Abstract
Two major spoilage yeasts in the wine industry, Brettanomyces bruxellensis and Zygosaccharomyces rouxii, produce off-flavors and gas, causing considerable economic losses. Traditionally, SO2 has been used in winemaking to prevent spoilage, but strict regulations are in place regarding its use due to its toxic and allergenic effects. To reduce its usage researchers have been searching for alternative techniques. One alternative is biocontrol, which can be used either independently or in a complementary way to chemical control (SO2). The present study analyzed 122 native non-Saccharomyces yeasts for their biocontrol activity and their ability to be employed under fermentation conditions, as well as certain enological traits. After the native non-Saccharomyces yeasts were assayed for their biocontrol activity, 10 biocontroller yeasts were selected and assayed for their ability to prevail in the fermentation medium, as well as with respect to their corresponding positive/negative contribution to the wine. Two yeasts that satisfy these characteristics were Wickerhamomyces anomalus BWa156 and Metschnikowia pulcherrima BMp29, which were selected for further research in application to mixed fermentations.
1. Introduction
Wine is the product of complex microbial interactions that start on the grape surface and continue throughout the fermentation [1]. Some yeasts generate metabolites that lead to wine faults that affect flavor, haze or CO2 production in the final product. One of the major spoilage yeasts is Brettanomyces bruxellensis [2]. Wines contaminated with this yeast are characterized by the presence of off-flavors [3]. Other spoilage yeasts frequently described in the food industry belong to the Zygosaccharomyces genus. They produce gas in food and beverages [4], and they are difficult to control chemically [5]. Spoilage resulting from this yeast is widespread and causes considerable economic losses in the food industry [6,7].
Traditionally, SO2 has been used in winemaking during non-fermentation stages to control microbial proliferation such as bacteria, yeasts and fungi. Nevertheless, there are strict regulations regarding its use due to its toxic and allergenic effects on human health [8]. International organizations such as the Organisation Internationale de la vigne et du vin encourage SO2 reduction [9]. Moreover, modern consumers prefer more natural and healthy foods and beverages that are minimally processed and free of preservatives [4,10].
Biocontrol is an alternative proposal that can be used either independently or in a complementary way to chemical control (SO2). Some Saccharomyces and non-Saccharomyces yeasts have the ability to biosuppress other yeasts through different mechanisms such as the production of toxic compounds [2], competition for limiting substrates [11] and/or cell to cell contact [1].
At present, a re-evaluation of the role of non-Saccharomyces yeasts in winemaking and their use as selected starters in mixed fermentations with Saccharomyces cerevisiae is being carried out [12,13]. Non-Saccharomyces yeasts are supposed to enhance the wine quality [14]. Nowadays there is a special interest in yeast strains associated with specific geographical locations as they may introduce a regional character or ‘terroir’ to the winemaking process [12,15].
Yeast growth parameters such as specific growth rate, lag phase duration, product yield and metabolic rates of substrates and products may provide useful information to understand their biocontrol mechanisms and how to use them during the fermentation process. Taking into account that yeast bio-suppression can be associated with substrate competition and secretion of toxic substances, it is important to understand the growth parameters of non-Saccharomyces yeasts during fermentation, in order to plan co-inoculation or sequential mixed inoculation with Saccharomyces [16,17,18,19].
Several authors have analyzed indirect values like “fermentation rate” (CO2 release) [20,21]. However, there are no reports related to selection of non-Saccharomyces yeast for vinification that have studied the prevalence of yeasts with clearly defined kinetic parameters. The aim of the present study was to analyze the biocontrol ability of 122 native non-Saccharomyces yeasts against two of the most relevant wine spoilage yeast species, Z. rouxii and B. bruxellensis. Subsequently, biocontrolling yeasts were characterized for their ability to be employed under fermentation conditions and their capacity to generate positive or negative enological traits, in order to reduce SO2 and improve the quality of regional wines.
2. Materials and Methods
2.1. Microorganisms
One hundred and twenty-two non-Saccharomyces yeasts (Table 1), previously isolated from enological environments from San Juan and Mendoza, Argentina (Cuyo region), were obtained from the Culture Collection of Autochthonous Microorganisms of the Institute of Biotechnology, School of Engineering, UNSJ, San Juan, Argentina, and used in the present study. The yeasts had been used in previous studies by our research group [22,23].

Table 1.
Non-Saccharomyces yeast isolates assayed.
Eight spoilage yeasts (4 Brettanomyces bruxellensis isolates and 4 Zygosaccharomyces rouxii isolates) were obtained from the EEA INTA culture collection, Lujan, Mendoza, Argentina, and used in the study [4,24]. Saccharomyces cerevisiae BSc114 [23] was used as positive control with regard to fermentative performance. Isolates were identified through biochemical, physiological and morphological methods [6] as well as molecular methods [25].
2.2. Culture Media
Propagation was carried out in YEPD broth (g/L): Yeast extract 10, peptone, glucose 20, pH: 4.5 adjusted with HCl 1N.
Viable yeast counting was carried out on YEPD-agar (g/L): Yeast extract 10, peptone 20, glucose 20, agar-agar 20, pH: 4.5.
Biocontrol was carried out on YMB-agar supplemented with 0.2 M citrate-phosphate buffer, pH 4.5 (g/L): Glucose 10, yeast extract 3, malt extract 3, peptone 5, NaCl 30, methylene blue 0.030, glycerol 10% v/v [26] (modified).
Kinetics and tolerance assays were carried out with concentrated grape must (65 °Brix), diluted at 21 °Brix with 1 g/L of yeast extract added, pH: 4.
Inocula for biocontrol and plate assays were obtained with YEPD broth pH: 4.5, 24 h incubation period.
Inocula for kinetic and tolerances assays were obtained with concentrated grape must (65 °Brix), diluted at 21 °Brix with 1 g/L of yeast extract added, pH: 4, 24 h incubation period.
Complex media was sterilized at 121 °C for 20 min and grape must media at 111 °C for 20 min.
2.3. Screening for Biocontrol Ability of Yeasts
Each spoilage yeast was incorporated at a concentration of 106 cells/mL in liquid YMB-agar biocontrol medium at 45 °C, mixed to uniform, and immediately poured into sterile petri dishes. Potential biocontrolling non-Saccharomyces yeasts were inoculated as a drop (20 µL) on the agar surface, and plates were incubated at 25 °C until a well-developed lawn. Killer activity was visualized as zone of growth inhibition (more than 1 mm) around the spotted killer yeast colony on plates [2].
Biocontrolling activity against the spoilage species was calculated in 2 ways: (a) Intraspecific inhibition: the proportion at which one biocontrolling strain inhibited the spoilage isolates belonging to one species. In addition, (b) Total inhibition: the proportion at which one biocontroller yeast strain inhibited the spoilage isolates belonging to both species.
2.4. Fermentative Performance
2.4.1. Growth Kinetics during Fermentation
Yeasts were separately cultured in Erlenmeyer flasks (250 mL) containing 200 mL of growth medium. Each isolate was seeded at a concentration of 106 cells/mL and incubated at 25 °C for 21 days under static conditions, according to [27] for growth determination by viable cell count. Samples were taken on days 1, 2, 3, 6, 15 and 21. Plates were used for viable cell counts and the experimental data was used to construct a growth curve, which was used to determine kinetic parameters. µmax was calculated as described Monod [28] and the lag phase as described Lodge and Hinshelwood [29], which are the most widespread methods according to [16,17].
2.4.2. Tolerance to Different Stress
Low temperature (15 °C), High concentrations of reducing sugar (30 °Brix), and different Ethanol concentrations (8, 10, 12 and 14% v/v) and different molecular SO2 concentrations (0.1, 0.15, 0.2, 0.3 and 0.4 ppm) were carried out according to Vazquez et al. [27] in tolerance medium for each strain. Growth was monitored with Durham tubes (CO2 production). Gas production in the Durham tubes was monitored one day after the positive control of each strain. Molecular SO2 was calculated according to [30,31]. Control was performed at 25 °C, 21 °Brix, 0% v/v ethanol and 0 ppm of SO2.
2.4.3. Plate Assays
SH2 production: Yeasts were spot-inoculated and evaluated as semi-quantitative over BigGy-agar (BBLTM, Becton, Dickinson and Company, Sparks, USA, Le Pont de Claix, France) following elaboration instructions. Incubation: 3 days at 25 °C. An arbitrary scale was used for the color of the colony from 1, white color (no production); 2, light brown; 3, brown; 4, dark brown; 5, dark brown/black (high production) [20].
β– glucosidase activity: was performed according to Strauss et al. [32]. Medium containing (g/L): yeast nitrogen base 6.7 (YNB, Difco™, Becton, Dickinson and Company, Sparks, USA), Arbutin 5 (SigmaTM, Sigma-Aldrich, Saint Louis, USA) and agar-agar 2, pH: 5, then autoclaved (121 ºC, 20 min). 2 mL of filtered 1% ammonium ferric citrate solution was added to 100 mL media before pouring into plates. Yeast was spot-inoculated. Incubation: 5 days at 30 °C. Activity was positive when a discolored halo of hydrolysis was observed.
Protease activity: was performed according to Comitini et al. [20]. The medium contained (g/L): yeast extract 3, malt extract 3, peptone 5, glucose 10, NaCl 5 and agar-agar 15. In a separate vessel, an equal volume of skimmed milk was prepared with sterile water at 10% p/v. After sterilization both solutions were mixed and poured into sterile petri dishes. Yeast was spot-inoculated. Incubation: 3 days at 25 °C. Activity was observed as a clear halo of hydrolysis.
Pectinase activity: was performed according to Fernandez-Salomäo et al. [33]. The medium contained (g/L): citrus pectin 2, yeast extract 1, KH2PO4 0.2, CaCl2 0.05, (NH4)2SO4 1, MgSO4.7H2O 0.8, MnSO4 0.05, agar-agar 20, pH: 4.5. After sterilization (121 °C, 20 min), it was poured into sterile petri dishes and yeast was spot-inoculated. Incubation: 3 days at 30 °C. After incubation, Lugol solution was added and pectin degradation was observed as a clear halo of hydrolysis.
Pathogenicity: hemolysin production of yeasts was performed according to Manns et al. [34] and Menezes et al. [35], which used Blood agar medium in petri dishes (Britania™, CABA, Argentina) for this purpose. Yeast was spot-inoculated. Incubation: 2 days at 37 °C. Positive activity was observed as a clear zone of hydrolysis.
All assays were carried out using Saccharomyces cerevisiae BSc114 as a positive control for biocontrol, sensitivity to inhibition of selected isolates, tolerance to low temperature, and high concentrations of reducing sugars, ethanol and SO2, and as a negative control for H2S, β–glucosidase, protease and pectinase activity [23]. The prokaryote Pseudomonas aeruginosa BPa987 was used as positive control for hemolysin production of the yeasts [36,37].
2.5. Data Analysis
Each assay was performed independently in triplicate and results are represented as the average of three determinations with the corresponding standard deviation (±SD). Data were tested for normality, homoscedasticity and independence. Parametrical data and significant differences were analyzed by Fisher test. Principal components analysis (PCA) was used to simplify interpretation of the yeast behavior data and is presented in a biplot graph. InfoStat™ -Professional software version 1.5 was used for data analysis.
3. Results and Discussion
To be used as co-inocula together with Saccharomyces cerevisiae in wine fermentations, biocontroller yeasts must possess a good specific growth rate and a short lag phase during anaerobiosis to predominate in the medium [18]. In addition, they should not produce any negative attributes to wine, but instead, they should contribute with positive attributes.
3.1. Biocontrol Screening
Non-Saccharomyces yeasts are considered to improve the wine complexity and enhance positive traits of regional wines. Several authors have reported that a rational selection of non-Saccharomyces yeasts as S. cerevisiae co-inoculum improves the quality of wines [21,23,38]. In the present study, 122 non-Saccharomyces yeasts belonging to 10 genera and isolated from different enological environments were screened to assess their ability to biocontrol wine spoilage yeasts belonging to Zygosaccharomyces rouxii (4 isolates) and Brettanomyces bruxellensis (4 isolates) species.
Bioassaying showed that 23 non-Saccharomyces yeasts belonging to 6 genera inhibited growth of at least one isolate of either Brettanomyces bruxellensis or Zygosaccharomyces rouxii (Table 2).

Table 2.
Biocontrol of spoilage yeasts by non-Saccharomyces yeast isolates.
None of the selected biocontrollers inhibited the control (BSc114) lawn development. This fact would allow the application of these yeast isolations in co-inocula with this strain of S. cerevisiae. Some of the species used in this work have already been used as biocontrollers of non-Saccharomyces yeasts and did not inhibit the development of S.cerevisiae [14].
Yeast species that showed biocontrol activity in our laboratories have also been cited in other studies as antagonists of different spoilage yeasts, with different mechanisms being involved. Pichia guilliermondii, associated with killer toxin production, has been proven to interact with Penicillium expansum [39]. Wickerhamomyces anomalus has been cited as a B. bruxellensis biocontroller, confirming the observations in the present study [40]. Different W. anomalus strains have been associated with three killer toxins [39]. This species has also been found to kill a broad range of organisms, including bacteria, hyphomycetes and yeasts [41].
Metschnikowia pulcherrima has been commented on by Oro et al. [14] because of its biocontrol capacity to a wide spectrum of genera like Hanseniaspora, Pichia, Torulaspora, Zygosaccharomyces, Saccharomycodes, Candida, Issatchenkia, Brettanomyces and Schizosaccharomyces, which also confirms our results. The biocontrol mechanism for M. pulcherrima would be iron depletion from the medium through binding to pulcherrimic acid [14].
These results can be analyzed from two perspectives: from the spoilage yeast or the antagonistic yeast point of view. Considering spoilage yeasts, the B. bruxellensis and Z. rouxii isolates analyzed in our study showed different sensitivity to Candida sake, Hanseniaspora uvarum, Metschnikowia pulcherrima, Pichia occidentalis and Starmerella bacillaris species. Oro et al. [14] reported a similar behavior of spoilage yeasts with different sensitivities to M. pulcherrima strains.
Regarding biocontrol isolates, intraspecific biocontrol was observed for BMp49 and BPg138 against all B. bruxellensis strains assayed. In the case of Z. rouxii, all 4 strains assayed were inhibited by BHu23, BMp145 and BPo108. BHu5, BHu27, BMp4, BMp29, BMp49, BPg138 and BWa156 showed an intraspecific inhibition higher than 0.5 against B. bruxellensis, whereas Cs88, Cs95, BHu5, BHu18, BHu23, BHu31, BHu32, BMp4, BMp29, BMp49, BMp145, BPo104, BPo108 and BWa156 demonstrated the same inhibition against Z. rouxii. The relevance of wide intraspecific inhibition is the possibility of avoiding adaptation of the spoilage yeast to a particular action mechanism by the antagonistic yeast [42]. In addition, wide interspecific/intergeneric inhibition is also considered a positive factor, because it may control other potential spoilage yeasts not detected in the spoilage analysis [43]. Interspecific/intergeneric biocontrol behavior against B. bruxellensis and Z. rouxii species was observed for BCi85, BCs95, BHu5, BHu27, BHu31, BHu32, BMp4, BMp29, BMp49, BMp145, BPo104, BPo108, BPg138, BSb57 and BWa156 yeasts; they biocontrolled at least one isolate from each species. Most of the yeasts with interspecific/intergeneric biocontrol showed an intraspecific inhibition of 0.5 or higher. This could be related to a common site of action of the killer toxin [42] or a common biocontrol mode of action affecting yeasts in general, like substrate competition [1].
Hanseniaspora uvarum isolates BHu5 and BHu23, Metschnikowia pulcherrima isolates BMp4, BMp29, BMp49 and BMp145, Pichia guilliermondii BPg138, Pichia occidentalis isolates BPo104 and BPo108 and Wickerhamomyces anomalus BWa156 were selected because they showed a total inhibition of 0.50 or more. Except for BHu23, all biocontroller yeasts inhibited at least one strain of both spoilage yeasts. In addition, the 10 isolates were evaluated for their enological characteristics.
3.2. Behavior of the Antagonistic Yeasts
3.2.1. Kinetic Parameters
When selecting non-Saccharomyces yeasts for oenological fermentations as a co-inoculum with S. cerevisiae, special attention should be paid to their beneficial characteristics to enhance wine quality besides their biocontrolling properties.
To achieve these goals, predominance of the selected non-Saccharomyces yeasts in the medium during the first stage of the fermentation is very important. Anaerobic growth kinetics of non-Saccharomyces yeasts possess important parameters to elucidate such predominance. Duration of the lag phase (or adaptation) and maximal growth rate are two relevant anaerobiosis parameters, which are described below [16,17,18,19].
The present study examined the kinetic parameters of each individual yeast. Nevertheless, in mixed fermentations with grape must, when limiting substrate availability is more prominent compared with the saturation constant, each species will growth at its maximum rate. This is the main parameter to ensure predominance [16], but only when the previous state of the culture (growth stage, age and size of the inoculum) is homogeneous for all experiments [44]. Moreover, this will be governed by the chemical and physical characteristics of the environment unless one of the interacting species produces inhibitory agents against the other [16]. It is also known that the yeast complexes behave differently because of competition, antagonism or cooperation and this could result in the predominance of different yeasts [45,46].
A fermentation growth curve of each antagonist yeast was performed. Viable cell data were recorded to calculate the specific maximal growth rate (µmax) and lag phase. Most of the non-Saccharomyces yeasts assayed reached a specific maximal growth rate near 0.04 h−1 (Figure 1). BMp29, BMp49 and BMp145 showed a higher µmax which was significantly different. BPg138 and BPo104 displayed a lower µmax which was also significantly different. High specific growth rates are desired because they are a relevant factor in the prevalence of an organism during fermentation [18]. M. pulcherrima isolates presented the highest specific growth rates. This behavior could be related to the fact that the mode of action of this species is through the competition of limited substrates and not through a killer factor [14]. The killer factor has been found to consume metabolic energy, reducing the fitness of the yeast that possesses the factor [47], and hence it could decrease the fitness of the other non-Saccharomyces yeasts. Growth rates of 0.29 h−1 [48], 0.31 h−1 [49] and even 0.5 h−1 have been found for Saccharomyces cerevisiae during anaerobiosis [50]. Therefore, prevalence of the non-Saccharomyces in the medium at the start of the fermentation should be considered for sequential co-inoculation with S. cerevisiae.
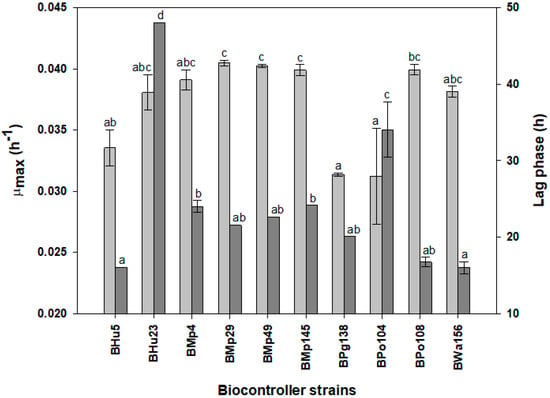
Figure 1.
Specific growth rate (light grey) and lag phase (dark grey) of the non-Saccharomyces yeasts assayed. Rates are means with standard deviation (n = 3). Values with different letters are significantly different.
A successful predominance of the biocontroller during the fermentation start should demonstrate a short lag phase [10]. Most strains showed a lag time of about 20 h (Figure 1). BHu5 and BWa156 showed the shortest lag phases, about 15 h, and they were significantly different. BPo104 and BHu23 showed a significant longer lag phase of about 35 h and 50 h, respectively. A reduced lag phase increments the possibility of non-Saccharomyces to predominate the medium, since the lag phase is defined as the period prior of reaching the specific growth rate [16]. The lag phase is also relevant for the non-Saccharomyces strains to achieve a constant number for a determined period of time prior to inoculation of S. cerevisiae in a sequential mixed fermentation.
3.2.2. Enological Characterization of Yeasts: Tolerance to Molecular SO2, Ethanol, High Reducing Sugar Concentrations and Low Temperature
The control yeast, S. cerevisiae BSc114, was able to ferment grape must at 30 °Brix and 15 °C and tolerated 14% v/v of ethanol and 0.4 ppm of molecular SO2 (Table 3). With respect to SO2, the non-Saccharomyces yeasts BMp29, BMp49, BMp145, BPg138, BPo104, BPo108 and BWa156 showed higher tolerance to SO2 (0.4 ppm) (Table 3). Although a reduction in SO2 is a goal of this study, it is relevant to evaluate resistance of the selected isolates to typical SO2 concentrations used in wineries at the beginning of the process. The chemical could be present after yeast production, but never more than 100 ppm of total SO2 [51]. Additionally, when non-Saccharomyces yeasts are used in integrated management (biocontrol yeasts—SO2 application) they should be able to tolerate certain SO2 concentrations. Typical winemaking generally uses at least 0.5 ppm of molecular SO2 and in order to avoid any microbial contamination, this can increase to a final molecular SO2 concentration of 0.8 ppm [52]. This means that BMp4, which showed the lowest tolerance and did not show any growth at molecular SO2 concentrations above 0.1 ppm, would not be suitable for integrated management. Typically, non-Saccharomyces yeasts have been cited to be low SO2 tolerant, but this sensitivity could also be linked to the combination of several factors such as ethanol, SO2 and temperature [53].

Table 3.
Tolerance of individual strains to high sugar concentration and low temperature, and different concentrations of molecular SO2 and ethanol.
Regarding ethanol tolerance, BPo104 was the least tolerant strain and did not present growth above 10% v/v ethanol (Table 3). The most tolerant strains were BMp29 and BMp49 (12% v/v of ethanol), and the remaining isolates tolerated 10% v/v. None of the isolates were able to grow at 14% v/v. Tolerance of the non-Saccharomyces yeasts to ethanol is especially important with increasing permanence in the fermentation medium, as the growing Saccharomyces sp. population produces high amounts of ethanol [1]. All isolates seemed to tolerate 8% v/v during the first fermentation stages [45]. However, high ethanol tolerance could be a problem, because S. cerevisiae uses this method to biocontrol other native microbiota [1,54]. The presence of some non-Saccharomyces species like killer yeasts for prolonged periods of time could negatively modify the sensory quality of wine and cause stuck fermentation [41]. Even, the effect of metabolic interactions between non-Saccharomyces and S. cerevisiae wine yeasts could affect the growth and fermentation behavior of S. cerevisiae during fermentation [22]. Despite the fact that the high ethanol tolerance and wide biocontrol spectrum described for M. pulcherrima could be a potential risk for the normal fermentation process of S. cerevisiae, Oro et al. [14] mentioned that Metschnikowia pulcherrima does not biocontrol S. cerevisiae.
BMp29, BMp145 and BWa156 were able to carry out fermentation at high sugar concentrations (Table 3) whereas the remaining isolates were not. Tolerance to high sugar concentrations is relevant, because must from the Cuyo region (San Juan and Mendoza provinces) usually possesses a high sugar concentration [13]. As Z. rouxii yeasts are highly osmotolerant, it is extra important that Z. rouxii antagonists develop well under similar conditions [4,55].
With regard to tolerance to low temperature, BHu5 was the only isolate that did not grow (-). The remaining isolates were considered tolerant to low temperatures at the start of the fermentation (+). This is also a relevant factor when the biocontroller yeast is used during white wine fermentations or fermentations carried out at low temperature to preserve aromas [56].
3.2.3. Enzyme and H2S Production
Control strain BSc114 reported low H2S production and did not present any of the desired enzymatic activities assayed (Table 4). All non-Saccharomyces isolates evaluated except for BMp4 demonstrated desired protease activity (Table 4). This activity contributes to the degradation of proteins that could cause haze in the wine, thus facilitating the process of clarification and filtration [2]. Only BWa156 showed pectinase activity. This activity is another positive attribute that enables degradation of structural grape polysaccharides, increasing juice extraction and improving wine clarification and filtration [57]. It facilitates the release of aromatic precursors from the cells of the skin, seeds and flesh of the grape to the must [22,58]. Pectinase activity could be linked to a substrate colonization role or a trophic role [59]. Regarding yeast development and sugar consumption, firstly, BWa156 could be able to obtain sugars from the intracellular matrix of plant cells. In red wine fermentations with BWa156, this could generate a competitive advantage of the strain in the grape skin layer. Secondly, the yeast could consume galacturonic acid [59] as an alternative to glucose, which is quickly consumed by S. cerevisiae [60]. This would extend the time of this energy source for BWa156 and therefore result in a long-term competitive advantage. Although the activity is strain-dependent [22], pectinase production has already been associated with W. anomalus [61,62]. Nevertheless, more research is needed. None of the assayed yeasts showed β- glucosidase activity [22].

Table 4.
Non-Saccharomyces attributes.
Regarding the possible contribution to negative wine characteristics, most of the assayed yeasts showed medium H2S production (3 or less on the scale in Table 3). Only BPg138 and BPo108 showed a higher production, 4, which is not desirable. Lowest production was produced by BWa156 (2 on the scale). H2S production is highly relevant in winemaking and thus very important for the yeast selection because it is associated with the negative persistent odor described as “rotten egg” [27,38].
None of the isolates displayed hemolysin production. This is an important phenotypic characteristic of pathogenicity because it is related to lysis of erythrocytes [34].
Principal components analysis explained 62% of the variation among components (Figure 2). Desirable and undesirable characteristics can be clearly differentiated on the main axis (explaining 36.4%). Desirable characteristics observed were: high growth rate, tolerance to low temperature and high concentrations of ethanol, SO2 and reducing sugars, and production of positive enzymes such as protease and pectinase. Prolonged adaptation time (Lag phase) and high H2S production were undesirable characteristics.
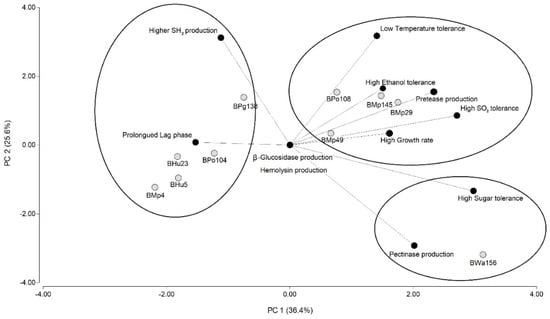
Figure 2.
Principal Components Analysis (PCA) of yeast characteristics. References: Ellipses represent clusters obtained from hierarchical cluster analysis (HCA).
In the biplot it can be observed that BWa156 has the ability to prevail in the medium during the early stage of the fermentation. Compared with the other non-Saccharomyces assayed, this yeast possesses a short lag phase and high growth rate. The latter characteristic is related to the cellular multiplication and enables the release of killer toxins that may be constitutive [63], incrementing the possibilities of the biocontrol yeast. BWa156 also releases enzymes that could allow utilization of alternative energy sources. Our study also showed its capacity to grow in adverse must conditions such as high sugar and high SO2 concentrations and the ability to grow at low temperature. In addition, it should be highlighted that the strain may positively contribute to the wine quality through the release of grape compounds because of its protease and pectinase production; these enzymes are not produced by BSc114. As a consequence, it could help intensify the color and enhance aromatic characteristics of the wine. Another advantage of the strain is the low H2S production.
The biplot demonstrates that BMp29 presents more possibilities to prevail in the medium compared with the other non-Saccharomyces isolates assayed, because of its high growth rate and short lag phase. The strain is also able to grow under adverse conditions of grape must such as high sugar and high SO2 concentrations and low temperature. Its high ethanol tolerance facilitates its growth and possible biocontrol during the fermentation. In the cluster, BMp49 presented a similar behavior to that of BMp29, but it did not develop at high reducing sugar concentrations. BMp145 and BPo108 also showed similar characteristics, but the first had a prolonged lag phase and the second strain the disadvantage of a higher potential to produce H2S.
BWa156 and BMp29 demonstrated a wide biocontrol spectrum. Wickerhamomyces anomalus and Metchnikowia pulcherrima have already been used in co-inocula with Saccharomyces cerevisiae by Comitini et al. [11] and Oro et al. [14]. Albertin et al. [19] described positive flavor attributes related to M. pulcherrima, which supports the possibility of using such species as co-inocula. However, further research is necessary to determine the biocontrol application of the two selected strains [41].
4. Conclusions
The selected non-Saccharomyces yeasts BWa156 and BMp29 are highly applicable antagonistic yeasts that positively contribute to the wine process. They are active against relevant spoilage yeasts in the wine industry and can be used to produce wines with reduced SO2 concentration. The present study is part of a comprehensive research project focusing on the application of non-Saccharomyces biocontroller yeasts. The biocontrolling sources and the conditions of implantation, prevalence and biocontrol kinetics is the projection of future research.
Author Contributions
Conceptualization, B.K., F.V., M.E.T., Y.P.M. and M.C.; methodology and experimental design, F.V., M.C. and Y.P.M.; software, B.K. and M.V.M.; validation, F.V. and M.E.T.; formal analysis, B.K., M.V.M. and F.V.; investigation, B.K., Y.P.M., M.V.V., M.C., M.E.T. and F.V.; data curation, B.K., writing—original draft preparation, B.K.; writing—review and editing, B.K., F.V., M.E.T. and Y.P.M.; visualization, B.K.; supervision, F.V., M.E.T. and M.C.; project administration, Y.P.M.; funding acquisition, Y.P.M. All authors discussed the results of the experiments and contributed to the final manuscript.
Funding
This work was supported by the Research Project IDeA 1400- SECITI 0041-2014.
Acknowledgments
The authors want to thank the Chemical Engineer Martha Dina Vallejo for the suggested corrections, the chemical donations and the accompaniment.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Ciani, M.; Capece, A.; Comitini, F.; Canonico, L.; Siesto, G.; Romano, P. Yeast interactions in inoculated wine fermentation. Front. Microbiol. 2016, 7, 555. [Google Scholar] [CrossRef] [PubMed]
- Mehlomakulu, N.N.; Setati, M.E.; Divol, B. Characterization of novel killer toxins secreted by wine-related non-Saccharomyces yeasts and their action on Brettanomyces spp. Int. J. Food Microbiol. 2014, 188, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Oelofse, A.; Lonvaud-Funel, A.; Du Toit, M. Molecular identification of Brettanomyces bruxellensis strains isolated from red wines and volatile phenol production. Food Microbiol. 2009, 26, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Rojo, M.C.; López, F.A.; Lerena, M.C.; Mercado, L.; Torres, A.; Combina, M. Evaluation of different chemical preservatives to control Zygosaccharomyces rouxii growth in high sugar culture media. Food Control. 2015, 50, 349–355. [Google Scholar] [CrossRef]
- Stratford, M.; Steels, H.; Nebe-von-Caron, G.; Avery, S.V.; Novodvorska, M.; Archer, D.B. Population heterogeneity and dynamics in starter culture and lag phase adaptation of the spoilage yeast Zygosaccharomyces bailii to weak acid preservatives. Int. J. Food Microbiol. 2014, 181, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G.H. Yeast Spoilage of Food and Beverages. In The Yeasts: A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier Science: London, UK; Burlington, MA, USA, 2011; pp. 53–63. [Google Scholar]
- Escott, C.; del Fresno, J.M.; Loira, I.; Morata, A.; Suárez-Lepe, J.A. Zygosaccharomyces rouxii: Control Strategies and Applications in Food and Winemaking. Fermentation 2018, 4, 69. [Google Scholar] [CrossRef]
- Cejudo-Bastante, M.J.; Sonni, F.; Chinnici, F.; Versari, A.; Perez-Coello, M.S.; Riponi, C. Fermentation of sulphite-free white musts with added lysozyme and oenological tannins: Nitrogen consumption and biogenic amines composition of final wines. LWT-Food Sci. Technol. 2010, 43, 1501–1507. [Google Scholar] [CrossRef]
- Ferrer-Gallego, R.; Puxeu, M.; Martín, L.; Nart, E.; Hidalgo, C.; Andorrà, I. Microbiological, Physical, and Chemical Procedures to Elaborate High-Quality SO2-Free Wines. In Grapes and Wines-Advances in Production, Processing, Analysis and Valorization; IntechOpen: Madrid, Spain, 2017; pp. 171–193. [Google Scholar]
- Swinnen, I.A.M.; Bernaerts, K.; Dens, E.J.; Geeraerd, A.H.; Van Impe, J.F. Predictive modelling of the microbial lag phase: A review. Int. J. Food Microbiol. 2004, 94, 137–159. [Google Scholar] [CrossRef]
- Comitini, F.; De Ingeniis, J.; Pepe, L.; Mannazzu, I.; Ciani, M. Pichia anomala and Kluyveromyces wickerhamii killer toxins as new tools against Dekkera/Brettanomyces spoilage yeasts. FEMS Microbiol. Lett. 2004, 238, 235–240. [Google Scholar] [CrossRef]
- Fleet, G.H. Wine yeasts for the future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef]
- Maturano, Y.P.; Mestre, M.V.; Kuchen, B.; Toro, M.E.; Mercado, L.A.; Vazquez, F.; Combina, M. Optimization of fermentation-relevant factors: A strategy to reduce ethanol in red wine by sequential culture of native yeasts. Int. J. Food Microbiol. 2019, 289, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Oro, L.; Ciani, M.; Comitini, F. Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 2014, 116, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Drumonde-Neves, J.; Franco-Duarte, R.; Lima, T.; Schuller, D.; Pais, C. Yeast biodiversity in vineyard environments is increased by human intervention. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Pirt, S.J. Parameters of growth and analysis of growth data. In Principles of Microbe and Cell Cultivation; Blackwell Scientific Publications: Hoboken, NJ, USA, 1975; pp. 4–14. [Google Scholar]
- Duarte, A. Introducción a la Ingeniería Bioquímica; Universidad Nacional de Colombia: Bogotá, Colombia, 1998; p. 198. [Google Scholar]
- Stanbury, P.F.; Whitaker, A.; Hall, S.J. Microbial growth kinetics. In Principles of Fermentation Technology; Elsevier Science: Kent, UK, 2014; pp. 21–74. [Google Scholar]
- Albertin, W.; Zimmer, A.; Miot-Sertier, C.; Bernard, M.; Coulon, J.; Moine, V.; Colonna-Ceccaldi, B.; Bely, M.; Marullo, P.; Masneuf-Pomarede, I. Combined effect of the Saccharomyces cerevisiae lag phase and the non-Saccharomyces consortium to enhance wine fruitiness and complexity. Appl. Microbiol. Biotechnol. 2017, 101, 7603–7620. [Google Scholar] [CrossRef] [PubMed]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [PubMed]
- Domizio, P.; Romani, C.; Lencioni, L.; Comitini, F.; Gobbi, M.; Mannazzu, I.; Ciani, M. Outlining a future for non-Saccharomyces yeasts: Selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 2011, 147, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Maturano, Y.P.; Rodriguez, L.A.; Toro, M.E.; Nally, M.C.; Vallejo, M.; Castellanos de Figueroa, L.I.; Combina, M.; Vazquez, F. Multi-enzyme production by pure and mixed cultures of Saccharomyces and non-Saccharomyces yeasts during wine fermentation. Int. J. Food Microbiol. 2012, 155, 43–50. [Google Scholar] [CrossRef]
- Mestre, V.; Maturano, Y.P.; Combina, M.; Mercado, L.A.; Toro, M.E.; Vazquez, F. Selection of non-Saccharomyces yeasts to be used in grape musts with high alcoholic potential: A strategy to obtain wines with reduced ethanol content. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef]
- Sturm, M.E.; Assof, M.; Fanzone, M.; Martinez, C.; Ganga, M.A.; Jofré, V.; Ramirez, M.L.; Combina, M. Relation between coumarate decarboxylase and vinylphenol reductase activity with regard to the production of volatile phenols by native Dekkera bruxellensis strains under ‘wine-like’ conditions. Int. J. Food Microbiol. 2015, 206, 51–55. [Google Scholar] [CrossRef]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8 S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Evol. Microbiol. 1999, 49, 329–337. [Google Scholar] [CrossRef]
- Santos, A.; San Mauro, M.; Bravo, E.; Marquina, D. PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiology 2009, 155, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, F.; Figueroa, L.; Toro, M. Enological characteristics of yeasts. In Methods in Biotechnology: Food Microbiology Protocols; Spencer, J.F.T., Spencer, A.L., Eds.; Humana Press: Totowa, NJ, USA, 2001; Volume 14, pp. 297–306. [Google Scholar]
- Monod, J. Recherches ser la Croissance des Cultures Bactériennes, 2nd ed.; Hermann: Paris, France, 1942. [Google Scholar]
- Lodge, R.M.; Hinshelwood, C.N. Physicochemical aspects of bacterial growth. Part IX. The lag phase of Bac. lactis aerogenes. J. Chem. Soc. 1943, 51, 213–219. [Google Scholar] [CrossRef]
- Sudraud, P.; Chauvet, S. Activite antilevure de l’anhydride sulfureux moleculaire. Conn. Vigne Vin. 1985, 19, 31–40. [Google Scholar] [CrossRef]
- Ribéreau-Gayon, J.; Peynaud, E.; Ribéreau-Gayon, P.; Sudraud, P. Sciences et Techniques du Vin. In Clarification et Stabilisation. Matériels et Installation; Dunod: Paris, France, 1977; Volume 4. [Google Scholar]
- Strauss, M.L.A.; Jolly, N.P.; Lambrechts, M.G.; Van Rensburg, P. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts. J. Appl. Microbiol. 2001, 91, 182–190. [Google Scholar] [CrossRef]
- Fernandez-Salomäo, T.; Amorim, A.C.; Chaves-Alves, V.; Coelho, J.; Silva, D.; Araujo, E. Isolation of pectinase hyperproducing mutants of Penicillium expansum. Rev. Microbiol. 1996, 27, 15–18. [Google Scholar]
- Manns, J.M.; Mosser, D.M.; Buckley, H.R. Production of a hemolytic factor by Candida albicans. Infect. Immun. 1994, 62, 5154–5156. [Google Scholar]
- Menezes, A.G.T.; Ramos, C.L.; Cenzi, G.; Melo, D.S.; Dias, D.R.; Schwan, R.F. Probiotic Potential, Antioxidant Activity, and Phytase Production of Indigenous Yeasts Isolated from Indigenous Fermented Foods. Probiotics Antimicrob. Prot. 2019. [Google Scholar] [CrossRef]
- Iglewsky, B.H. Medical Microbiology, 4th ed.; Chapter 27: Pseudomonas; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Luo, G.; Samaranayake, L.P.; Yau, J.Y. Candida species exhibit differential in vitro hemolytic activities. J. Clin. Microbiol. 2001, 39, 2971–2974. [Google Scholar] [CrossRef]
- Massera, A.; Assof, M.; Sturm, M.E.; Sari, S.; Jofré, V.; Cordero-Otero, R.; Combina, M. Selection of indigenous Saccharomyces cerevisiae strains to ferment red musts at low temperature. Ann. Microbiol. 2012, 62, 367–380. [Google Scholar] [CrossRef]
- Liu, G.L.; Chi, Z.; Wang, G.Y.; Wang, Z.P.; Li, Y.; Chi, Z.M. Yeast killer toxins, molecular mechanisms of their action and their applications. Crit. Rev. Biotechnol. 2013, 35, 222–234. [Google Scholar] [CrossRef]
- De Ingeniis, J.; Raffaelli, N.; Ciani, M.; Mannazzu, I. Pichia anomala DBVPG 3003 secretes a ubiquitin-like protein that has antimicrobial activity. Appl. Environ. Microbiol. 2009, 75, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Starmer, W.T.; Lachance, M.A. Yeast Ecology. In The Yeasts: A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier Science: London, UK; Burlington, MA, USA, 2011; pp. 65–83. [Google Scholar]
- De Vries, S.; von Dahlen, J.K.; Schnake, A.; Ginschel, S.; Schulz, B.; Rose, L.E. Broad-spectrum inhibition of Phytophthora infestans by fungal endophytes. FEMS Microbiol. Ecol. 2018, 94, fiy037. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C.; Brown, A.J.P.; Gow, N.A.R. Antifungal agents: Mechanisms of action. Trends Microbiol. 2003, 11, 272–279. [Google Scholar] [CrossRef]
- Ginovart, M.; Prats, C.; Portell, X.; Silbert, M. Exploring the lag phase and growth initiation of a yeast culture by means of an individual-based model. Food Microbial. 2011, 28, 810–817. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Combina, M.; Elía, A.; Mercado, L.; Catania, C.; Ganga, A.; Martinez, C. Dynamics of indigenous yeast populations during spontaneous fermentation of wines from Mendoza, Argentina. Int. J. Food Microbiol. 2005, 99, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Rivero, D.; Berná, L.; Stefanini, I.; Baruffini, E.; Bergerat, A.; Csikász-Nagy, A.; De Filippo, C.; Cavalieri, D. Hsp12p and PAU genes are involved in ecological interactions between natural yeast strains. Environ. Microbiol. 2015, 17, 3069–3081. [Google Scholar] [CrossRef] [PubMed]
- Nally, M.C. Función del carácter killer en la competencia por el dominio del substrato: Caracterización de mutantes de cepas de levadura Saccharomyces cerevisiae. In Tesis de Licenciatura en Biología; Universidad Nacional de San Juan: San Juan, Argentina, 2005. [Google Scholar]
- Auling, G.; Bellgardt, K.H.; Diekmann, M.; Thoma, M. Dynamics of the growth in batch and continuous culture of Saccharomyces cerevisiae during a shift from aerobiosis to anaerobiosis and reverse. Appl. Microbiol. Biotechnol. 1984, 19, 353–357. [Google Scholar] [CrossRef]
- Verduyn, C.; Postma, E.; Scheffers, W.A.; van Dijken, J.P. Physiology of Saccharomyces cerevisiae in anaerobic glucoselimited chemostat cultures. J. Gen. Microbiol. 1990, 136, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Jain, V.K. Relationship between energy metabolism and growth. I. Glucose dependence of the exponential growth rate of Saccharomyces cerevisiae. Arch. Microbiol. 1970, 72, 252–259. [Google Scholar]
- Andorrà, I.; Martín, L.; Nart, E.; Puxeu, M.; Hidalgo, C.; Ferrer-Gallego, R. Effect of grape juice composition and nutrient supplementation on the production of sulfur dioxide and carboxylic compounds by Saccharomyces cerevisiae. Aust. J. Grape Wine Res. 2018, 24, 260–266. [Google Scholar] [CrossRef]
- Milheiro, J.; Filipe-Ribeiro, L.; Vilela, A.; Cosme, F.; Nunes, F.M. 4-Ethylphenol, 4-ethylguaiacol and 4-ethylcatechol in red wines: Microbial formation, prevention, remediation and overview of analytical approaches. Crit. Rev. Food Sci. Nutr. 2017, 59, 1367–1391. [Google Scholar] [CrossRef] [PubMed]
- Jolly, N.P.; Augustyn, O.P.H.; Pretorius, I.S. The role and use of non-Saccharomyces yeasts in wine production. S. Afr. J. Enol. Vitic. 2006, 27, 15–29. [Google Scholar] [CrossRef]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Wang, H.; Hu, Z.; Long, F.; Guo, C.; Niu, C.; Yuan, Y.; Yue, T. Combined effect of sugar content and pH on the growth of a wild strain of Zygosaccharomyces rouxii and time for spoilage in concentrated apple juice. Food Control. 2016, 59, 298–305. [Google Scholar] [CrossRef]
- Molina, A.M.; Swiegers, J.H.; Varela, C.; Pretorius, I.S.; Agosin, E. Influence of wine fermentation temperature on the synthesis of yeast-derived volatile aroma compounds. Appl. Microbiol. Biotechnol. 2007, 77, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Charoenchai, C.; Fleet, G.H.; Henschke, P.A.; Todd, B.E.N. Screening of non-Saccharomyces wine yeasts for the presence of extracellular hydrolytic enzymes. Aust. J. Grape Wine Res. 1997, 3, 2–8. [Google Scholar] [CrossRef]
- Bautista-Ortín, A.B.; Fernández-Fernández, J.I.; López-Roca, J.M.; Gómez-Plaza, E. The effects of enological practices in anthocyanins, phenolic compounds and wine colour and their dependence on grape characteristics. J. Food Compos. Anal. 2007, 20, 546–552. [Google Scholar] [CrossRef]
- Blanco, P.; Sieiro, C.; Villa, T.G. Production of pectic enzymes in yeasts. FEMS Microbiol. Lett. 1999, 175, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Apel, A.R.; Ouellet, M.; Szmidt-Middleton, H.; Keasling, J.D.; Mukhopadhyay, A. Evolved hexose transporter enhances xylose uptake and glucose/xylose co-utilization in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, 19512. [Google Scholar] [CrossRef]
- Masoud, W.; Jespersen, L. Pectin degrading enzymes in yeasts involved in fermentation of Coffea arabica in East Africa. Int. J. Food Microbiol. 2006, 110, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Martos, M.A.; Butiuk, A.P.; Rojas, N.L.; Hours, R.A. Cultivo por lote de Wickerhamomyces anomalus en un biorreactor a escala laboratorio para la producción de una poligalacturonasa. Rev. Colomb. Biotecnol. 2014, 16, 68–73. [Google Scholar] [CrossRef]
- Schmidt, S.; Zimmermann, S.Y.; Tramsen, L.; Koehl, U.; Lehrnbecher, T. Natural killer cells and antifungal host response. Clin. Vaccine Immunol. 2013, 20, 452–458. [Google Scholar] [CrossRef] [PubMed][Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).