Augmentation of Granular Anaerobic Sludge with Algalytic Bacteria Enhances Methane Production from Microalgal Biomass
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Algalytic Bacteria
2.2. Qualitative and Quantitative Assessments of Algalytic Activity
2.3. Source of Inoculum and Substrate for Bioaugmetation
2.4. Specific Methanogenic Activity (SMA) Test
2.5. Polymerase Chain Reaction (PCR) Analysis
2.6. 16SrRNA Gene Sequencing and Analysis
2.7. Statistical and Diversity Analysis
3. Results
3.1. Isolation, Identification, and Characterization of Algalytic Bacteria
3.1.1. Isolation and Identification
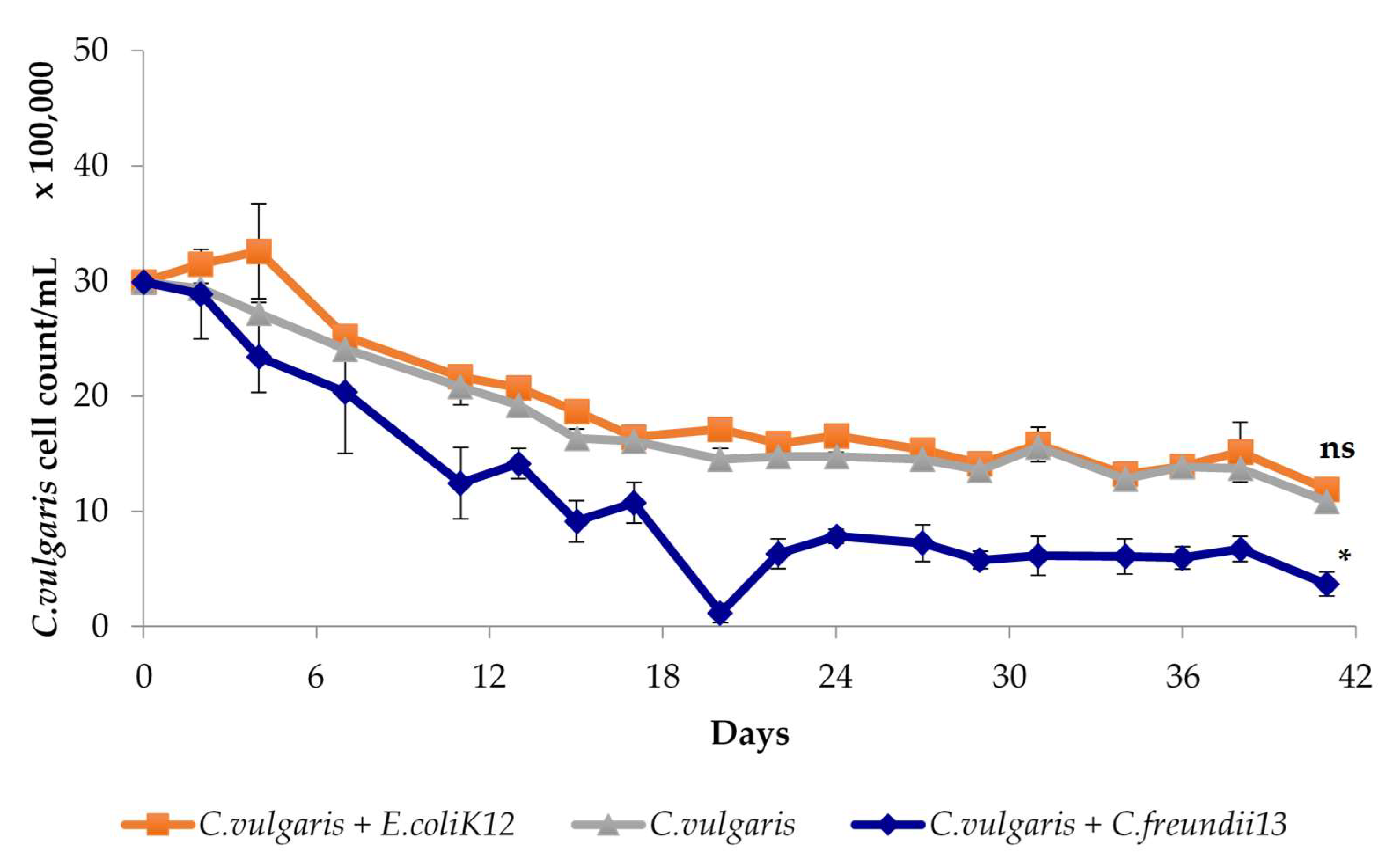
3.1.2. Characterization of Algalytic Activity
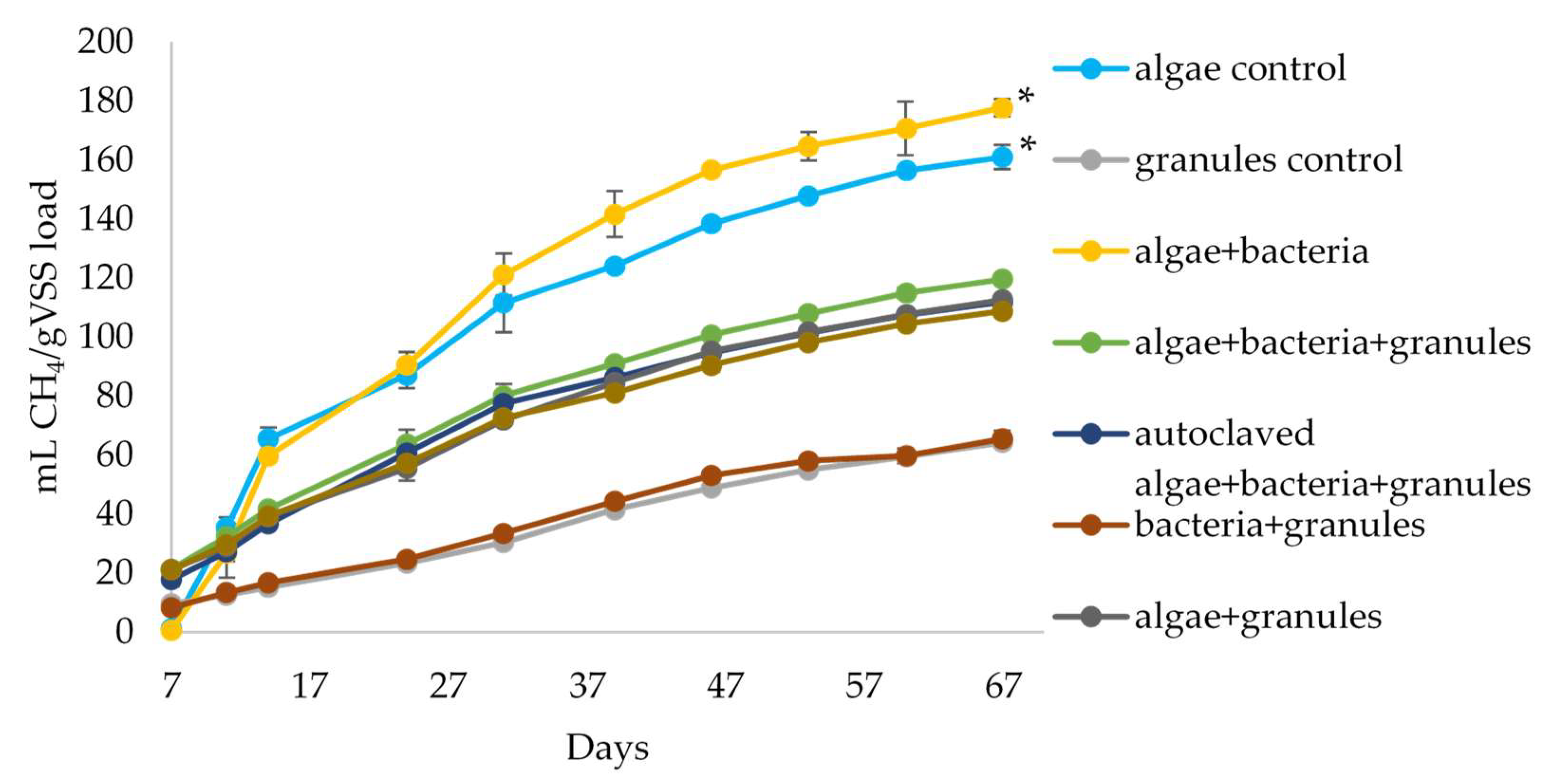
3.2. Anaerobic Digestion of Algal Biomass by Intact and Bioaugmeted Granular Sludge
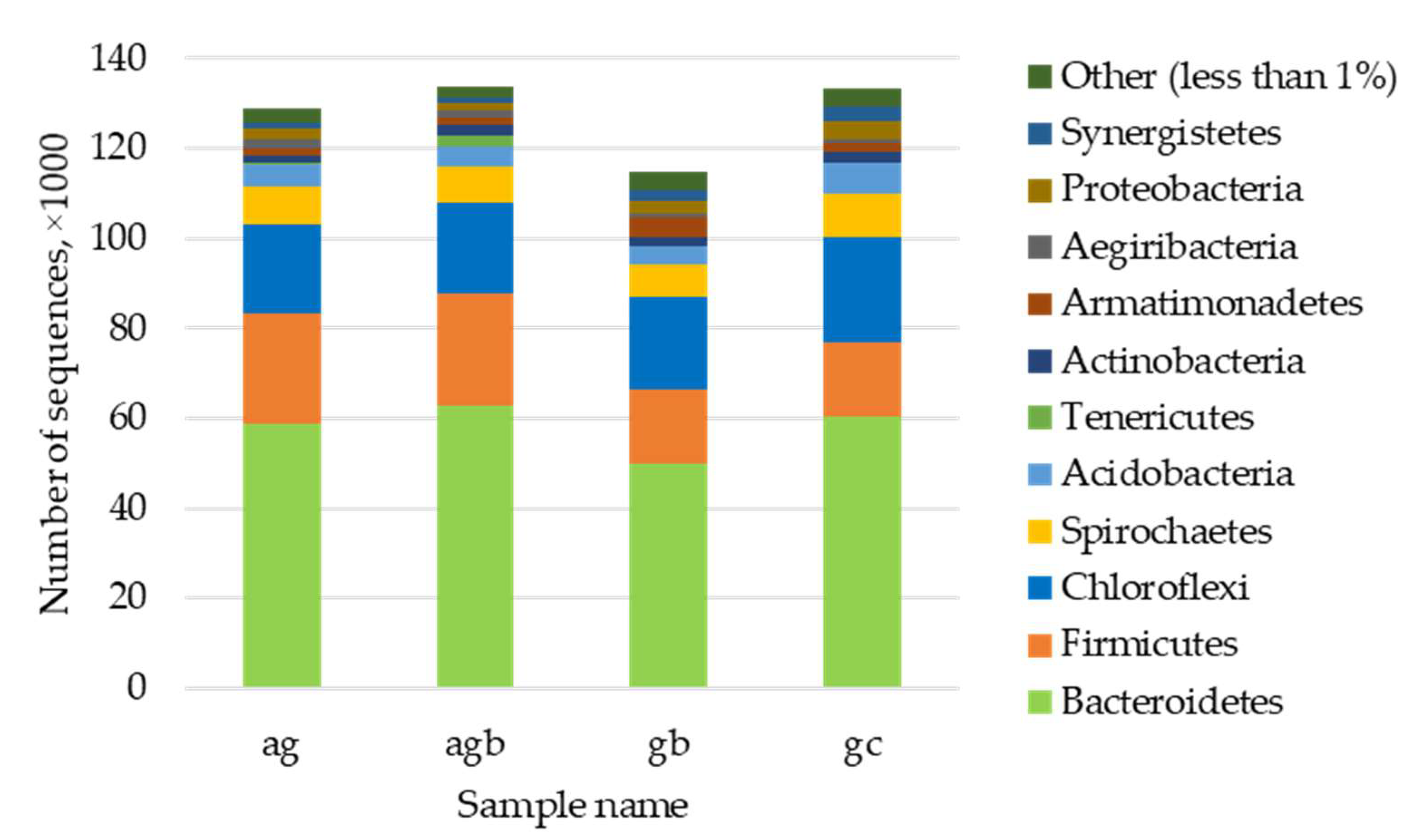
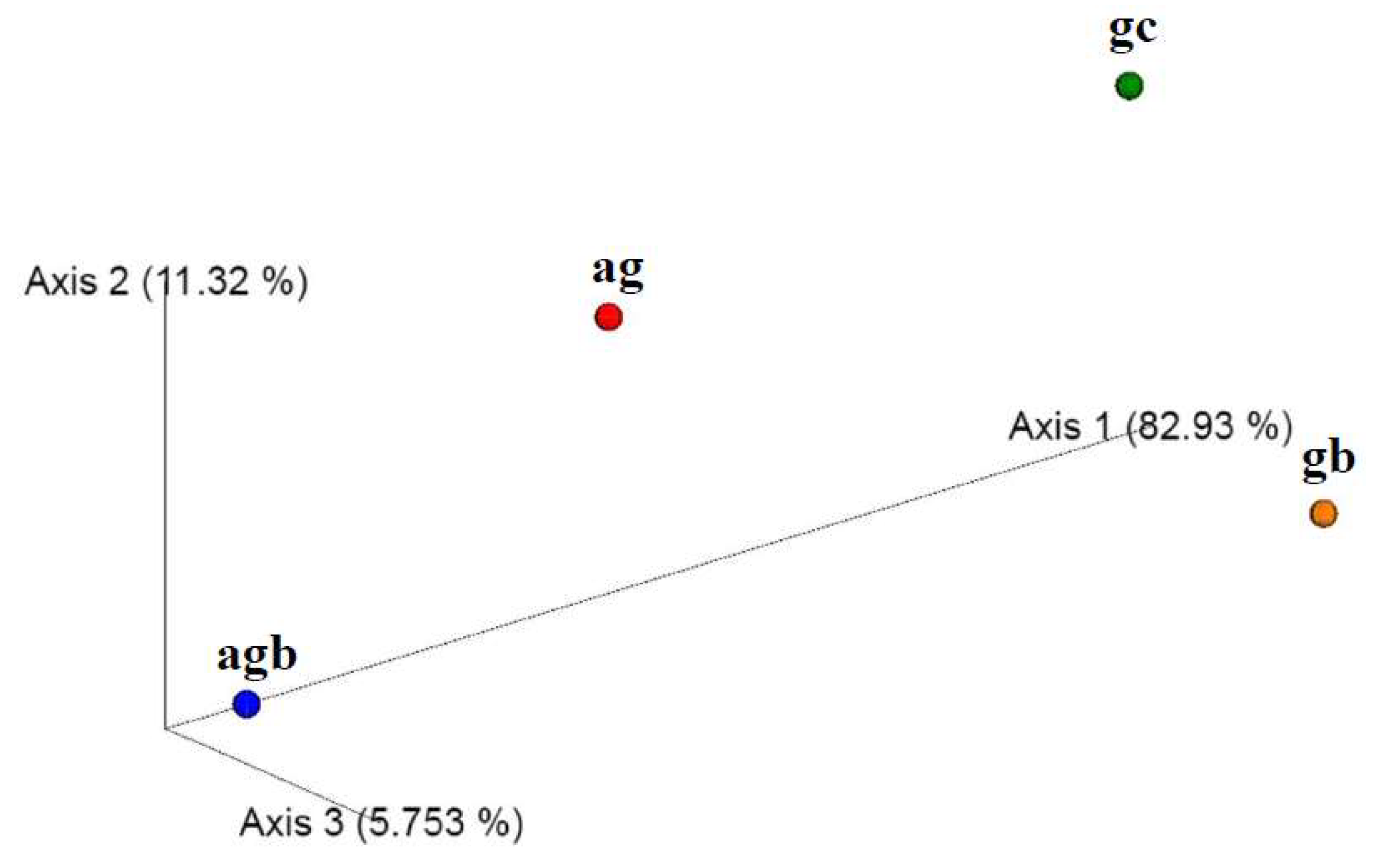
3.3. Assessment of the Presence and Influence of Algalytic Bacteria on the Anaerobic Digestion
4. Discussion
4.1. Bioaugmentation of Granular Sludge Does not Require a UASB-Like System
4.2. Bioaugmenting Granular Sludge Community Leads to Its Specialization toward Algae Digestion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
References
- Hajji, K.T.; Lepine, F.; Bisaillon, J.G.; Beaudet, R.; Hawari, J.; Guiot, S.R. Effects of bioaugmentation strategies in UASB reactors with a methanogenic consortium for removal of phenolic compounds. Biotechnol. Bioeng. 2000, 67, 417–423. [Google Scholar] [CrossRef]
- Cater, M.; Fanedl, L.; Malovrh, S.; Logar, R.M. Biogas production from brewery spent grain enhanced by bioaugmentation with hydrolytic anaerobic bacteria. Bioresour. Technol. 2015, 186, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Horber, C.; Christiansen, N.; Arvin, E.; Ahring, B.K. Improved dechlorinating performance of upflow anaerobic sludge blanket reactors by incorporation of Dehalospirillum multivorans into granular sludge. Appl. Environ. Microbol. 1998, 64, 1860–1863. [Google Scholar]
- Fotidis, I.A.; Karakashev, D.; Angelidaki, I. Bioaugmentation with an acetate-oxidising consortium as a tool to tackle ammonia inhibition of anaerobic digestion. Bioresour. Technol. 2013, 146, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Bagi, Z.; Acs, N.; Balint, B.; Horvath, L.; Dobo, K.; Perei, K.R.; Rakhely, G.; Kovacs, K.L. Biotechnological intensification of biogas production. Appl. Microbiol. Biotechnol. 2007, 76, 473–482. [Google Scholar] [CrossRef]
- Wei, S.Z. The application of biotechnology on the enhancing of biogas production from lignocellulosic waste. Appl. Microbiol. Biotechnol. 2016, 100, 9821–9836. [Google Scholar] [CrossRef]
- Yang, Z.M.; Guo, R.B.; Xu, X.H.; Wang, L.; Dai, M. Enhanced methane production via repeated batch bioaugmentation pattern of enriched microbial consortia. Bioresour. Technol. 2016, 216, 471–477. [Google Scholar] [CrossRef]
- Debowski, M.; Zielinski, M.; Grala, A.; Dudek, M. Algae biomass as an alternative substrate in biogas production technologies—Review. Renew. Sustain. Energy Rev. 2013, 27, 596–604. [Google Scholar] [CrossRef]
- Ramos-Suarez, J.L.; Carreras, N. Use of microalgae residues for biogas production. Chem. Eng. J. 2014, 242, 86–95. [Google Scholar] [CrossRef]
- Montingelli, M.E.; Tedesco, S.; Olabi, A.G. Biogas production from algal biomass: A review. Renew. Sustain. Energy Rev. 2015, 43, 961–972. [Google Scholar] [CrossRef]
- Doloman, A.; Soboh, Y.; Walters, A.J.; Sims, R.C.; Miller, C.D. Qualitative Analysis of Microbial Dynamics during Anaerobic Digestion of Microalgal Biomass in a UASB Reactor. Int. J. Microbiol. 2017, 2017, 5291283. [Google Scholar] [CrossRef] [PubMed]
- Soboh, Y.M.; Sorensen, D.L.; Sims, R.C. Upflow Anaerobic Sludge Blanket Reactor Codigestion of Algae and Acetate to Produce Methane. Water Environ. Res. 2016, 88, 2094–2103. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Yasumoto, H. Colony Formation by Algicidal Saprospira Sp on Marine Agar Plates. Nippon Suisan Gakk 1991, 57, 2139–2143. [Google Scholar] [CrossRef]
- Krueger, C.L.; Sheikh, W. A New Selective Medium for Isolating Pseudomonas Spp from Water. Appl. Environ. Microbol. 1987, 53, 895–897. [Google Scholar]
- De Lipthay, J.R.; Johnsen, K.; Albrechtsen, H.-J.; Rosenberg, P.; Aamand, J. Bacterial diversity and community structure of a sub-surface aquifer exposed to realistic low herbicide concentrations. FEMS Microbiol. Ecol. 2004, 49, 59–69. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaclikova, E.; Krascsenicsova, K.; Pangallo, D.; Kuchta, T. Detection and quantification of Citrobacter freundii and C. braakii by 5’-nuclease polymerase chain reaction. Curr. Microbiol. 2005, 51, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, J.L.; Meints, R.H.; Kuczmarski, D.; Burbank, D.E.; Lee, K. Viruses of symbiotic Chlorella-like algae isolated from Paramecium bursaria and Hydra viridis. Proc. Natl. Acad. Sci. USA 1982, 79, 3867–3871. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Cha, M.; Guss, A.M.; Westpheling, J. Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc. Natl. Acad. Sci. USA 2014, 111, 8931–8936. [Google Scholar] [CrossRef]
- Jung, S.W.; Kim, B.H.; Katano, T.; Kong, D.S.; Han, M.S. Pseudomonas fluorescens HYK0210-SK09 offers species-specific biological control of winter algal blooms caused by freshwater diatom Stephanodiscus hantzschii. J. Appl. Microbiol. 2008, 105, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K. Chemical Kinetics and Reaction Dynamics; Springer: Dordrecht, The Netherlands, 2006. [Google Scholar] [CrossRef]
- Nopharatana, A.; Clarke, W.P.; Pullammanappallil, P.C.; Silvey, P.; Chynoweth, D.P. Evaluation of methanogenic activities during anaerobic digestion of municipal solid waste. Bioresour. Technol. 1998, 64, 169–174. [Google Scholar] [CrossRef]
- Ince, O.; Ince, B.K.; Yenigun, O. Determination of potential methane production capacity of a granular sludge from a pilot-scale upflow anaerobic sludge blanket reactor using a specific methanogenic activity test. J. Chem. Technol. Biotechnol. 2001, 76, 573–578. [Google Scholar] [CrossRef]
- Wolfe, R.S. Techniques for cultivating methanogens. Methods Enzymol. 2011, 494, 1–22. [Google Scholar] [CrossRef]
- Huws, S.A.; Edwards, J.E.; Kim, E.J.; Scollan, N.D. Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems. J. Microbiol. Methods 2007, 70, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef]
- Joshi, N.A.; Fass, J.N. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ files; Version 1.33; Software; GitHub: San Francisco, CA, USA, 2011. [Google Scholar]
- Buffalo, V. Scythe—A Bayesian Adapter Trimmer; Software; GitHub: San Francisco, CA, USA, 2014. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- QIIME2. Quantitative Insights into Microbial Ecology. Available online: https://qiime2.org (accessed on 1 September 2018).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Passos, F.; Uggetti, E.; Carrère, H.; Ferrer, I. Pretreatment of microalgae to improve biogas production: A review. Bioresour. Technol. 2014, 172, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.K.; Guha, S.; Harendranath, C.S.; Tripathi, S. Influence of extrinsic factors on granulation in UASB reactor. Appl. Microbiol. Biotechnol. 2006, 71, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Tartakovsky, B.; Guiot, S.R. Modeling and analysis of layered stationary anaerobic granular biofilms. Biotechnol. Bioeng. 1997, 54, 122–130. [Google Scholar] [CrossRef]
- Sekiguchi, Y.; Kamagata, Y.; Nakamura, K.; Ohashi, A.; Harada, H. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl. Environ. Microbol. 1999, 65, 1280–1288. [Google Scholar]
- Opazo, R.; Ortuzar, F.; Navarrete, P.; Espejo, R.; Romero, J. Reduction of Soybean Meal Non-Starch Polysaccharides and alpha-Galactosides by Solid-State Fermentation Using Cellulolytic Bacteria Obtained from Different Environments. PLoS ONE 2012, 7, e0044783. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Somitsch, W.; Klymiuk, I.; Trajanoski, S.; Guebitz, G.M. Comparison of biogas sludge and raw crop material as source of hydrolytic cultures for anaerobic digestion. Bioresour. Technol. 2016, 207, 244–251. [Google Scholar] [CrossRef]
- Yin, L.J.; Jiang, S.T.; Pon, S.H.; Lin, H.H. Hydrolysis of Chlorella by Cellulomonas sp YJ5 Cellulases and Its Biofunctional Properties. J. Food Sci. 2010, 75, H317–H323. [Google Scholar] [CrossRef]
- Shiratori, H.; Ohiwa, H.; Ikeno, H.; Ayame, S.; Kataoka, N.; Miya, A.; Beppu, T.; Ueda, K. Lutispora thermophila gen. nov., sp nov., a thermophilic, spore-forming bacterium isolated from a thermophilic methanogenic bioreactor digesting municipal solid wastes. Int. J. Syst. Evol. Microbiol. 2008, 58, 964–969. [Google Scholar] [CrossRef]
- Liu, Y.; Qiao, J.T.; Yuan, X.Z.; Guo, R.B.; Qiu, Y.L. Hydrogenispora ethanolica gen. nov., sp nov., an anaerobic carbohydrate-fermenting bacterium from anaerobic sludge. Int. J. Syst. Evol. Microbiol. 2014, 64, 1756–1762. [Google Scholar] [CrossRef]
- Traore, A.S.; Hatchikian, C.E.; Le Gall, J.; Belaich, J.P. Microcalorimetric studies of the growth of sulfate-reducing bacteria: Comparison of the growth parameters of some Desulfovibrio species. J. Bacteriol. 1982, 149, 606–611. [Google Scholar]
- Bouanane-Darenfed, A.; Ben Hania, W.; Hacene, H.; Cayol, J.L.; Ollivier, B.; Fardeau, M.L. Caldicoprobacter guelmensis sp. nov., a thermophilic, anaerobic, xylanolytic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 2049–2053. [Google Scholar] [CrossRef] [PubMed]
- Gobet, A.; Mest, L.; Perennou, M.; Dittami, S.M.; Caralp, C.; Coulombet, C.; Huchette, S.; Roussel, S.; Michel, G.; Leblanc, C. Seasonal and algal diet-driven patterns of the digestive microbiota of the European abalone Haliotis tuberculata, a generalist marine herbivore. Microbiome 2018, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Antunes, A. The Family Haloplasmataceae. In The Prokaryotes, 4th ed.; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doloman, A.; Pererva, Y.; Cortez, M.H.; Sims, R.C.; Miller, C.D. Augmentation of Granular Anaerobic Sludge with Algalytic Bacteria Enhances Methane Production from Microalgal Biomass. Fermentation 2019, 5, 88. https://doi.org/10.3390/fermentation5040088
Doloman A, Pererva Y, Cortez MH, Sims RC, Miller CD. Augmentation of Granular Anaerobic Sludge with Algalytic Bacteria Enhances Methane Production from Microalgal Biomass. Fermentation. 2019; 5(4):88. https://doi.org/10.3390/fermentation5040088
Chicago/Turabian StyleDoloman, Anna, Yehor Pererva, Michael H. Cortez, Ronald C. Sims, and Charles D. Miller. 2019. "Augmentation of Granular Anaerobic Sludge with Algalytic Bacteria Enhances Methane Production from Microalgal Biomass" Fermentation 5, no. 4: 88. https://doi.org/10.3390/fermentation5040088
APA StyleDoloman, A., Pererva, Y., Cortez, M. H., Sims, R. C., & Miller, C. D. (2019). Augmentation of Granular Anaerobic Sludge with Algalytic Bacteria Enhances Methane Production from Microalgal Biomass. Fermentation, 5(4), 88. https://doi.org/10.3390/fermentation5040088




