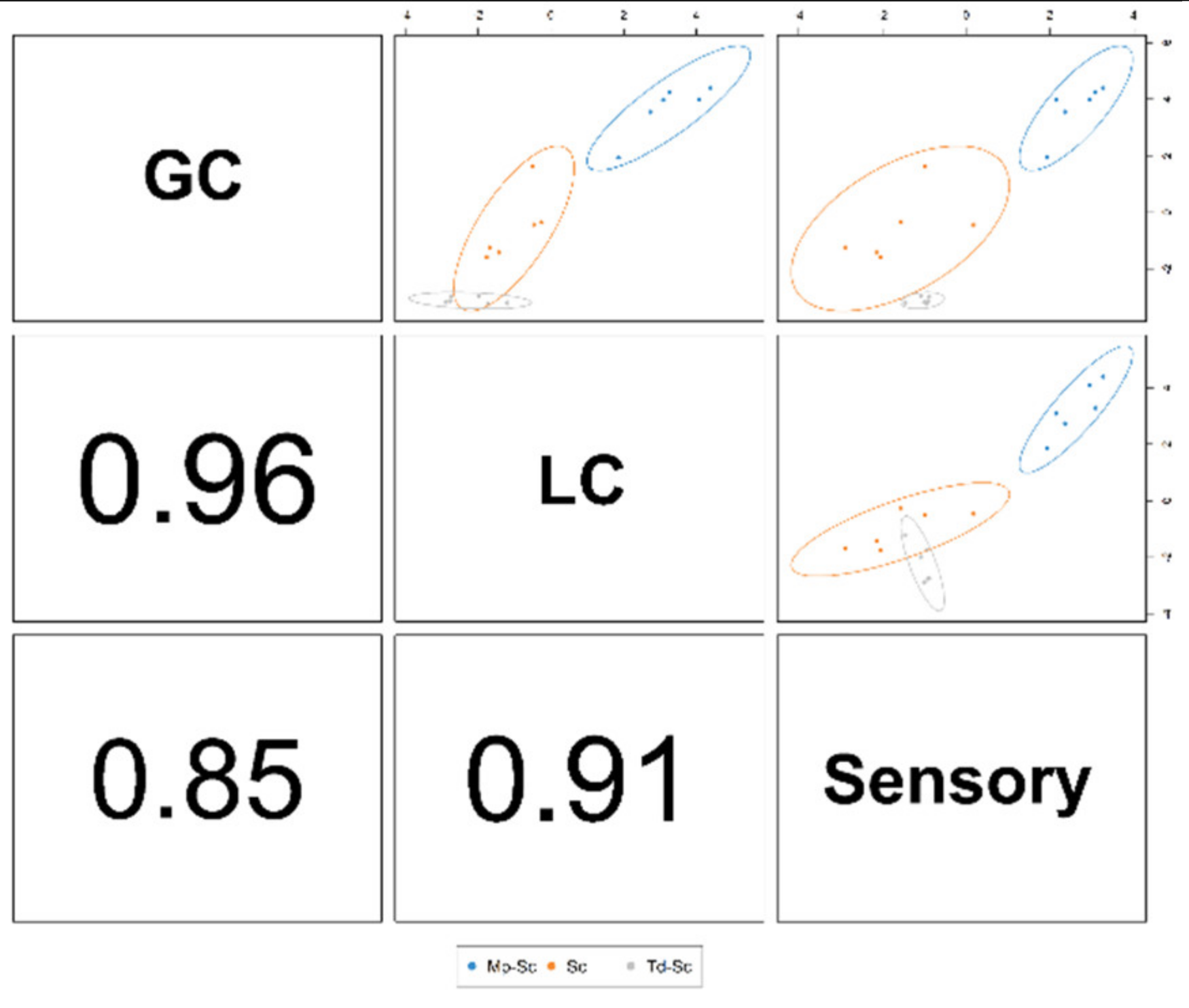
3.4. Data Integration: Sparse Generalised Canonical Correlation Analysis Discriminant Analysis (sGCC-DA) Approach
3.5. Partial Least Squares (PLS) Regression
3.6. Data Integration: Regularised Generalised Canonical Correlation Analysis (RGCCA) Approach
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegrist, M.; Cousin, M.-E. Expectations Influence Sensory Experience in a Wine Tasting. Appetite 2009, 52, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Sherman, E.; Coe, M.; Grose, C.; Martin, D.; Greenwood, D.R. Metabolomics Approach to Assess the Relative Contributions of the Volatile and Non-Volatile Composition to Expert Quality Ratings of Pinot Noir Wine Quality. J. Agric. Food Chem. 2020, 68, 13380–13396. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lu, Y.; Guo, Y.; Cao, H.; Wang, Q.; Shui, W. Comprehensive Evaluation of Untargeted Metabolomics Data Processing Software in Feature Detection, Quantification and Discriminating Marker Selection. Anal. Chim. Acta 2018, 1029, 50–57. [Google Scholar] [CrossRef]
- Arapitsas, P.; Ugliano, M.; Marangon, M.; Piombino, P.; Rolle, L.; Gerbi, V.; Versari, A.; Mattivi, F. Use of Untargeted Liquid Chromatography–Mass Spectrometry Metabolome to Discriminate Italian Monovarietal Red Wines, Produced in Their Different Terroirs. J. Agric. Food Chem. 2020, 68, 13353–13366. [Google Scholar] [CrossRef]
- Šuklje, K.; Carlin, S.; Stanstrup, J.; Antalick, G.; Blackman, J.W.; Meeks, C.; Deloire, A.; Schmidtke, L.M.; Vrhovsek, U. Unravelling Wine Volatile Evolution during Shiraz Grape Ripening by Untargeted HS-SPME-GC × GC-TOFMS. Food Chem. 2019, 277, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R. Grape and Wine Metabolomics to Develop New Insights Using Untargeted and Targeted Approaches. Fermentation 2018, 4, 92. [Google Scholar] [CrossRef]
- Rocchetti, G.; Gatti, M.; Bavaresco, L.; Lucini, L. Untargeted Metabolomics to Investigate the Phenolic Composition of Chardonnay Wines from Different Origins. J. Food Compos. Anal. 2018, 71, 87–93. [Google Scholar] [CrossRef]
- Whitener, M.E.B.; Stanstrup, J.; Panzeri, V.; Carlin, S.; Divol, B.; Du Toit, M.; Vrhovsek, U. Untangling the Wine Metabolome by Combining Untargeted SPME–GCxGC-TOF-MS and Sensory Analysis to Profile Sauvignon Blanc Co-Fermented with Seven Different Yeasts. Metabolomics 2016, 12, 53. [Google Scholar] [CrossRef]
- Arbulu, M.; Sampedro, M.C.; Gómez-Caballero, A.; Goicolea, M.A.; Barrio, R.J. Untargeted Metabolomic Analysis Using Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry for Non-Volatile Profiling of Wines. Anal. Chim. Acta 2015, 858, 32–41. [Google Scholar] [CrossRef]
- Castro, C.C.; Martins, R.C.; Teixeira, J.A.; Ferreira, A.C.S. Application of a High-Throughput Process Analytical Technology Metabolomics Pipeline to Port Wine Forced Ageing Process. Food Chem. 2014, 143, 384–391. [Google Scholar] [CrossRef]
- Goodacre, R.; Broadhurst, D.; Smilde, A.K.; Kristal, B.S.; Baker, J.D.; Beger, R.; Bessant, C.; Connor, S.; Capuani, G.; Craig, A. Proposed Minimum Reporting Standards for Data Analysis in Metabolomics. Metabolomics 2007, 3, 231–241. [Google Scholar] [CrossRef]
- Johnsen, L.G.; Skou, P.B.; Khakimov, B.; Bro, R. Gas Chromatography–Mass Spectrometry Data Processing Made Easy. J. Chromatogr. A 2017, 1503, 57–64. [Google Scholar] [CrossRef]
- Amigo, J.M.; Skov, T.; Bro, R.; Coello, J.; Maspoch, S. Solving GC-MS Problems with Parafac2. TrAC Trends Anal. Chem. 2008, 27, 714–725. [Google Scholar] [CrossRef]
- Mahieu, N.G.; Genenbacher, J.L.; Patti, G.J. A Roadmap for the XCMS Family of Software Solutions in Metabolomics. Curr. Opin. Chem. Biol. 2016, 30, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Considine, E.C.; Salek, R.M. A Tool to Encourage Minimum Reporting Guideline Uptake for Data Analysis in Metabolomics. Metabolites 2019, 9, 43. [Google Scholar] [CrossRef]
- Ren, S.; Hinzman, A.A.; Kang, E.L.; Szczesniak, R.D.; Lu, L.J. Computational and Statistical Analysis of Metabolomics Data. Metabolomics 2015, 11, 1492–1513. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.-A. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Wanichthanarak, K.; Fahrmann, J.F.; Grapov, D. Genomic, Proteomic, and Metabolomic Data Integration Strategies. Biomark. Insights 2015, 10, BMI–S29511. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Libiseller, G.; Dvorzak, M.; Kleb, U.; Gander, E.; Eisenberg, T.; Madeo, F.; Neumann, S.; Trausinger, G.; Sinner, F.; Pieber, T.; et al. IPO: A Tool for Automated Optimization of XCMS Parameters. BMC Bioinform. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An Integrated Strategy for Compound Spectra Extraction and Annotation of LC/MS Data Sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef]
- Arapitsas, P.; Della Corte, A.; Gika, H.; Narduzzi, L.; Mattivi, F.; Theodoridis, G. Studying the Effect of Storage Conditions on the Metabolite Content of Red Wine Using HILIC LC–MS Based Metabolomics. Food Chem. 2016, 197, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Puertas, B.; Jimenez-Hierro, M.J.; Cantos-Villar, E.; Marrufo-Curtido, A.; Carbú, M.; Cuevas, F.J.; Moreno-Rojas, J.M.; González-Rodríguez, V.E.; Cantoral, J.M.; Ruiz-Moreno, M.J. The Influence of Yeast on Chemical Composition and Sensory Properties of Dry White Wines. Food Chem. 2018, 253, 227–235. [Google Scholar] [CrossRef]
- Kokla, M.; Klåvus, A.; Noerman, S.; Koistinen, V.M.; Tuomainen, M.; Zarei, I.; Meuronen, T.; Häkkinen, M.R.; Rummukainen, S.; Babu, A.F. “NoTaMe”: Workflow for Non-Targeted LC-MS Metabolic Profiling. Metabolites 2020, 10, 135. [Google Scholar]
- Kokla, M.; Virtanen, J.; Kolehmainen, M.; Paananen, J.; Hanhineva, K. Random Forest-Based Imputation Outperforms Other Methods for Imputing LC-MS Metabolomics Data: A Comparative Study. BMC Bioinform. 2019, 20, 492. [Google Scholar] [CrossRef] [PubMed]
- Noonan, M.J.; Tinnesand, H.V.; Buesching, C.D. Normalizing Gas-Chromatography–Mass Spectrometry Data: Method Choice Can Alter Biological Inference. BioEssays 2018, 40, 1700210. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-Check: Validation of Diagnostic Statistics for PLS-DA Models in Metabolomics Studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef]
- Muñoz-Redondo, J.M.; Ruiz-Moreno, M.J.; Puertas, B.; Cantos-Villar, E.; Moreno-Rojas, J.M. Multivariate Optimization of Headspace Solid-Phase Microextraction Coupled to Gas Chromatography-Mass Spectrometry for the Analysis of Terpenoids in Sparkling Wines. Talanta 2020, 208, 120483. [Google Scholar] [CrossRef]
- Shi, L.; Westerhuis, J.A.; Rosén, J.; Landberg, R.; Brunius, C. Variable Selection and Validation in Multivariate Modelling. Bioinformatics 2019, 35, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Sadoudi, M.; Tourdot-Maréchal, R.; Rousseaux, S.; Steyer, D.; Gallardo-Chacón, J.-J.; Ballester, J.; Vichi, S.; Guérin-Schneider, R.; Caixach, J.; Alexandre, H. Yeast–Yeast Interactions Revealed by Aromatic Profile Analysis of Sauvignon Blanc Wine Fermented by Single or Co-Culture of Non-Saccharomyces and Saccharomyces Yeasts. Food Microbiol. 2012, 32, 243–253. [Google Scholar] [CrossRef]
- Renault, P.; Miot-Sertier, C.; Marullo, P.; Hernández-Orte, P.; Lagarrigue, L.; Lonvaud-Funel, A.; Bely, M. Genetic Characterization and Phenotypic Variability in Torulaspora Delbrueckii Species: Potential Applications in the Wine Industry. Int. J. Food Microbiol. 2009, 134, 201–210. [Google Scholar] [CrossRef]
- Benito, S. The Impact of Torulaspora Delbrueckii Yeast in Winemaking. Appl. Microbiol. Biotechnol. 2018, 102, 3081–3094. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, R.; Zamora, E.; Álvarez, M.L.; Ramírez, M. Using Torulaspora Delbrueckii Killer Yeasts in the Elaboration of Base Wine and Traditional Sparkling Wine. Int. J. Food Microbiol. 2019, 289, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Sumby, K.M.; Grbin, P.R.; Jiranek, V. Microbial Modulation of Aromatic Esters in Wine: Current Knowledge and Future Prospects. Food Chem. 2010, 121, 1–16. [Google Scholar] [CrossRef]
- Oliveira, I.; Ferreira, V. Modulating Fermentative, Varietal and Aging Aromas of Wine Using Non-Saccharomyces Yeasts in a Sequential Inoculation Approach. Microorganisms 2019, 7, 164. [Google Scholar] [CrossRef] [PubMed]
- Renault, P.; Coulon, J.; de Revel, G.; Barbe, J.-C.; Bely, M. Increase of Fruity Aroma during Mixed T. Delbrueckii/S. Cerevisiae Wine Fermentation Is Linked to Specific Esters Enhancement. Int. J. Food Microbiol. 2015, 207, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Tourdot-Maréchal, R.; Morge, C.; Sparrow, C.; Liu, Y.; Quintanilla-Casas, B.; Vichi, S.; Alexandre, H. Non-Saccharomyces Yeasts Nitrogen Source Preferences: Impact on Sequential Fermentation and Wine Volatile Compounds Profile. Front. Microbiol. 2017, 8, 2175. [Google Scholar] [CrossRef]
- King, A.; Richard Dickinson, J. Biotransformation of Monoterpene Alcohols by Saccharomyces Cerevisiae, Torulaspora Delbrueckii and Kluyveromyces Lactis. Yeast 2000, 16, 499–506. [Google Scholar] [CrossRef]
- Jiang, B.; Zhang, Z. Volatile Compounds of Young Wines from Cabernet Sauvignon, Cabernet Gernischet and Chardonnay Varieties Grown in the Loess Plateau Region of China. Molecules 2010, 15, 9184–9196. [Google Scholar] [CrossRef] [PubMed]
- Coelho, E.; Coimbra, M.A.; Nogueira, J.M.F.; Rocha, S.M. Quantification Approach for Assessment of Sparkling Wine Volatiles from Different Soils, Ripening Stages, and Varieties by Stir Bar Sorptive Extraction with Liquid Desorption. Anal. Chim. Acta 2009, 635, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Soetaert, W.; Vanhooren, P.T.; Vandamme, E.J. The production of mannitol by fermentation. In Carbohydrate Biotechnology Protocols; Springer: Berlin/Heidelberg, Germany, 1999; pp. 261–275. [Google Scholar]
- Azzolini, M.; Tosi, E.; Lorenzini, M.; Finato, F.; Zapparoli, G. Contribution to the Aroma of White Wines by Controlled Torulaspora Delbrueckii Cultures in Association with Saccharomyces Cerevisiae. World J. Microbiol. Biotechnol. 2015, 31, 277–293. [Google Scholar] [CrossRef] [PubMed]




| Analytical Platform | Model | Mean Overall BER (1) | Ncomp | Class | Mean Class Error (2) | p-Value (3) |
|---|---|---|---|---|---|---|
| GC-MS | Allvariables | 0.11 ± 0.08 | 3 | Sc-Td | 0.01 ± 0.03 | BER: <0.001 |
| Sc-Mp | 0.15 ± 0.16 | NMC: 0.024 | ||||
| Sc | 0.18 ± 0.17 | AUROC: 0.020 | ||||
| GC-MS | Variable reduction | 0.08 ± 0.06 | 2 | Sc-Td | 0.00 ± 0.00 | BER: <0.001 |
| Sc-Mp | 0.12 ± 0.11 | NMC: 0.007 | ||||
| Sc | 0.14 ± 0.11 | AUROC: 0.003 | ||||
| LC-MS | Allvariables | 0.17 ± 0.10 | 4 | Sc-Td | 0.14 ± 0.09 | BER: <0.001 |
| Sc-Mp | 0.07 ± 0.13 | NMC: 0.120 | ||||
| Sc | 0.29 ± 0.21 | AUROC: 0.035 | ||||
| LC-MS | Variable reduction | 0.02 ± 0.04 | 2 | Sc-Td | 0.00 ± 0.00 | BER: <0.001 |
| Sc-Mp | 0.05 ± 0.11 | NMC: 0.027 | ||||
| Sc | 0.00 ± 0.02 | AUROC: 0.002 |
| Sensory Descriptor | Sc-Td1 | Sc-Mp 2 | Sc 3 | p-Value 4 | CS 5 | GT 6 | p-Value | Interactions (p-Value) |
|---|---|---|---|---|---|---|---|---|
| Scent intensity | 6.0b | 6.2ab | 6.6a | * | 6.4 | 6.1 | ns | *** |
| Red fruit | 1.35b | 1.97a | 2.01a | *** | 1.90 | 1.66 | ns | ns |
| Black fruit | 1.47b | 1.99a | 2.03a | * | 2.95a | 0.71b | *** | *** |
| Citrus fruit | 0.86b | 1.65a | 1.84a | *** | 1.00b | 1.90a | *** | *** |
| Tree fruit | 1.10b | 1.78a | 2.12a | *** | 1.58 | 1.75 | ns | ** |
| Taste intensity | 5.8 | 5.5 | 5.6 | ns | 5.9a | 5.4b | ** | ns |
| Acidity | 4.5b | 4.9a | 5.0a | ** | 4.6b | 5.0a | * | ns |
| Alcohol | 4.7b | 5.0a | 4.9a | ** | 4.8 | 4.9 | ns | ns |
| Complexity | 4.1b | 4.6a | 4.7a | *** | 4.6a | 4.3b | * | ns |
| Balance | 4.1c | 5.0b | 5.7a | *** | 4.5b | 5.3a | *** | ** |
| Persistence | 5.1a | 4.2b | 4.3b | *** | 4.7 | 4.4 | ns | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz-Redondo, J.M.; Puertas, B.; Pereira-Caro, G.; Ordóñez-Díaz, J.L.; Ruiz-Moreno, M.J.; Cantos-Villar, E.; Moreno-Rojas, J.M. A Statistical Workflow to Evaluate the Modulation of Wine Metabolome and Its Contribution to the Sensory Attributes. Fermentation 2021, 7, 72. https://doi.org/10.3390/fermentation7020072
Muñoz-Redondo JM, Puertas B, Pereira-Caro G, Ordóñez-Díaz JL, Ruiz-Moreno MJ, Cantos-Villar E, Moreno-Rojas JM. A Statistical Workflow to Evaluate the Modulation of Wine Metabolome and Its Contribution to the Sensory Attributes. Fermentation. 2021; 7(2):72. https://doi.org/10.3390/fermentation7020072
Chicago/Turabian StyleMuñoz-Redondo, José Manuel, Belén Puertas, Gema Pereira-Caro, José Luis Ordóñez-Díaz, María José Ruiz-Moreno, Emma Cantos-Villar, and José Manuel Moreno-Rojas. 2021. "A Statistical Workflow to Evaluate the Modulation of Wine Metabolome and Its Contribution to the Sensory Attributes" Fermentation 7, no. 2: 72. https://doi.org/10.3390/fermentation7020072
APA StyleMuñoz-Redondo, J. M., Puertas, B., Pereira-Caro, G., Ordóñez-Díaz, J. L., Ruiz-Moreno, M. J., Cantos-Villar, E., & Moreno-Rojas, J. M. (2021). A Statistical Workflow to Evaluate the Modulation of Wine Metabolome and Its Contribution to the Sensory Attributes. Fermentation, 7(2), 72. https://doi.org/10.3390/fermentation7020072