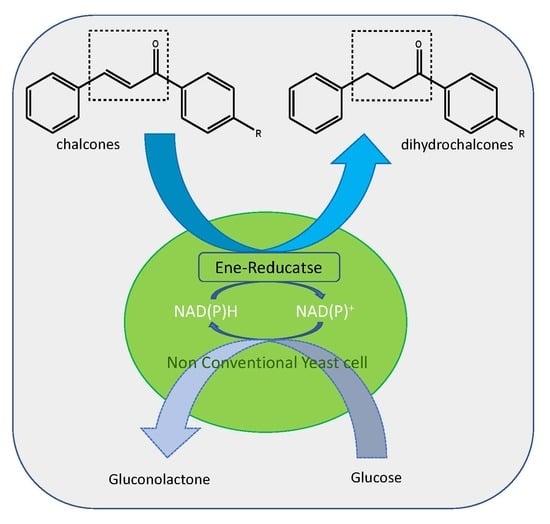
Non-Conventional Yeasts as Sources of Ene-Reductases for the Bioreduction of Chalcones
Abstract
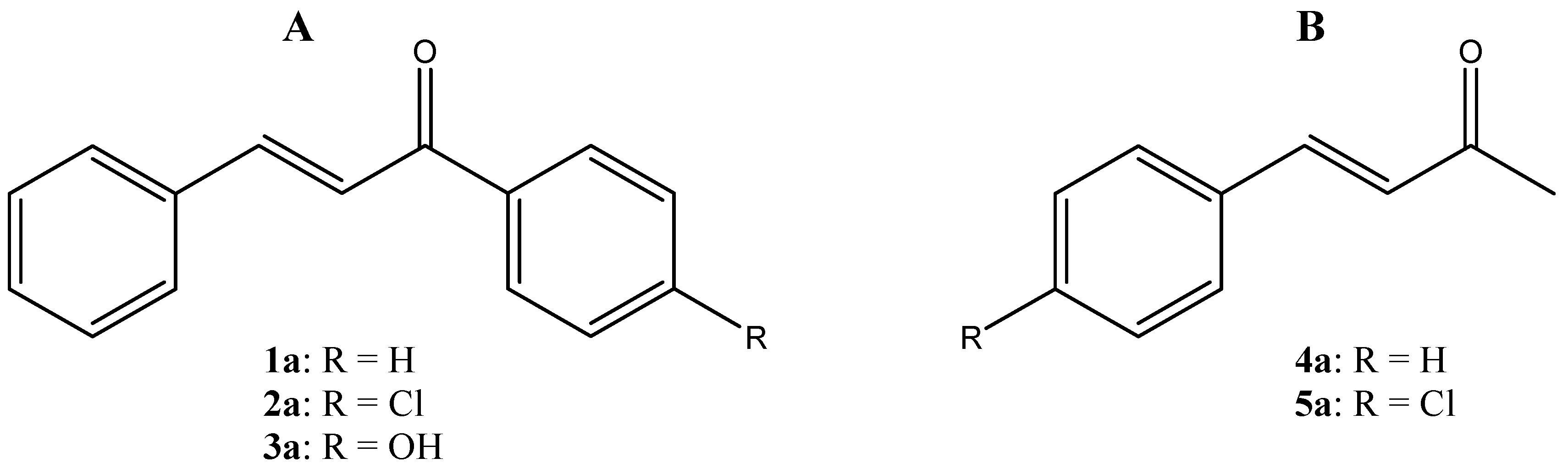
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Culture Media
2.2. Yeast Strain
2.3. Preparation of the Lyophilized NCYs Whole-Cells Biocatalyst
2.4. Bio-Reduction Reactions
2.5. GC–MS Analyses
2.6. LogP Calculation
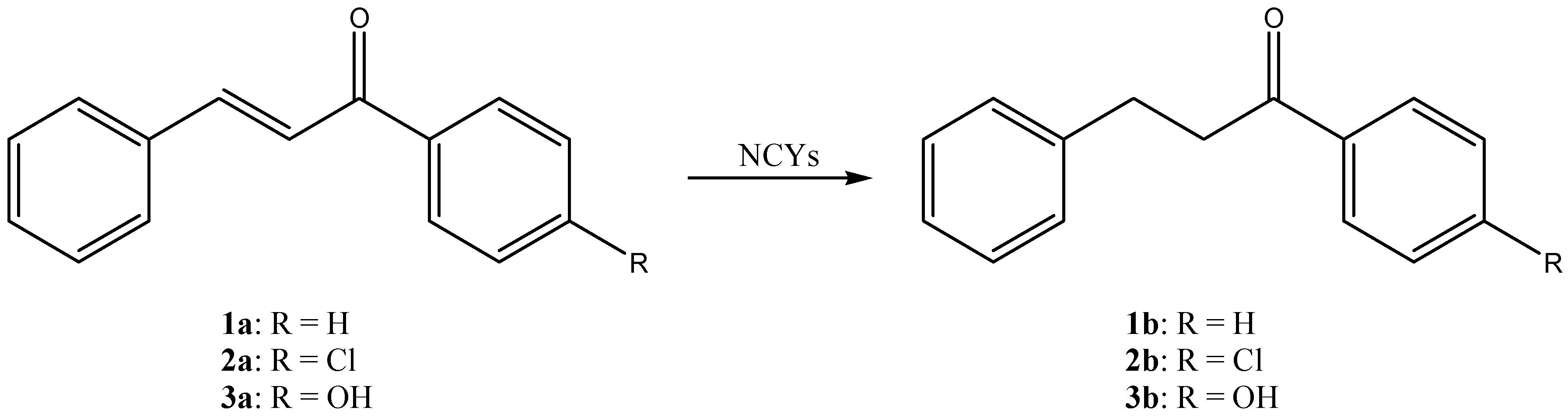
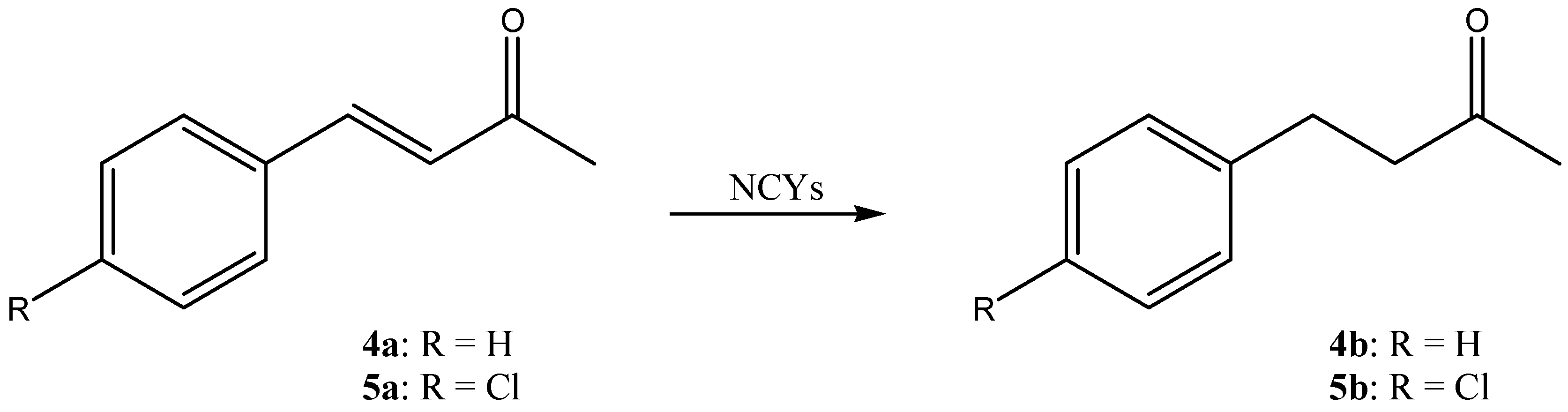
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brenna, E.; Gatti, F.G.; Manfredi, A.; Monti, D.; Parmeggiani, F. Enoate Reductase-Mediated Preparation of Methyl (S)-2-Bromobutanoate, a Useful Key Intermediate for the Synthesis of Chiral Active Pharmaceutical Ingredients. Org. Process. Res. Dev. 2012, 16, 262–268. [Google Scholar] [CrossRef]
- Winkler, C.K.; Tasnadi, G.; Clay, D.; Hall, M.; Faber, K. Asymmetric bioreduction of activated alkenes to industrially relevant optically active compounds. J. Biotechnol. 2012, 162, 381–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waller, J.; Toogood, H.S.; Karuppiah, V.; Rattray, N.J.W.; Mansell, D.J.; Leys, D.; Gardiner, J.M.; Fryszkowska, A.; Ahmed, S.T.; Bandichhor, R.; et al. Structural insights into the ene-reductase synthesis of profens. Org. Biomol. Chem. 2017, 15, 4440–4448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swiderska, M.A.; Stewart, J.D. Asymmetric Bioreductions of β-nitro acrylates as a route to chiral β2-Amino Acids. Org. Lett. 2006, 8, 6131–6133. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.J.; Stuekler, C.; Hauer, B.; Baudendistel, N.; Housden, H.; Bruce, N.C.; Faber, K. The substrate spectra of Pentaerythritol tetranitrate reductase, Morphinone reductase, N-Ethylmaleimide reductase and estrogen-binding protein in the asymmetric bioreduction of activated alkenes. Adv. Synth. Catal. 2010, 352, 387–394. [Google Scholar] [CrossRef]
- Yanto, Y.; Winkler, C.K.; Lohr, S.; Hall, M.; Faber, K.; Bommarius, A.S. Asymmetric bioreduction of alkenes using ene-reductases YersER and KYE1 and effects of organic solvents. Org. Lett. 2011, 13, 2540–2543. [Google Scholar] [CrossRef]
- Brenna, E.; Gatti, F.G.; Monti, D.; Parmeggiani, F.; Sacchetti, A. Cascade coupling of ene reductases with alcohol dehydrogenases: Enantioselective reduction of prochiral unsaturated aldehydes. ChemCatChem 2012, 4, 653–659. [Google Scholar] [CrossRef]
- Durchschein, K.; Wallner, S.; Macheroux, P.; Schwab, W.; Winkler, T.; Kreis, W.; Faber, K. Nicotinamide-dependent ene reductases as alternative biocatalysts for the Reduction of Activated Alkenes. Eur. J. Org. Chem. 2012, 4963–4968. [Google Scholar] [CrossRef]
- Toogood, H.S.; Scrutton, N.S. New developments in ‘ene’-reductase catalysed biological hydrogenations. Curr. Opin. Chem. Biol. 2014, 19, 107–115. [Google Scholar] [CrossRef]
- Reß, T.; Hummel, W.; Hanlon, S.P.; Iding, H.; Gröger, H. The organic-synthetic potential of recombinant ene reductases: Substrate-scope evaluation and process optimization. ChemCatChem 2015, 7, 1302–1311. [Google Scholar] [CrossRef]
- Turrini, N.G.; Hall, M.; Faber, K. Enzymatic synthesis of optically active lactones via asymmetric bioreduction using ene-reductases from the old yellow enzyme family. Adv. Synth. Catal. 2015, 357, 1861–1871. [Google Scholar] [CrossRef]
- Reich, S.; Nestl, B.M.; Hauer, B. Loop-grafted old yellow enzymes in the bienzymatic cascade reduction of allylic alcohols. ChemBioChem 2016, 17, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, K.; Fu, Y.; Polte, I.; Leupold, S.; Meo, A.; Weuster-Botz, D. Asymmetric whole-cell bioreduction of (R)-carvone by recombinant Escherichia coli with in situ substrate supply and product removal. Biochem. Eng. J. 2017, 117, 102–111. [Google Scholar] [CrossRef]
- Turrini, N.G.; Cioc, R.C.; van der Niet, D.J.H.; Ruijter, E.; Orru, R.V.A.; Hall, M.; Faber, K. Biocatalytic access to nonracemic γ-oxo esters via stereoselective reduction using ene-reductases. Green Chem. 2017, 19, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Hall, M.; Stueckler, C.; Kroutil, W.; Macheroux, P.; Faber, K. Asymmetric bioreduction of activated alkenes using cloned 12-oxophytodienoate reductase isoenzymes OPR-1 and OPR-3 from Lycopersicon esculentum (Tomato): A striking change of stereoselectivity. Angew. Chem. Int. Ed. 2007, 46, 3934–3937. [Google Scholar] [CrossRef]
- Mueller, A.; Stuermer, R.; Hauer, B.; Rosche, B. Asymmetric alkene reduction by yeast old yellow enzymes and by a novel Zymomonas mobilis reductase. Biotechnol. Bioeng. 2007, 98, 22–29. [Google Scholar] [CrossRef]
- Fu, Y.; Hoelsch, K.; Weuster-Botz, D. A novel ene-reductase from Synechococcus sp. PCC 7942 for the asymmetric reduction of alkenes. Proc. Biochem. 2012, 47, 1988–1997. [Google Scholar] [CrossRef]
- Fu, Y.; Castiglione, K.; Weuster-Botz, D. Comparative characterization of novel ene-reductases from cyanobacteria. Biotechnol. Bioeng. 2013, 110, 1293–1301. [Google Scholar] [CrossRef]
- Peters, C.; Kölzsch, R.; Kadow, M.; Skalden, L.; Rudroff, F.F.; Michovilovic, M.D.; Bornsceuer, U. Identification, characterization, and application of three enoate reductases from Pseudomonas putida in vitro enzyme cascade reactions. ChemCatChem 2014, 6, 1021–1027. [Google Scholar] [CrossRef]
- Romagnolo, A.; Spina, F.; Brenna, E.; Crotti, M.; Parmeggiani, F.; Varese, G.C. Identification of fungal ene-reductase activity by means of a functional screening. Fungal Biol. 2015, 119, 487–493. [Google Scholar] [CrossRef]
- Riedel, A.; Mehnert, M.; Paul, C.E.; Westphal, A.H.; van Berkel, W.J.H.; Tischler, D. Functional characterization and stability improvement of a ‘thermophilic-like’ ene-reductase from Rhodococcus opacus 1CP. Front. Microbiol. 2015, 6, 1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, X.Q.; Xu, M.Y.; Wu, Z.L. Two classical old yellow enzymes from Chryseobacterium sp. CA49: Broad substrate specificity of Chr-OYE1 and limited activity of Chr-OYE2. J. Mol. Catal. B Enzym. 2016, 23, 91–99. [Google Scholar] [CrossRef]
- Zhang, B.; Zheng, L.; Lin, J.; Wei, D. Characterization of an ene-reductase from Meyerozyma guilliermondii for asymmetric bioreduction of α,β-unsaturated compounds. Biotechnol. Lett. 2016, 38, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Scholtissek, A.; Tischler, D.; Westphal, A.H.; van Berkel, W.J.H.; Paul, C.E. Old yellow enzyme-catalysed asymmetric hydrogenation: Linking family roots with improved catalysis. Catalysts 2017, 7, 130. [Google Scholar] [CrossRef]
- Magallanes-Noguera, C.; Cecati, F.M.; Mascotti, M.L.; Reta, G.F.; Agostini, E.; Orden, A.A.; Kurina-Sanz, M. Plant tissue culture as sources of new ene- and ketoreductase activities. J. Biotechnol. 2017, 251, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivakumar, P.M.; Seenivasan, S.P.; Kumar, V.; Doble, M. Synthesis, antimycobacterial activity evaluation, and QSAR studies of chalcone derivatives. Bioorg. Med. Chem. Lett. 2007, 17, 1695–1700. [Google Scholar] [CrossRef]
- Gacche, R.N.; Dhole, N.A.; Kamble, S.G.; Bandgar, B.P. In-vitro evaluation of selected chalcones for antioxidant activity. J. Enzyme Inhib. Med. Chem. 2008, 23, 28–31. [Google Scholar] [CrossRef]
- Tomar, V.; Bhattacharjee, G.; Kumar, A. Synthesis and antimicrobial evaluation of new chalcone containing piperazine or 2,5-dicholothiophene moiety. Bioorg. Med. Chem. Lett. 2007, 17, 5321–5324. [Google Scholar] [CrossRef]
- Shibata, S. Antitumorigenic chalcones. Stem Cells 1994, 12, 44–52. [Google Scholar] [CrossRef]
- Gall, M.; Thomsen, M.; Peters, C.; Pavlidis, I.V.; Jonczyk, P.; Grunert, P.P.; Beutel, S.; Scheper, T.; Gross, E.; Beckes, M.; et al. Enzymatic conversion of flavonoids using bacterial chalcone isomerase and enoate reductase. Angew. Chem. Int. Ed. 2014, 53, 1439–1442. [Google Scholar] [CrossRef]
- Janeczko, T.; Gladkowski, W.; Kostrzewa-Suslow, E. Microbial transformations of chalcones to produce food sweetener derivatives. J. Mol. Catal. B Enzym. 2013, 98, 55–61. [Google Scholar] [CrossRef]
- Zyska, B.; Aniol, M.; Lipok, J. Highly effective, regiospecific reduction of chalcone by cyanobacteria leads to the formation of dihydrochalcone: Two steps towards natural sweetness. Microb. Cell Fact. 2017, 16, 136. [Google Scholar] [CrossRef] [PubMed]
- Kouamé, F.P.B.K.; Silvestre, V.; Bedi, G.; Loquet, D.; Robins, R.J.; Tea, I. Phytochemical investigation of the leaves of Leptoderris fasciculata. Phytochem. Lett. 2013, 6, 253–256. [Google Scholar] [CrossRef]
- Burkill, H.M. The useful plants of west tropical Africa—Entry for Leptoderris fasciculata (Benth.) Dunn [family Leguminosae-Papilionoideae]. JSTOR Plant Sci. 1985. Available online: http://plants.jstor.org/upwta/3_622 (accessed on 14 February 2020).
- Sibirny, A.A.; Scheffers, L. Thematic section Biochemistry, Genetics, Biotechnology and Ecology of Non-conventional Yeasts. FEMS Yeast Res. 2002, 2, 293. [Google Scholar] [CrossRef]
- Wolf, K.; Breunig, K.; Barth, G. Non Conventional Yeasts in Genetics, Biochemistry and Biotechnology; Springer: Berlin, Germany, 2003; p. 464. ISBN1 978-3-540-44215-8 (softcover). ISBN2 978-3-642-55758-3 (eBook). [Google Scholar]
- Buzzini, P.; Vaughan-Martini, A. Yeast biodiversity and biotechnology. In Biodiversity and Ecophysiology of Yeasts; Rosa, C.A., Peter, G., Eds.; Springer: Berlin, Germany, 2006; pp. 533–559. ISBN1 978-3-540-26100-1 (hardcover). ISBN2 978-3-642-06552-1 (softcover). ISBN3 978-3-540-30985-7 (eBook). [Google Scholar]
- Forti, L.; Di Mauro, S.; Cramarossa, M.R.; Filippucci, S.; Turchetti, B.; Buzzini, P. Non-conventional yeasts whole cells as efficient biocatalysts for the production of flavors and fragrances. Molecules 2015, 20, 10377–10398. [Google Scholar] [CrossRef] [Green Version]
- Goretti, M.; Ponzoni, C.; Caselli, E.; Marchigiani, E.; Cramarossa, M.R.; Turchetti, B.; Buzzini, P.; Forti, L. Biotransformation of electron-poor alkenes by yeasts: Asymmetric reduction of (4S)-(+)-carvone by yeast enoate reductases. Enz. Microb. Technol. 2009, 45, 463–468. [Google Scholar] [CrossRef]
- Goretti, M.; Ponzoni, C.; Caselli, E.; Marchegiani, E.; Cramarossa, M.R.; Turchetti, B.; Forti, L.; Buzzini, P. Bioreduction of α,β-unsaturated ketones and aldehydes by non-conventional yeast (NCY) whole-cells. Bioresour. Technol. 2011, 102, 3993–3998. [Google Scholar] [CrossRef]
- Goretti, M.; Branda, E.; Turchetti, B.; Cramarossa, M.R.; Onofri, A.; Forti, L.; Buzzini, P. Response surface methodology as optimization strategy for asymmetric bioreduction of (4S)-(+)-carvone by Cryptococcus gastricus. Bioresour. Technol. 2012, 121, 290–297. [Google Scholar] [CrossRef]
- Goretti, M.; Turchetti, B.; Cramarossa, M.R.; Forti, L.; Buzzini, P. Production of flavours and fragrances via bioreduction of (4R)-(-)-carvone and (1R)-(-)-myrtenal by non-conventional yeast whole-cells. Molecules 2013, 18, 5736–5748. [Google Scholar] [CrossRef]
- Lavandera, I.; Oberdorfer, G.; Gross, J.; Wildeman, S.; Kroutil, W. Stereocomplementary asymmetric reduction of bulky–bulky ketones by biocatalytic hydrogen transfer. Eur. J. Org. Chem. 2008, 2539–2543. [Google Scholar] [CrossRef]
- Garzon-Posse, F.; Becerra-Figueroa, L.; Hernàndez-Arias, J.; Gamba-Sànchez, D. Whole cells as biocatalysts in organic transformations. Molecules 2018, 23, 1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Oliveira Lopes, R.; Benzaquem Ribeiro, J.; de Souza Ramos, A.; Miranda, L.S.M.; Leal, I.C.R.; Leite, S.G.F.; Alves de Souza, R.O.M. Highly enantioselective bioreduction of 4-bromoacetophenone. Tetrahedron Asymmetry 2011, 22, 1763–1766. [Google Scholar] [CrossRef]
- Rocha, L.C.; de Souza, A.L.; Rodrigues Filho, U.P.; Campana Filho, S.P.; Sette, L.D.; Porto, A.L.M. Immobilization of marine fungi on silica gel, silica xerogel and chitosan for biocatalytic reduction of ketones. J. Mol. Catal. B Enzym. 2012, 84, 160–165. [Google Scholar] [CrossRef]
- Yang, Z.H.; Luo, L.; Chang, X.; Zhou, W.; Chen, G.H.; Zhao, Y.; Wang, Y.J. Production of chiral alcohols from prochiral ketones by microalgal photo-biocatalytic asymmetric reduction reactions. J. Ind. Microb. Biotech. 2012, 39, 835–841. [Google Scholar] [CrossRef]
- Gašo-Sokač, D.; Nujić, M.; Bušić, V.; Habuda-Satanić, M. Biocatalytic reductions by plant tissue—Green alternative to alcohol production. Croat. J. Food Sci. Technol. 2014, 6, 51–60. Available online: https://hrcak.srce.hr/125421 (accessed on 14 February 2020).
- Walker, G.M. Yeast nutrition. In Yeast Physiology and Biotechnology; Walker, G.M., Ed.; Wiley: Chichester, UK, 2000; pp. 51–99. ISBN 978-0-471-96446-9 (softcover). [Google Scholar]
- Fickers, P.; Benedetti, P.H.; Waché, Y.; Marty, A.; Mauersberger, S.; Smit, M.S.; Nicaud, J.-M. Hydrophobic substrate utilization by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 2005, 5, 527–543. [Google Scholar] [CrossRef] [Green Version]
- Rapoport, A.I.; Khroustalyova, G.M.; Crowe, L.M.; Crowe, J.H. Anhydrobiosis in yeast: Stabilization by exogenous lactose. Mikrobiologiia 2009, 78, 690–694. [Google Scholar] [CrossRef]
- Rapoport, A.I. Anhydrobiosis and Dehydration of Yeasts. In Biotechnology of Yeasts and Filamentous Fungi; Sibirny, A.A., Ed.; Springer: Berlin, Germany, 2017; pp. 87–116. ISBN1 978-3-319-58828-5 (hardcover). ISBN2 978-3-319-86485-3 (softcover). ISBN3 978-3-319-58829-2 (eBook). [Google Scholar]
- Molinari, F.; Gandolfi, R.; Villa, R.; Occhiato, E.G. Lyophilized yeasts: Easy-to-handle biocatalysts for stereoselective reduction of ketones. Tetrahedron Asymmetry 1999, 10, 3515–3520. [Google Scholar] [CrossRef]
- Stuermer, R.; Hauer, B.; Hall, M.; Faber, K. Asymmetric bioreduction of activated C=C bonds using enoate reductases from the old yellow enzyme family. Curr. Opin. Chem.Biol. 2007, 11, 203–213. [Google Scholar] [CrossRef]
| Species | Origin | Location |
|---|---|---|
| Candida freyschussii DBVPG 6208 | Wood pulp | Sweden |
| Cyberlindnera amylophila DBVPG 6346 | Frass of Pinus taeda (loblolly pine) | USA |
| Goffeauzyma gastrica DBVPG 4709 | Sub-glacial debris of the Sforzellina glacier | Italy |
| Goffeauzyma gilvescens DBVPG 4712 | Supra-glacial debris of the Sforzellina glacier | Italy |
| Hanseniaspora guilliermondii DBVPG 6790 | Trachea of bee | France |
| Kazachstania exigua DBVPG 6469 | Soil | South Africa |
| Kazachstania naganishii DBVPG 7133 | Decaying leaves | Japan |
| Kazachstania spencerorum DBVPG 6746 | Soil | South Africa |
| Kluyveromyces lactis DBVPG 6854 | Rain forest drosophilids | Brazil |
| Naganishia diffluens DBVPG 6237 | Soil of vineyard | Hungary |
| Pichia kluyveri DBVPG 5826 | Soil close to plum tree | Algeria |
| Scheffersomyces shehatae DBVPG 6850 | Rain forest drosophilids | Brazil |
| Wickerhamomyces canadensis DBVPG 6211 | Ground wood pulp | Sweden |
| DBVPG Accession Numbers | Species | 1a Conversion mol% (± SD) | 2a Conversion mol% (± SD) | 3a Conversion mol% (± SD) | 4a Conversion mol% (± SD) | 5a Conversion mol% (± SD) |
|---|---|---|---|---|---|---|
| 6208 | Candida freyschussii | 2.9 ± 1.5 | 23.2 ± 13.5 | 0.0 ± 0.0 | 0.0 ± 0.0 | 3.1 ± 0.5 |
| 6346 | Cyberlindnera amylophila | 94.0 ± 2.8 | 88.3 ± 10.2 | 100.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 4709 | Goffeauzyma gastrica | 100.0 ± 0.0 | 65.2 ± 56.5 | 0.0 ± 0.0 | 3.2 ± 0.6 | 2.7 ± 1.0 |
| 4712 | Goffeauzyma gilvescens | 11. 5 ± 5.6 | 78.0 ± 6.1 | 47.8 ± 11.0 | 7.4 ± 4.0 | 16.9 ± 1.4 |
| 6790 | Hanseniaspora guilliermondii | 96.2 ± 1.6 | 76.4 ± 22.2 | 0.0 ± 0.0 | 0.0 ± 0.0 | 1.8 ± 1.1 |
| 6469 | Kazachstania exigua | 96.3 ± 0.8 | 17.4 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 7133 | Kazachstania naganishii | 0.0 ± 0.0 | 83.5 ± 3.6 | 0.0 ± 0.0 | 3.2 ± 0.4 | 2.7 ± 1.2 |
| 6746 | Kazachstania spencerorum | 95.2 ± 2.0 | 48.4 ± 4.3 | 100.0 ± 0.0 | 46.9 ± 7.4 | 4.7 ± 0.4 |
| 6854 | Kluyveromyces lactis | 100.0 ± 0.0 | 73.3 ± 24.3 | 100.0 ± 0.0 | 99.1 ± 0.8 | 0.0 ± 0.0 |
| 6237 | Naganishia diffluens | 99.1 ± 1.6 | 20.9 ± 20.9 | 0.0 ± 0.0 | 15.6 ± 8.3 | 21.2 ± 4.7 |
| 5826 | Pichia kluyveri | 0.7 ± 0.7 | 98.1 ± 3.4 | 0.0 ± 0.0 | 0.0 ± 0.0 | 1.5 ± 0.5 |
| 6850 | Scheffersomyces shehatae | 97.5 ± 4.3 | 51.6 ± 11.5 | 0.0 ± 0.0 | 13.5 ± 3.9 | 0.0 ± 0.0 |
| 6211 | Wickerhamomyces canadensis | 100.0 ± 0.0 | 8.0 ± 11.3 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filippucci, S.; Tasselli, G.; Kenza Labbani, F.-Z.; Turchetti, B.; Cramarossa, M.R.; Buzzini, P.; Forti, L. Non-Conventional Yeasts as Sources of Ene-Reductases for the Bioreduction of Chalcones. Fermentation 2020, 6, 29. https://doi.org/10.3390/fermentation6010029
Filippucci S, Tasselli G, Kenza Labbani F-Z, Turchetti B, Cramarossa MR, Buzzini P, Forti L. Non-Conventional Yeasts as Sources of Ene-Reductases for the Bioreduction of Chalcones. Fermentation. 2020; 6(1):29. https://doi.org/10.3390/fermentation6010029
Chicago/Turabian StyleFilippucci, Sara, Giorgia Tasselli, Fatima-Zohra Kenza Labbani, Benedetta Turchetti, Maria Rita Cramarossa, Pietro Buzzini, and Luca Forti. 2020. "Non-Conventional Yeasts as Sources of Ene-Reductases for the Bioreduction of Chalcones" Fermentation 6, no. 1: 29. https://doi.org/10.3390/fermentation6010029
APA StyleFilippucci, S., Tasselli, G., Kenza Labbani, F.-Z., Turchetti, B., Cramarossa, M. R., Buzzini, P., & Forti, L. (2020). Non-Conventional Yeasts as Sources of Ene-Reductases for the Bioreduction of Chalcones. Fermentation, 6(1), 29. https://doi.org/10.3390/fermentation6010029