PTR-ToF-MS for the Online Monitoring of Alcoholic Fermentation in Wine: Assessment of VOCs Variability Associated with Different Combinations of Saccharomyces/Non-Saccharomyces as a Case-Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and the Determination of Microbial Population
2.2. Micro-Vinifications and Wine Analysis
2.3. Samples Preparation and PTR-ToF-MS (Proton Transfer Reaction-Time of Flight-Mass Spectrometry) Analysis
2.4. Data Analysis
3. Results
3.1. Alcoholic Fermentation Kinetics and Yeast Dynamics
3.2. Automated Monitoring Volatile Organic Compound (VOC) Evolution in Red Grape Must and Juice Fermentation by Different Yeast Mixed Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, M.V. Microbial Contribution to Wine Aroma and Its Intended Use for Wine Quality Improvement. Mol. Basel Switz. 2017, 22, 189. [Google Scholar] [CrossRef] [PubMed]
- Benito, Á.; Calderón, F.; Benito, S. The Influence of Non-Saccharomyces Species on Wine Fermentation Quality Parameters. Fermentation 2019, 5, 54. [Google Scholar] [CrossRef]
- Lebrón-Aguilar, R.; Soria, A.C.; Quintanilla-López, J.E. Comprehensive evaluation of direct injection mass spectrometry for the quantitative profiling of volatiles in food samples. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2016, 374, 20150375. [Google Scholar] [CrossRef]
- Pinu, F.R. Grape and Wine Metabolomics to Develop New Insights Using Untargeted and Targeted Approaches. Fermentation 2018, 4, 92. [Google Scholar] [CrossRef]
- Tufariello, M.; Maiorano, G.; Rampino, P.; Spano, G.; Grieco, F.; Perrotta, C.; Capozzi, V.; Grieco, F. Selection of an autochthonous yeast starter culture for industrial production of Primitivo “Gioia del Colle” PDO/DOC in Apulia (Southern Italy). LWT 2019, 99, 188–196. [Google Scholar] [CrossRef]
- Garofalo, C.; Tristezza, M.; Grieco, F.; Spano, G.; Capozzi, V. From grape berries to wine: Population dynamics of cultivable yeasts associated to “Nero di Troia” autochthonous grape cultivar. World J. Microbiol. Biotechnol. 2016, 32, 59. [Google Scholar] [CrossRef]
- Capozzi, V.; Garofalo, C.; Chiriatti, M.A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2015, 181, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Comitini, F.; Mannazzu, I.; Domizio, P. Controlled mixed culture fermentation: A new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res. 2010, 10, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef]
- Tronchoni, J.; Curiel, J.A.; Morales, P.; Torres-Pérez, R.; Gonzalez, R. Early transcriptional response to biotic stress in mixed starter fermentations involving Saccharomyces cerevisiae and Torulaspora delbrueckii. Int. J. Food Microbiol. 2017, 241, 60–68. [Google Scholar] [CrossRef]
- Ciani, M.; Morales, P.; Comitini, F.; Tronchoni, J.; Canonico, L.; Curiel, J.A.; Oro, L.; Rodrigues, A.J.; Gonzalez, R. Non-conventional Yeast Species for Lowering Ethanol Content of Wines. Front. Microbiol. 2016, 7, 642. [Google Scholar] [CrossRef] [PubMed]
- Rossouw, D.; Bauer, F.F. Exploring the phenotypic space of non-Saccharomyces wine yeast biodiversity. Food Microbiol. 2016, 55, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Beckner Whitener, M.E.; Stanstrup, J.; Panzeri, V.; Carlin, S.; Divol, B.; Du Toit, M.; Vrhovsek, U. Untangling the wine metabolome by combining untargeted SPME–GCxGC-TOF-MS and sensory analysis to profile Sauvignon blanc co-fermented with seven different yeasts. Metabolomics 2016, 12, 53. [Google Scholar] [CrossRef]
- Lu, Y.; Huang, D.; Lee, P.R.; Liu, S.Q. Assessment of volatile and non-volatile compounds in durian wines fermented with four commercial non-Saccharomyces yeasts. J. Sci. Food Agric. 2016, 96, 1511–1521. [Google Scholar] [CrossRef]
- Medina, K.; Boido, E.; Dellacassa, E.; Carrau, F. Growth of non-Saccharomyces yeasts affects nutrient availability for Saccharomyces cerevisiae during wine fermentation. Int. J. Food Microbiol. 2012, 157, 245–250. [Google Scholar] [CrossRef]
- Petruzzi, L.; Capozzi, V.; Berbegal, C.; Corbo, M.R.; Bevilacqua, A.; Spano, G.; Sinigaglia, M. Microbial Resources and Enological Significance: Opportunities and Benefits. Front. Microbiol. 2017, 8, 995. [Google Scholar] [CrossRef]
- Dutraive, O.; Benito, S.; Fritsch, S.; Beisert, B.; Patz, C.D.; Rauhut, D. Effect of Sequential Inoculation with Non-Saccharomyces and Saccharomyces Yeasts on Riesling Wine Chemical Composition. Fermentation 2019, 5, 79. [Google Scholar] [CrossRef]
- Du Plessis, H.; Du Toit, M.; Nieuwoudt, H.; Van der Rijst, M.; Hoff, J.; Jolly, N. Modulation of Wine Flavor using Hanseniaspora uvarum in Combination with Different Saccharomyces cerevisiae, Lactic Acid Bacteria Strains and Malolactic Fermentation Strategies. Fermentation 2019, 5, 64. [Google Scholar] [CrossRef]
- Morata, A.; Loira, I.; Escott, C.; del Fresno, J.M.; Bañuelos, M.A.; Suárez-Lepe, J.A. Applications of Metschnikowia pulcherrima in Wine Biotechnology. Fermentation 2019, 5, 63. [Google Scholar] [CrossRef]
- Garofalo, C.; Russo, P.; Beneduce, L.; Massa, S.; Spano, G.; Capozzi, V. Non-Saccharomyces biodiversity in wine and the ‘microbial terroir’: A survey on Nero di Troia wine from the Apulian region, Italy. Ann. Microbiol. 2016, 66, 143–150. [Google Scholar] [CrossRef]
- Sgouros, G.; Chalvantzi, I.; Mallouchos, A.; Paraskevopoulos, Y.; Banilas, G.; Nisiotou, A. Biodiversity and Enological Potential of Non-Saccharomyces Yeasts from Nemean Vineyards. Fermentation 2018, 4, 32. [Google Scholar] [CrossRef]
- Roudil, L.; Russo, P.; Berbegal, C.; Albertin, W.; Spano, G.; Capozzi, V. Non-Saccharomyces Commercial Starter Cultures: Scientific Trends, Recent Patents and Innovation in the Wine Sector. Recent Pat. Food Nutr. Agric. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lindinger, W.; Jordan, A. Proton-transfer-reaction mass spectrometry (PTR–MS): On-line monitoring of volatile organic compounds at pptv levels. Chem. Soc. Rev. 1998, 27, 347–375. [Google Scholar] [CrossRef]
- Jordan, A.; Haidacher, S.; Hanel, G.; Hartungen, E.; Märk, L.; Seehauser, H.; Schottkowsky, R.; Sulzer, P.; Märk, T.D. A high resolution and high sensitivity proton-transfer-reaction time-of-flight mass spectrometer (PTR-TOF-MS). Int. J. Mass Spectrom. 2009, 286, 122–128. [Google Scholar] [CrossRef]
- Capozzi, V.; Yener, S.; Khomenko, I.; Farneti, B.; Cappellin, L.; Gasperi, F.; Scampicchio, M.; Biasioli, F. PTR-ToF-MS Coupled with an Automated Sampling System and Tailored Data Analysis for Food Studies: Bioprocess Monitoring, Screening and Nose-space Analysis. JoVE J. Vis. Exp. 2017, e54075. [Google Scholar] [CrossRef]
- Cappellin, L.; Loreto, F.; Aprea, E.; Romano, A.; Del Pulgar, J.S.; Gasperi, F.; Biasioli, F. PTR-MS in Italy: A Multipurpose Sensor with Applications in Environmental, Agri-Food and Health Science. Sensors 2013, 13, 11923–11955. [Google Scholar] [CrossRef]
- Pico, J.; Khomenko, I.; Capozzi, V.; Navarini, L.; Bernal, J.; Gómez, M.; Biasioli, F. Analysis of volatile organic compounds in crumb and crust of different baked and toasted gluten-free breads by direct PTR-ToF-MS and fast-GC-PTR-ToF-MS. J. Mass Spectrom. 2018, 53, 893–902. [Google Scholar] [CrossRef]
- Deuscher, Z.; Andriot, I.; Sémon, E.; Repoux, M.; Preys, S.; Roger, J.M.; Boulanger, R.; Labouré, H.; Quéré, J.L.L. Volatile compounds profiling by using proton transfer reaction-time of flight-mass spectrometry (PTR-ToF-MS). The case study of dark chocolates organoleptic differences. J. Mass Spectrom. 2019, 54, 92–119. [Google Scholar] [CrossRef]
- van Ruth, S.M.; Koot, A.; Akkermans, W.; Araghipour, N.; Rozijn, M.; Baltussen, M.; Wisthaler, A.; Märk, T.D.; Frankhuizen, R. Butter and butter oil classification by PTR-MS. Eur. Food Res. Technol. 2008, 227, 307–317. [Google Scholar] [CrossRef]
- Capozzi, V.; Lonzarich, V.; Khomenko, I.; Cappellin, L.; Navarini, L.; Biasioli, F. Unveiling the Molecular Basis of Mascarpone Cheese Aroma: VOCs analysis by SPME-GC/MS and PTR-ToF-MS. Molecules 2020, 25, 1242. [Google Scholar] [CrossRef]
- Makhoul, S.; Yener, S.; Khomenko, I.; Capozzi, V.; Cappellin, L.; Aprea, E.; Scampicchio, M.; Gasperi, F.; Biasioli, F. Rapid non-invasive quality control of semi-finished products for the food industry by direct injection mass spectrometry headspace analysis: The case of milk powder, whey powder and anhydrous milk fat. J. Mass Spectrom. 2016, 51, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Capozzi, V.; Spano, G.; Biasioli, F. Proton transfer reaction–mass spectrometry: Online and rapid determination of volatile organic compounds of microbial origin. Appl. Microbiol. Biotechnol. 2015, 99, 3787–3795. [Google Scholar] [CrossRef] [PubMed]
- Benozzi, E.; Romano, A.; Capozzi, V.; Makhoul, S.; Cappellin, L.; Khomenko, I.; Aprea, E.; Scampicchio, M.; Spano, G.; Märk, T.D.; et al. Monitoring of lactic fermentation driven by different starter cultures via direct injection mass spectrometric analysis of flavour-related volatile compounds. Food Res. Int. 2015, 76, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Richter, T.M.; Silcock, P.; Algarra, A.; Eyres, G.T.; Capozzi, V.; Bremer, P.J.; Biasioli, F. Evaluation of PTR-ToF-MS as a tool to track the behavior of hop-derived compounds during the fermentation of beer. Food Res. Int. 2018, 111, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Khomenko, I.; Stefanini, I.; Cappellin, L.; Cappelletti, V.; Franceschi, P.; Cavalieri, D.; Märk, T.D.; Biasioli, F. Non-invasive real time monitoring of yeast volatilome by PTR-ToF-MS. Metab. Off. J. Metab. Soc. 2017, 13, 118. [Google Scholar] [CrossRef]
- Capozzi, V.; Makhoul, S.; Aprea, E.; Romano, A.; Cappellin, L.; Sanchez Jimena, A.; Spano, G.; Gasperi, F.; Scampicchio, M.; Biasioli, F. PTR-MS Characterization of VOCs Associated with Commercial Aromatic Bakery Yeasts of Wine and Beer Origin. Molecules 2016, 21, 483. [Google Scholar] [CrossRef]
- Makhoul, S.; Romano, A.; Capozzi, V.; Spano, G.; Aprea, E.; Cappellin, L.; Benozzi, E.; Scampicchio, M.; Märk, T.D.; Gasperi, F.; et al. Volatile Compound Production During the Bread-Making Process: Effect of Flour, Yeast and Their Interaction. Food Bioprocess Technol. 2015, 8, 1925–1937. [Google Scholar] [CrossRef]
- Makhoul, S.; Romano, A.; Cappellin, L.; Spano, G.; Capozzi, V.; Benozzi, E.; Märk, T.D.; Aprea, E.; Gasperi, F.; El-Nakat, H.; et al. Proton-transfer-reaction mass spectrometry for the study of the production of volatile compounds by bakery yeast starters. J. Mass Spectrom. JMS 2014, 49, 850–859. [Google Scholar] [CrossRef]
- Campbell-Sills, H.; Capozzi, V.; Romano, A.; Cappellin, L.; Spano, G.; Breniaux, M.; Lucas, P.; Biasioli, F. Advances in wine analysis by PTR-ToF-MS: Optimization of the method and discrimination of wines from different geographical origins and fermented with different malolactic starters. Int. J. Mass Spectrom. 2016, 397–398, 42–51. [Google Scholar] [CrossRef]
- Romano, A.; Fischer, L.; Herbig, J.; Campbell-Sills, H.; Coulon, J.; Lucas, P.; Cappellin, L.; Biasioli, F. Wine analysis by FastGC proton-transfer reaction-time-of-flight-mass spectrometry. Int. J. Mass Spectrom. 2014, 369, 81–86. [Google Scholar] [CrossRef]
- Sémon, E.; Arvisenet, G.; Guichard, E.; Quéré, J.L.L. Modified proton transfer reaction mass spectrometry (PTR-MS) operating conditions for in vitro and in vivo analysis of wine aroma. J. Mass Spectrom. 2018, 53, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-González, C.; Canon, F.; Feron, G.; Guichard, E.; Pozo-Bayón, M.A. Assessment Wine Aroma Persistence by Using an in Vivo PTR-ToF-MS Approach and Its Relationship with Salivary Parameters. Mol. Basel Switz. 2019, 24, 1277. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; El Khoury, M.; Lucas, P.; Bely, M.; Russo, P.; Spano, G.; Capozzi, V. Autochthonous starter cultures and indigenous grape variety for regional wine production. J. Appl. Microbiol. 2015, 118, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Cappellin, L.; Biasioli, F.; Fabris, A.; Schuhfried, E.; Soukoulis, C.; Märk, T.D.; Gasperi, F. Improved mass accuracy in PTR-TOF-MS: Another step towards better compound identification in PTR-MS. Int. J. Mass Spectrom. 2010, 290, 60–63. [Google Scholar] [CrossRef]
- Cappellin, L.; Biasioli, F.; Granitto, P.M.; Schuhfried, E.; Soukoulis, C.; Costa, F.; Märk, T.D.; Gasperi, F. On data analysis in PTR-TOF-MS: From raw spectra to data mining. Sens. Actuators B Chem. 2011, 155, 183–190. [Google Scholar] [CrossRef]
- Lindinger, W.; Hansel, A.; Jordan, A. On-line monitoring of volatile organic compounds at pptv levels by means of proton-transfer-reaction mass spectrometry (PTR-MS) medical applications, food control and environmental research. Int. J. Mass Spectrom. Ion Process. 1998, 173, 191–241. [Google Scholar] [CrossRef]
- Bisson, L.F.; Waterhouse, A.L.; Ebeler, S.E.; Walker, M.A.; Lapsley, J.T. The present and future of the international wine industry. Nature 2002, 418, 696–699. [Google Scholar] [CrossRef]
- Berbegal, C.; Fragasso, M.; Russo, P.; Bimbo, F.; Grieco, F.; Spano, G.; Capozzi, V. Climate Changes and Food Quality: The Potential of Microbial Activities as Mitigating Strategies in the Wine Sector. Fermentation 2019, 5, 85. [Google Scholar] [CrossRef]
- Bisson, L.F. Stuck and Sluggish Fermentations. Am. J. Enol. Vitic. 1999, 50, 107–119. [Google Scholar]
- Fleet, G.H. Wine yeasts for the future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Nardi, T. Microbial Resources as a Tool for Enhancing Sustainability in Winemaking. Microorganisms 2020, 8, 507. [Google Scholar] [CrossRef]
- Sablayrolles, J.M. Control of alcoholic fermentation in winemaking: Current situation and prospect. Food Res. Int. 2009, 42, 418–424. [Google Scholar] [CrossRef]
- Molina, A.M.; Swiegers, J.H.; Varela, C.; Pretorius, I.S.; Agosin, E. Influence of wine fermentation temperature on the synthesis of yeast-derived volatile aroma compounds. Appl. Microbiol. Biotechnol. 2007, 77, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Berbegal, C.; De Ceglie, C.; Grieco, F.; Spano, G.; Capozzi, V. Pesticide Residues and Stuck Fermentation in Wine: New Evidences Indicate the Urgent Need of Tailored Regulations. Fermentation 2019, 5, 23. [Google Scholar] [CrossRef]
- Di Egidio, V.; Sinelli, N.; Giovanelli, G.; Moles, A.; Casiraghi, E. NIR and MIR spectroscopy as rapid methods to monitor red wine fermentation. Eur. Food Res. Technol. 2010, 230, 947–955. [Google Scholar] [CrossRef]
- Mouret, J.R.; Perez, M.; Angenieux, M.; Nicolle, P.; Farines, V.; Sablayrolles, J.M. Online-Based Kinetic Analysis of Higher Alcohol and Ester Synthesis During Winemaking Fermentations. Food Bioprocess Technol. 2014, 7, 1235–1245. [Google Scholar] [CrossRef]
- Mallouchos, A.; Komaitis, M.; Koutinas, A.; Kanellaki, M. Investigation of Volatiles Evolution during the Alcoholic Fermentation of Grape Must Using Free and Immobilized Cells with the Help of Solid Phase Microextraction (SPME) Headspace Sampling. J. Agric. Food Chem. 2002, 50, 3840–3848. [Google Scholar] [CrossRef]
- Morales, M.L.; Fierro-Risco, J.; Callejón, R.M.; Paneque, P. Monitoring volatile compounds production throughout fermentation by Saccharomyces and non-Saccharomyces strains using headspace sorptive extraction. J. Food Sci. Technol. 2017, 54, 538–557. [Google Scholar] [CrossRef]
- Silva Ferreira, A.C.; Monforte, A.R.; Teixeira, C.S.; Martins, R.; Fairbairn, S.; Bauer, F.F. Monitoring Alcoholic Fermentation: An Untargeted Approach. J. Agric. Food Chem. 2014, 62, 6784–6793. [Google Scholar] [CrossRef]
- Buratti, S.; Benedetti, S. Chapter 28-Alcoholic Fermentation Using Electronic Nose and Electronic Tongue. In Electronic Noses and Tongues in Food Science; Rodríguez Méndez, M.L., Ed.; Academic Press: San Diego, CA, USA, 2016; pp. 291–299. ISBN 978-0-12-800243-8. [Google Scholar]
- Aguilera, T.; Lozano, J.; Paredes, J.A.; Álvarez, F.J.; Suárez, J.I. Electronic Nose Based on Independent Component Analysis Combined with Partial Least Squares and Artificial Neural Networks for Wine Prediction. Sensors 2012, 12, 8055–8072. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Zhang, Z. Volatile Compounds of Young Wines from Cabernet Sauvignon, Cabernet Gernischet and Chardonnay Varieties Grown in the Loess Plateau Region of China. Molecules 2010, 15, 9184–9196. [Google Scholar] [CrossRef] [PubMed]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Berbegal, C.; Spano, G.; Tristezza, M.; Grieco, F.; Capozzi, V. Microbial Resources and Innovation in the Wine Production Sector. S. Afr. J. Enol. Vitic. 2017, 38, 156–166. [Google Scholar] [CrossRef]
- Lambrechts, M.G.; Pretorius, I.S. Yeast and its Importance to Wine Aroma—A Review. S. Afr. J. Enol. Vitic. 2000, 21, 97–129. [Google Scholar] [CrossRef]
- Escribano, R.; González-Arenzana, L.; Garijo, P.; Berlanas, C.; López-Alfaro, I.; López, R.; Gutiérrez, A.R.; Santamaría, P. Screening of enzymatic activities within different enological non-Saccharomyces yeasts. J. Food Sci. Technol. 2017, 54, 1555–1564. [Google Scholar] [CrossRef]
- Escribano, R.; González-Arenzana, L.; Portu, J.; Garijo, P.; López-Alfaro, I.; López, R.; Santamaría, P.; Gutiérrez, A.R. Wine aromatic compound production and fermentative behaviour within different non-Saccharomyces species and clones. J. Appl. Microbiol. 2018, 124, 1521–1531. [Google Scholar] [CrossRef]
- Padilla, B.; Gil, J.V.; Manzanares, P. Past and Future of Non-Saccharomyces Yeasts: From Spoilage Microorganisms to Biotechnological Tools for Improving Wine Aroma Complexity. Front. Microbiol. 2016, 7, 411. [Google Scholar] [CrossRef]
- Ciani, M.; Beco, L.; Comitini, F. Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 2006, 108, 239–245. [Google Scholar] [CrossRef]
- Taillandier, P.; Lai, Q.P.; Julien-Ortiz, A.; Brandam, C. Interactions between Torulaspora delbrueckii and Saccharomyces cerevisiae in wine fermentation: Influence of inoculation and nitrogen content. World J. Microbiol. Biotechnol. 2014, 30, 1959–1967. [Google Scholar] [CrossRef]
- Contreras, A.; Hidalgo, C.; Henschke, P.A.; Chambers, P.J.; Curtin, C.; Varela, C. Evaluation of Non-Saccharomyces Yeasts for the Reduction of Alcohol Content in Wine. Appl. Environ. Microbiol. 2014, 80, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Tristezza, M.; Tufariello, M.; Capozzi, V.; Spano, G.; Mita, G.; Grieco, F. The Oenological Potential of Hanseniaspora uvarum in Simultaneous and Sequential Co-fermentation with Saccharomyces cerevisiae for Industrial Wine Production. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Tufariello, M.; Renna, R.; Tristezza, M.; Taurino, M.; Palombi, L.; Capozzi, V.; Rizzello, C.G.; Grieco, F. New Insights into the Oenological Significance of Candida zemplinina: Impact of Selected Autochthonous Strains on the Volatile Profile of Apulian Wines. Microorganisms 2020, 8, 628. [Google Scholar] [CrossRef]
- González-Royo, E.; Pascual, O.; Kontoudakis, N.; Esteruelas, M.; Esteve-Zarzoso, B.; Mas, A.; Canals, J.M.; Zamora, F. Oenological consequences of sequential inoculation with non-Saccharomyces yeasts (Torulaspora delbrueckii or Metschnikowia pulcherrima) and Saccharomyces cerevisiae in base wine for sparkling wine production. Eur. Food Res. Technol. 2015, 240, 999–1012. [Google Scholar] [CrossRef]
- Renault, P.E.; Albertin, W.; Bely, M. An innovative tool reveals interaction mechanisms among yeast populations under oenological conditions. Appl. Microbiol. Biotechnol. 2013, 97, 4105–4119. [Google Scholar] [CrossRef]
- Gobert, A.; Tourdot-Maréchal, R.; Morge, C.; Sparrow, C.; Liu, Y.; Quintanilla-Casas, B.; Vichi, S.; Alexandre, H. Non-Saccharomyces Yeasts Nitrogen Source Preferences: Impact on Sequential Fermentation and Wine Volatile Compounds Profile. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Hansen, E.H.; Nissen, P.; Sommer, P.; Nielsen, J.C.; Arneborg, N. The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J. Appl. Microbiol. 2001, 91, 541–547. [Google Scholar] [CrossRef]
- Vasserot, Y.; Mornet, F.; Jeandet, P. Acetic acid removal by Saccharomyces cerevisiae during fermentation in oenological conditions. Metabolic consequences. Food Chem. 2010, 119, 1220–1223. [Google Scholar] [CrossRef]
- Jackowetz, J.N.; Dierschke, S.; Mira de Orduña, R. Multifactorial analysis of acetaldehyde kinetics during alcoholic fermentation by Saccharomyces cerevisiae. Food Res. Int. 2011, 44, 310–316. [Google Scholar] [CrossRef]
- Legras, J.L.; Karst, F. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterization. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef]
- Garofalo, C.; Berbegal, C.; Grieco, F.; Tufariello, M.; Spano, G.; Capozzi, V. Selection of indigenous yeast strains for the production of sparkling wines from native Apulian grape varieties. Int. J. Food Microbiol. 2018, 285, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Chen, W.; Chen, H.; Chen, W.; Zhong, Q. Combination of Lactobacillus plantarum and Saccharomyces cerevisiae DV10 as Starter Culture to Produce Mango Slurry: Microbiological, Chemical Parameters and Antioxidant Activity. Molecules 2019, 24, 4349. [Google Scholar] [CrossRef] [PubMed]
- Seguinot, P.; Bloem, A.; Brial, P.; Meudec, E.; Ortiz-Julien, A.; Camarasa, C. Analyzing the impact of the nature of the nitrogen source on the formation of volatile compounds to unravel the aroma metabolism of two non-Saccharomyces strains. Int. J. Food Microbiol. 2020, 316, 108441. [Google Scholar] [CrossRef] [PubMed]
- Chua, J.Y.; Lu, Y.; Liu, S.Q. Evaluation of five commercial non-Saccharomyces yeasts in fermentation of soy (tofu) whey into an alcoholic beverage. Food Microbiol. 2018, 76, 533–542. [Google Scholar] [CrossRef]
- Prior, K.J.; Bauer, F.F.; Divol, B. The utilization of nitrogenous compounds by commercial non-Saccharomyces yeasts associated with wine. Food Microbiol. 2019, 79, 75–84. [Google Scholar] [CrossRef]
- Ivit, N.N.; Kemp, B. The Impact of Non-Saccharomyces Yeast on Traditional Method Sparkling Wine. Fermentation 2018, 4, 73. [Google Scholar] [CrossRef]
- Nardi, T.; Panero, L.; Petrozziello, M.; Guaita, M.; Tsolakis, C.; Cassino, C.; Vagnoli, P.; Bosso, A. Managing wine quality using Torulaspora delbrueckii and Oenococcus oeni starters in mixed fermentations of a red Barbera wine. Eur. Food Res. Technol. 2019, 245, 293–307. [Google Scholar] [CrossRef]
- Chandra, M.; Oro, I.; Ferreira-Dias, S.; Malfeito-Ferreira, M. Effect of Ethanol, Sulfur Dioxide and Glucose on the Growth of Wine Spoilage Yeasts Using Response Surface Methodology. PLoS ONE 2015, 10, e0128702. [Google Scholar] [CrossRef]
- Ciani, M.; Capece, A.; Comitini, F.; Canonico, L.; Siesto, G.; Romano, P. Yeast Interactions in Inoculated Wine Fermentation. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Pinu, F.R.; Edwards, P.J.B.; Gardner, R.C.; Villas-Boas, S.G. Nitrogen and carbon assimilation by Saccharomyces cerevisiae during Sauvignon blanc juice fermentation. FEMS Yeast Res. 2014, 14, 1206–1222. [Google Scholar] [CrossRef]
- Barbosa, C.; Lage, P.; Esteves, M.; Chambel, L.; Mendes-Faia, A.; Mendes-Ferreira, A. Molecular and Phenotypic Characterization of Metschnikowia pulcherrima Strains from Douro Wine Region. Fermentation 2018, 4, 8. [Google Scholar] [CrossRef]
- Escribano-Viana, R.; Portu, J.; Garijo, P.; López, R.; Santamaría, P.; López-Alfaro, I.; Gutiérrez, A.R.; González-Arenzana, L. Effect of the Sequential Inoculation of Non-Saccharomyces/Saccharomyces on the Anthocyans and Stilbenes Composition of Tempranillo Wines. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Benito, Á.; Calderón, F.; Palomero, F.; Benito, S. Combine Use of Selected Schizosaccharomyces pombe and Lachancea thermotolerans Yeast Strains as an Alternative to theTraditional Malolactic Fermentation in Red Wine Production. Molecules 2015, 20, 9510–9523. [Google Scholar] [CrossRef] [PubMed]
- Benito, Á.; Calderón, F.; Benito, S. The Combined Use of Schizosaccharomyces pombe and Lachancea thermotolerans—Effect on the Anthocyanin Wine Composition. Molecules 2017, 22, 739. [Google Scholar] [CrossRef] [PubMed]
- Benito, Á.; Calderón, F.; Benito, S. Combined Use of S. pombe and L. thermotolerans in Winemaking. Beneficial Effects Determined Through the Study of Wines’ Analytical Characteristics. Molecules 2016, 21, 1744. [Google Scholar] [CrossRef] [PubMed]
- King, E.S.; Kievit, R.L.; Curtin, C.; Swiegers, J.H.; Pretorius, I.S.; Bastian, S.E.P.; Leigh Francis, I. The effect of multiple yeasts co-inoculations on Sauvignon Blanc wine aroma composition, sensory properties and consumer preference. Food Chem. 2010, 122, 618–626. [Google Scholar] [CrossRef]
- Gustafsson, F.S.; Jiranek, V.; Neuner, M.; Scholl, C.M.; Morgan, S.C.; Durall, D.M. The Interaction of Two Saccharomyces cerevisiae Strains Affects Fermentation-Derived Compounds in Wine. Fermentation 2016, 2, 9. [Google Scholar] [CrossRef]
- Berbegal, C.; Garofalo, C.; Russo, P.; Pati, S.; Capozzi, V.; Spano, G. Use of Autochthonous Yeasts and Bacteria in Order to Control Brettanomyces bruxellensis in Wine. Fermentation 2017, 3, 65. [Google Scholar] [CrossRef]
- Berbegal, C.; Spano, G.; Fragasso, M.; Grieco, F.; Russo, P.; Capozzi, V. Starter cultures as biocontrol strategy to prevent Brettanomyces bruxellensis proliferation in wine. Appl. Microbiol. Biotechnol. 2018, 102, 569–576. [Google Scholar] [CrossRef]
- Capozzi, V.; Fragasso, M.; Russo, P. Microbiological Safety and the Management of Microbial Resources in Artisanal Foods and Beverages: The Need for a Transdisciplinary Assessment to Conciliate Actual Trends and Risks Avoidance. Microorganisms 2020, 8, 306. [Google Scholar] [CrossRef]
- Berbegal, C.; Borruso, L.; Fragasso, M.; Tufariello, M.; Russo, P.; Brusetti, L.; Spano, G.; Capozzi, V. A Metagenomic-Based Approach for the Characterization of Bacterial Diversity Associated with Spontaneous Malolactic Fermentations in Wine. Int. J. Mol. Sci. 2019, 20, 3980. [Google Scholar] [CrossRef] [PubMed]
- Englezos, V.; Cachón, D.C.; Rantsiou, K.; Blanco, P.; Petrozziello, M.; Pollon, M.; Giacosa, S.; Río Segade, S.; Rolle, L.; Cocolin, L. Effect of mixed species alcoholic fermentation on growth and malolactic activity of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2019, 103, 7687–7702. [Google Scholar] [CrossRef] [PubMed]
- Balmaseda, A.; Bordons, A.; Reguant, C.; Bautista-Gallego, J. Non-Saccharomyces in Wine: Effect Upon Oenococcus oeni and Malolactic Fermentation. Front. Microbiol. 2018, 9, 534. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Berbegal, C.; Tufariello, M.; Grieco, F.; Spano, G.; Grieco, F. Impact of co-inoculation of Saccharomyces cerevisiae, Hanseniaspora uvarum and Oenococcus oeni autochthonous strains in controlled multi starter grape must fermentations. LWT 2019, 109, 241–249. [Google Scholar] [CrossRef]
- Willaert, R.; Kasas, S.; Devreese, B.; Dietler, G. Yeast Nanobiotechnology. Fermentation 2016, 2, 18. [Google Scholar] [CrossRef]
- Botstein, D.; Fink, G.R. Yeast: An Experimental Organism for 21st Century Biology. Genetics 2011, 189, 695–704. [Google Scholar] [CrossRef]
- Walker, R.S.K.; Pretorius, I.S. Applications of Yeast Synthetic Biology Geared towards the Production of Biopharmaceuticals. Genes 2018, 9, 340. [Google Scholar] [CrossRef]
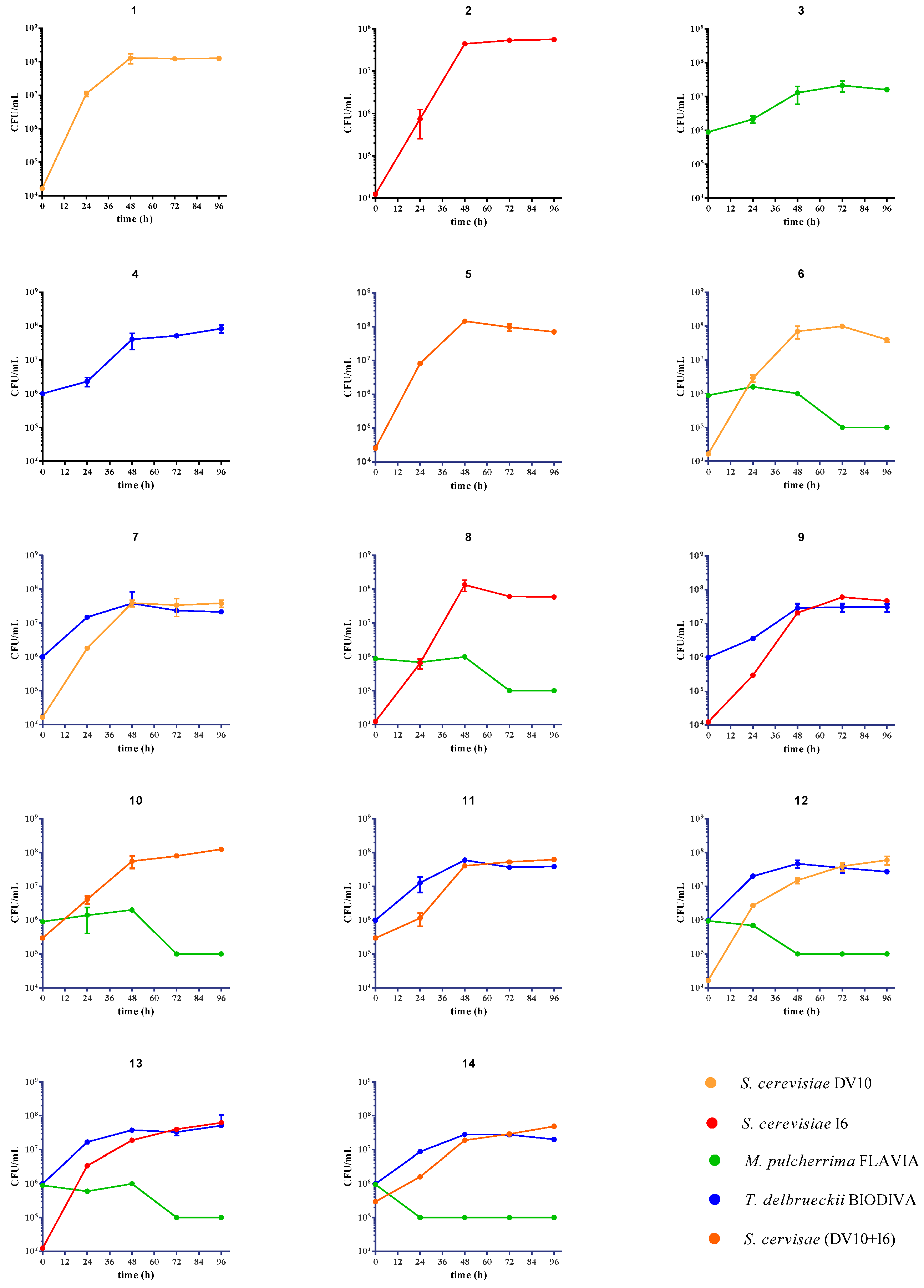
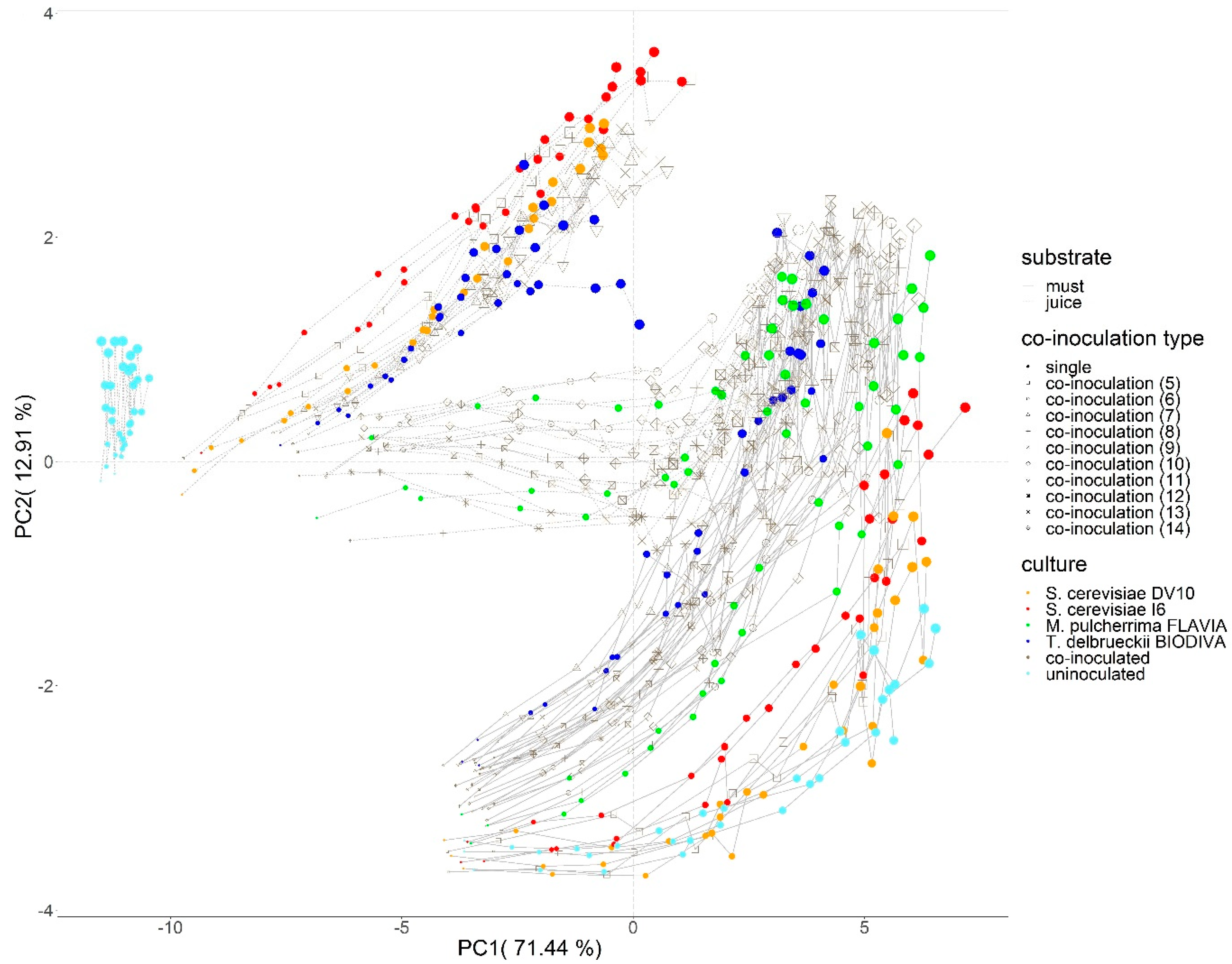
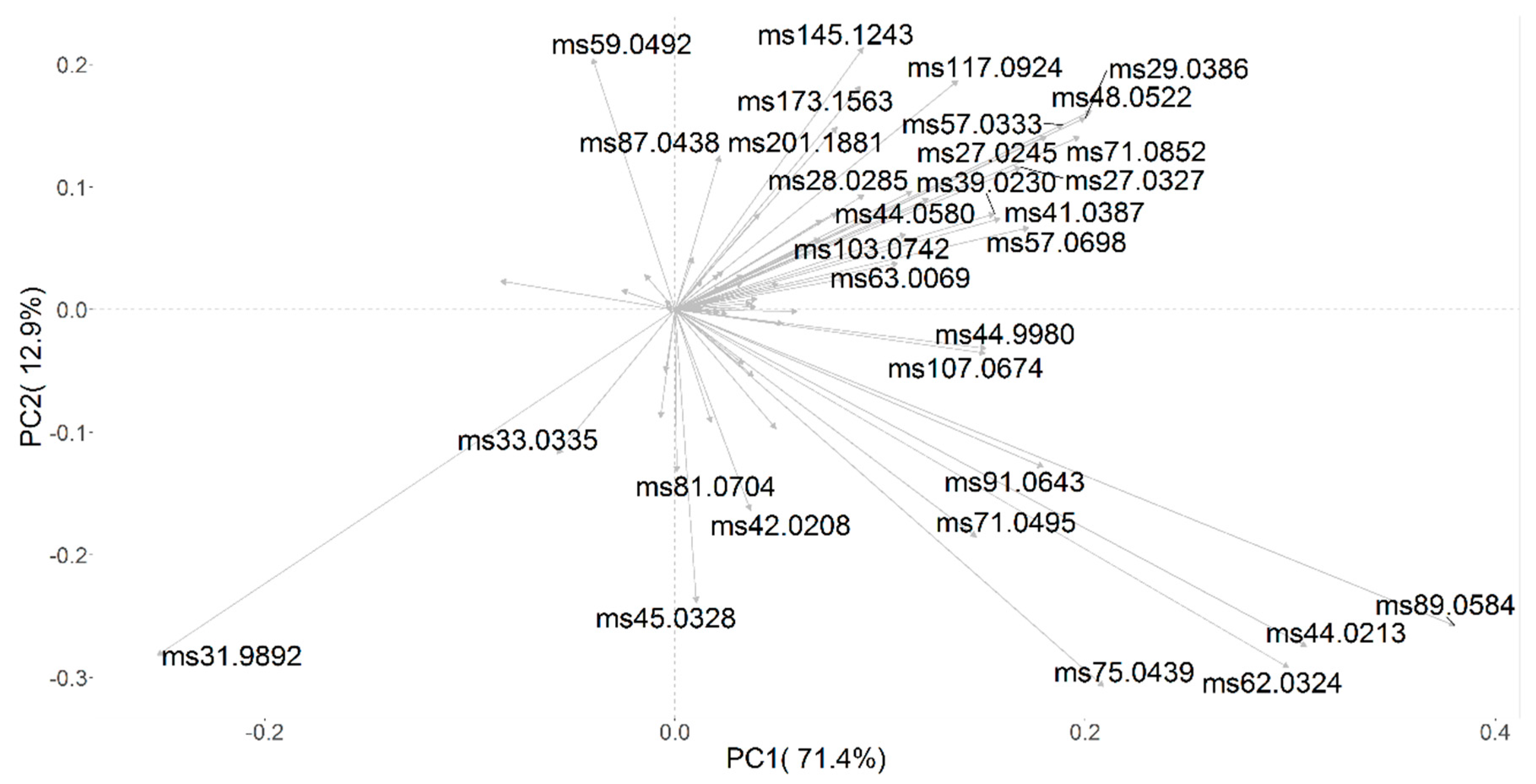
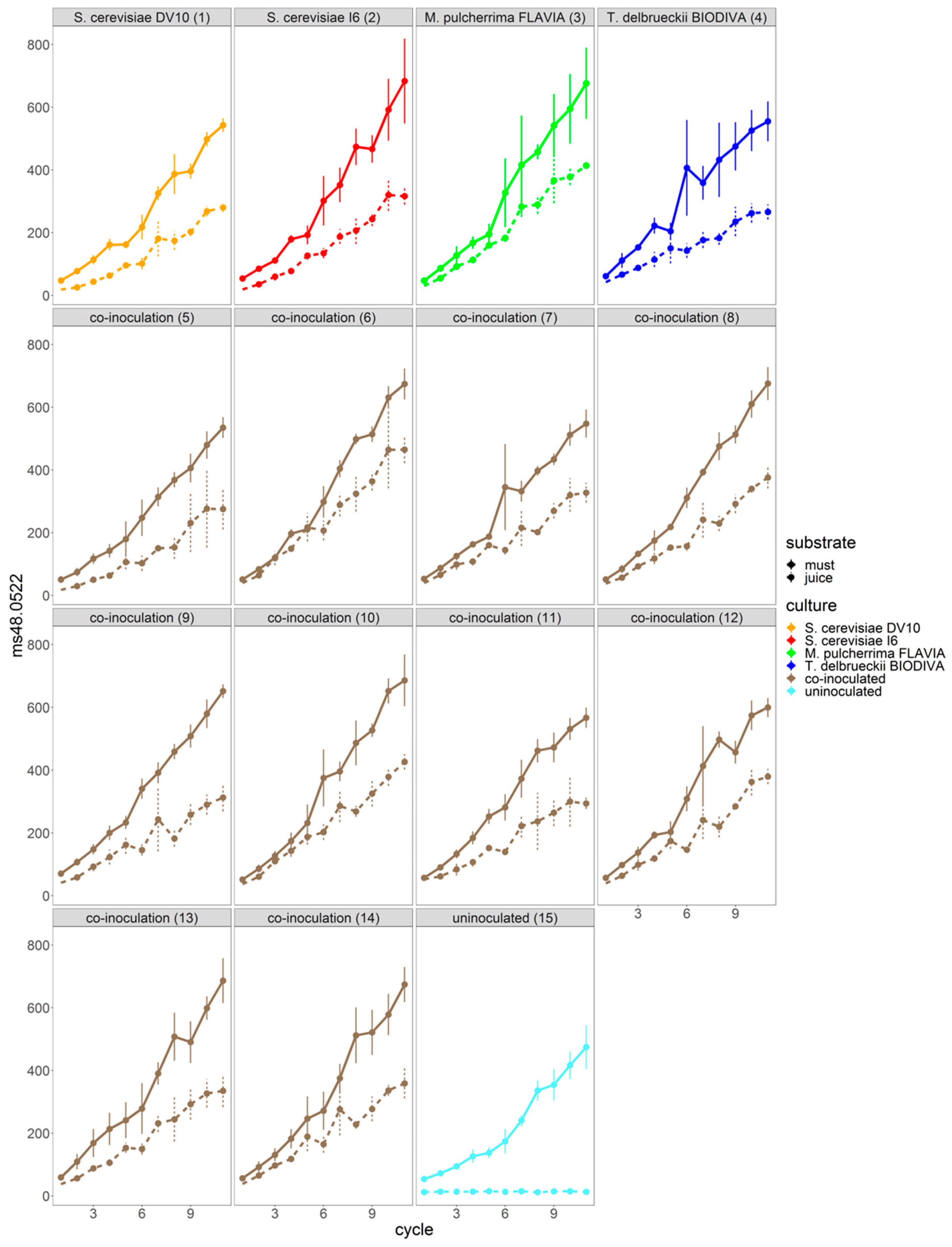
| Sample Code | Inoculated Yeast Cultures |
|---|---|
| 1 | S. cerevisiae DV10 |
| 2 | S. cerevisiae I6 |
| 3 | M. pulcherrima FLAVIA |
| 4 | T. delbrueckii BIODIVA |
| 5 | S. cerevisiae DV10 + S. cerevisiae I6 |
| 6 | S. cerevisiae DV10 + M. pulcherrima FLAVIA |
| 7 | S. cerevisiae DV10 + T. delbrueckii BIODIVA |
| 8 | S. cerevisiae I6 + M. pulcherrima FLAVIA |
| 9 | S. cerevisiae I6 + T. delbrueckii BIODIVA |
| 10 | S. cerevisiae DV10 + S. cerevisiae I6 + M. pulcherrima FLAVIA |
| 11 | S. cerevisiae DV10 + S. cerevisiae I6 + T. delbrueckii BIODIVA |
| 12 | S. cerevisiae DV10 + M. pulcherrima FLAVIA + T. delbrueckii BIODIVA |
| 13 | S. cerevisiae I6 + M. pulcherrima FLAVIA + T. delbrueckii BIODIVA |
| 14 | S. cerevisiae DV10 + S. cerevisiae I6 + M. pulcherrima FLAVIA + T. delbrueckii BIODIVA |
| 15 | Uninoculated grape must/juice |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berbegal, C.; Khomenko, I.; Russo, P.; Spano, G.; Fragasso, M.; Biasioli, F.; Capozzi, V. PTR-ToF-MS for the Online Monitoring of Alcoholic Fermentation in Wine: Assessment of VOCs Variability Associated with Different Combinations of Saccharomyces/Non-Saccharomyces as a Case-Study. Fermentation 2020, 6, 55. https://doi.org/10.3390/fermentation6020055
Berbegal C, Khomenko I, Russo P, Spano G, Fragasso M, Biasioli F, Capozzi V. PTR-ToF-MS for the Online Monitoring of Alcoholic Fermentation in Wine: Assessment of VOCs Variability Associated with Different Combinations of Saccharomyces/Non-Saccharomyces as a Case-Study. Fermentation. 2020; 6(2):55. https://doi.org/10.3390/fermentation6020055
Chicago/Turabian StyleBerbegal, Carmen, Iuliia Khomenko, Pasquale Russo, Giuseppe Spano, Mariagiovanna Fragasso, Franco Biasioli, and Vittorio Capozzi. 2020. "PTR-ToF-MS for the Online Monitoring of Alcoholic Fermentation in Wine: Assessment of VOCs Variability Associated with Different Combinations of Saccharomyces/Non-Saccharomyces as a Case-Study" Fermentation 6, no. 2: 55. https://doi.org/10.3390/fermentation6020055
APA StyleBerbegal, C., Khomenko, I., Russo, P., Spano, G., Fragasso, M., Biasioli, F., & Capozzi, V. (2020). PTR-ToF-MS for the Online Monitoring of Alcoholic Fermentation in Wine: Assessment of VOCs Variability Associated with Different Combinations of Saccharomyces/Non-Saccharomyces as a Case-Study. Fermentation, 6(2), 55. https://doi.org/10.3390/fermentation6020055








