Abstract
Hanseniaspora uvarum is one of the predominant non-Saccharomyces yeast species found on grapes and in juice, but its effect on lactic acid bacteria (LAB) growth and wine flavor has not been extensively studied. Therefore, the interaction between H. uvarum, two Saccharomyces cerevisiae yeast strains, two LAB species (Lactobacillus plantarum and Oenococcus oeni) in combination with two malolactic fermentation (MLF) strategies was investigated in Shiraz wine production trials. The evolution of the different microorganisms was monitored, non-volatile and volatile compounds were measured, and the wines were subjected to sensory evaluation. Wines produced with H. uvarum in combination with S. cerevisiae completed MLF in a shorter period than wines produced with only S. cerevisiae. Sequential MLF wines scored higher for fresh vegetative and spicy aroma than wines where MLF was induced as a simultaneous inoculation. Wines produced with H. uvarum had more body than wines produced with only S. cerevisiae. The induction of MLF using L. plantarum also resulted in wines with higher scores for body. H. uvarum can be used to reduce the duration of MLF, enhance fresh vegetative aroma and improve the body of a wine.
1. Introduction
The contribution of yeasts to wine composition and quality is well-known [1,2]. The Saccharomyces yeasts drive alcoholic fermentation by converting the grape sugar to alcohol, carbon dioxide, and other compounds affecting the wine aroma and taste [3,4]. The other group of yeasts important to winemaking are the non-Saccharomyces yeasts, also known as “wild yeast”, which have different oenological characteristics to S. cerevisiae and can be used to improve wine quality through enhanced wine aroma and complexity [2,5]. Non-Saccharomyces yeast species such as Hanseniaspora uvarum (Kloeckera apiculata), frequently found on grapes and in grape must, are known to dominate the initial phases of spontaneous fermentations [6,7,8]. Certain H. uvarum strains can produce high levels of acetic acid and ethyl acetate, although there is great variability among strains [9,10,11]. It has also been reported that H. uvarum can produce increased levels of desirable compounds such as esters, higher alcohols, and carbonyl compounds [2,11,12]. Mendoza et al. [13] and Tristezza et al. [11] showed that mixed culture fermentations of H. uvarum and S. cerevisiae could be used to enhance wine aroma and quality.
Another process that plays an important role with regard to wine flavor and quality is malolactic fermentation (MLF), which decreases acidity by converting L-malic acid to L-lactic acid and CO2. Malolactic fermentation can affect wine flavor by modifying the concentrations of aroma impact compounds such as diacetyl, esters, higher alcohols, and volatile acids [14,15]. Previously, Oenococcus oeni has been the lactic acid bacteria (LAB) of choice as a MLF starter [16], but recently more Lactobacillus plantarum starters have become available. L. plantarum produces a broader range of extracellular enzymes than O. oeni, which enhances flavor development [17,18,19]. Different MLF inoculation strategies, i.e., simultaneous inoculation (at the start of alcoholic fermentation) and sequential inoculation (after alcoholic fermentation), have been shown to affect the flavor profiles of wines [20,21,22].
A better understanding of how wine production methods can be manipulated to enhance wine attributes, such as aroma, flavor, body, or mouth-feel, is important for the production of a targeted wine style [23]. Du Plessis et al. [24] reported that the MLF strategy had a greater impact on the chemical and sensory profiles of Shiraz wines than yeast combinations. Only one S. cerevisiae strain and one LAB species were used in that study, therefore we wanted to investigate whether the same trend would be observed if different S. cerevisiae strains and LAB species were used. The H. uvarum strain in that study was shown to be compatible with MLF, had potential to enhance wine flavor and is one of the non-Saccharomyces yeast species frequently found on grapes and in must. The aims were to investigate the interactions between H. uvarum, two commercial S. cerevisiae strains, two LAB species (L. plantarum and O. oeni) and three MLF strategies, as well as to determine how these interactions affect shiraz wine composition and flavor.
2. Materials and Methods
2.1. Cultivation and Enumeration of Micro-Organisms
The selected yeast and LAB strains used are listed in Table 1. Similar culturing conditions and procedures were followed as described in Du Plessis et al. [24]. Eight hundred milliliters of the H. uvarum yeast culture was inoculated, at a concentration of ~1 × 106 cells/mL, into the Shiraz juice and skin mixture (30 kg). Commercial S. cerevisiae yeast and LAB cultures (O. oeni and L. plantarum) were inoculated according to the manufacturers’ recommendations.

Table 1.
Yeasts and lactic acid bacteria used in this study.
Non-Saccharomyces and total yeast counts for the shiraz juice and wine were obtained by plating out on Wallerstein Laboratory(WL) medium and Lysine medium, respectively (Biolab, Merck, South Africa). Bacterial counts were obtained by plating out on de Man, Rogosa and Sharpe (MRS) agar (Biolab, Merck) supplemented with 25% (v/v) grape juice and 100 mg/L Natamycin (Danisco A/S, Denmark). Yeasts were grown aerobically for 2–3 days at 28 °C, while bacteria were cultivated under facultative anaerobic conditions at the same temperature for 2–7 days. The respective control wines produced with Sc1 and Sc2, which only received a S. cerevisiae inoculum, were used to determine the levels of naturally occurring non-Saccharomyces yeasts during fermentation [24]. The naturally occurring Saccharomyces yeasts were only determined on day 0 and 1, and counts were obtained from the treatments without a S. cerevisiae inoculum, i.e., H. uvarum treatments. The development of the naturally occurring LAB during fermentation was monitored by sampling the treatments that were not inoculated with LAB and the sequential MLF treatments until day 5, because the commercial LAB cultures were added to induce sequential MLF after that.
2.2. Wine Production
Shiraz grapes were obtained from the Nietvoorbij research farm (Stellenbosch, South Africa). A standardized small-scale (20 L) winemaking procedure was followed as described by Du Plessis et al. [24]. The treatments that were applied are listed in Table 2. Four different yeast treatments (Sc1, Sc2, Hu + Sc1 and Hu + Sc2) were investigated. Each yeast treatment was used in combination with LAB1 and LAB2 (O. oeni and L. plantarum), respectively. Additionally, the two MLF strategies were applied: (1) Simultaneous inoculation of LAB (hereafter referred to as simultaneous MLF) and (2) sequential inoculation of LAB (hereafter referred to as sequential MLF). Wines that did not undergo MLF will be referred to as non-MLF wines, while wines that underwent MLF will be referred to as MLF wines. Sixty wines were produced, which included 20 different treatments with three replicates each.

Table 2.
Oenological parameters and duration of malolactic fermentation (MLF) of shiraz juice 1 and wines produced with Saccharomyces cerevisiae (Sc1 or Sc2) only or in combination with Hanseniaspora uvarum (Hu), two lactic acid bacteria strains (LAB1 or LAB2) and two MLF strategies (simultaneous or sequential inoculation). Values are means of three replicate fermentations, and the standard deviations are also shown.
The S. cerevisiae strains, Sc1 and Sc2, were inoculated on day 0 for the control treatments. H. uvarum was inoculated on day 0, and Sc1 and Sc2, respectively, were inoculated after 24 h (day 1) for all the mixed yeast fermentations. The LAB in the simultaneous MLF treatments was added 25 hours after the initial yeast inoculations on day 0. Fermentations were carried out at ca. 24 °C and after completion of the alcoholic fermentation, the sequential MLF treatments were inoculated with LAB1 or LAB2. All treatments were racked, fined, cold stabilized, and bottled as described by Minnaar et al. [25]. After bottling, all wines were stored at 15 °C until required for evaluation.
2.3. Juice and Wine Analysis
The following parameters of the grape must were measured, i.e., sugar (°Balling), free and total SO2 (Ripper method), pH and titratable acidity (Mettler titrator) analyses as described in the South African Wine Laboratories Association (SAWLA) Manual [26]. Alcoholic fermentation was monitored using an OenoFoss™ Fourier transform infrared (FTIR) spectrometer (FOSS Analytical A/S, Denmark) and was considered to be complete when the residual sugar concentrations were below 4 g/L. The progression of MLF was monitored with the OenoFoss™ FTIR spectrometer until the malic acid levels were below 0.2 g/L, the point where MLF was considered to be complete. Oenological parameters of the wines, such as residual sugar (glucose + fructose), pH, malic and lactic acids, total acidity (TA), alcohol, volatile acidity (VA), and glycerol, were determined with a WineScanTM FT120 instrument (FOSS Analytical A/S) as described by Du Plessis et al. [24] and Louw et al. [27]. The method described by Louw et al. [27] using gas chromatography with a flame ionization detector (GC-FID) was applied to analyze the volatile compounds in the wines.
2.4. Sensory Evaluation of Wines
A panel consisting of 22 experienced wine judges (13 men and nine women, aged 22 to 50 years) evaluated the wines four months after bottling. The same panelists and procedures were used as described by Du Plessis et al. [24]. The panelists were asked to rate the intensity of the aroma and taste descriptors on a 100 mm unstructured line scale. The intensity of aroma descriptors: Berry, fruity, fresh vegetative, cooked vegetative, floral, sweet associated, and spicy were rated from undetectable to prominent, while the taste descriptors were rated from low to high for acid balance, thin to full for body (mouth-feel) and undetectable to prominent for astringency and bitterness. The descriptors were scored by measuring where the mark was made on the line and expressing the value as a percentage. Each judge had a separate tasting booth and ca. 30 mL of the wine sample were presented in a randomized order in a standard wine glass, labeled with a three digit code. Research Randomizer (Version 4.0, http://randomizer.org) was used to generate the three digit code and to randomize the order in which the wines were presented to each panelist.
2.5. Data and Statistical Analysis
Chemical and sensory data were tested for deviation from normality by the Shapiro–Wilk test and then subjected to analysis of variance (ANOVA) using the general linear means procedure of SAS version 9.4 (SAS Institute Inc., Cary, North Carolina, USA). Fisher’s least significant difference (LSD) values were calculated at the 5% probability level (p = 0.05) to facilitate comparison between treatment means. Principal component analysis (PCA) was performed using XLSTAT software (Version 18.07.39157, Addinsoft, New York, USA) to examine the correlation between treatments and the volatile chemical variables.
2.6. Verification of H. uvarum Implantations
Yeasts were isolated from juice and wine (day 2) samples to verify successful implantation. From WL plates with a colony count of 30 to 300, five colonies were selected randomly per replicate. Subsequently, yeast DNA was extracted using the method described by Lõoke et al. [28]. Yeast identification to the species level was carried out by amplification of the 5.8S-internal transcribed spacer (ITS) ribosomal region, using primers, ITS1 and ITS4, followed by enzyme restriction with CfoI, as described by Esteve-Zarzoso et al. [29]. Restriction profiles of the isolates were compared to those of known yeast species. Successful implantation of the H. uvarum strain was verified with random amplified polymorphic DNA (RAPD), using primer 1283 and conditions described by Pfliegler et al. [30]. Amplification products (ITS-RFLP and RAPD) were separated on 2% agarose gels, and banding patterns were visualized on a Bio-Rad image analyzer, following staining with 0.01% (v/v) ethidium bromide (Bio-Rad Laboratories, Inc., USA).
3. Results and Discussion
3.1. Yeast Development
The naturally occurring Saccharomyces and non-Saccharomyces yeast populations in the shiraz juice were ca. 4.2 × 105 and 4.1 × 105 colony forming units/mL (CFU/mL), respectively (Figure 1). The naturally occurring non-Saccharomyces yeast populations decreased notably on day 1, in treatments inoculated with the commercial S. cerevisiae yeasts, before increasing again on day 2. Thereafter the naturally occurring non-Saccharomyces yeast populations remained at levels of ca. 1 × 105 CFU/mL in wines fermented with Sc2 or decreased to ca. 1 × 104 CFU/mL in wines inoculated with Sc1. S. cerevisiae strain Sc1 had a negative effect on the growth of naturally occurring non-Saccharomyces yeasts because after five days of the Sc1 treatment, the non-Saccharomyces yeast levels were lower than for wines fermented with Sc2.
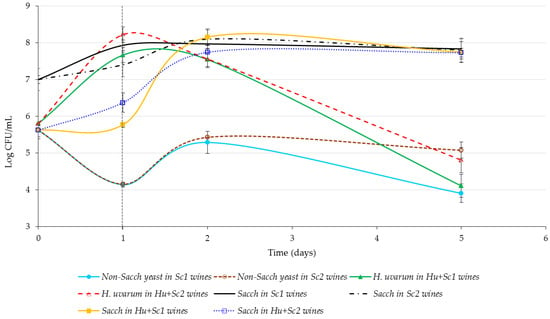
Figure 1.
Cell counts (colony forming units/milliliters, CFU/mL) of naturally occurring and inoculated Saccharomyces cerevisiae (Sacch), naturally occurring non-Saccharomyces (Non-Sacch), and inoculated Hanseniaspora uvarum (H. uvarum) yeasts during alcoholic fermentation. The dashed vertical line at day 1 indicates when commercial S. cerevisiae yeasts were added. Abbreviations: Sc1 = commercial S. cerevisiae strain 1, Sc2 = commercial S. cerevisiae strain 2, Hu = inoculated H. uvarum yeasts. Values are means of three replicates and error bars indicate standard deviation.
Initial yeast counts of the wines inoculated with H. uvarum were just below 1 × 106 CFU/mL, but increased to levels >10 million CFU/mL after 24 h. However, this trend changed after inoculation of commercial S. cerevisiae yeasts (day 1, Figure 1), which resulted in the decrease of H. uvarum numbers. The same trend was found with regard to the inhibitory activity of Sc1 on non-Saccharomyces yeast viability. At the end of alcoholic fermentation, inoculated and naturally occurring non-Saccharomyces yeast populations were at a similar level.
The naturally occurring Saccharomyces yeast populations were present at moderately high numbers (4 × 105 CFU/mL), which increased after 24 h, but the inoculated H. uvarum yeasts were present at higher numbers (8 × 107 to 1 × 108 CFU/mL). However, both aforementioned populations were dominated by the inoculated S. cerevisiae yeasts, following their addition after 24 h. These results indicate that the inoculated S. cerevisiae strains were responsible for completing the alcoholic fermentations. However, the inoculated H. uvarum populations were present at high levels (107 to 108 CFU/mL) and long enough to potentially make a contribution to wine flavor. A similar trend was observed by Du Plessis et al. [24].
3.2. Yeast Verification
A selection of yeast colonies 2 was identified by amplification of the ITS-5.8S region, followed by subsequent restriction analysis. Isolate profiles were compared to profiles of known yeast species. The dominant non-Saccharomyces yeasts isolated from the Hu + Sc1 and Hu + Sc2 wines were identified as H. uvarum. DNA of these isolates were subsequently amplified using primer 1283 and the products were compared to the reference H. uvarum strain (Table 1). All wine isolates had similar banding patterns as the H. uvarum reference strain (Figure 2), indicating successful implantation. The banding patterns of H. uvarum juice isolates (naturally occurring strains) differed from the H. uvarum reference strain and were not detected in any of the implanted wines during the first two days of alcoholic fermentation. These results indicate that the inoculated H. uvarum dominated the naturally occurring H. uvarum population.
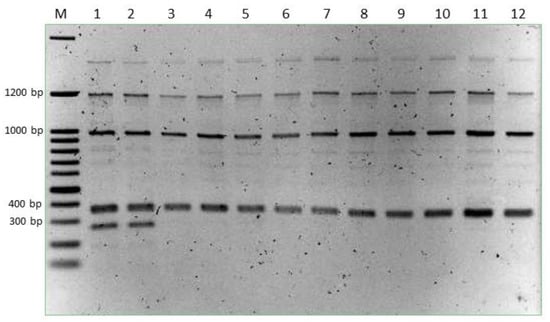
Figure 2.
Random amplified polymorphic DNA products of selected Hanseniaspora uvarum isolates from shiraz wines produced with Saccharomyces cerevisiae Sc1 or Sc2 in combination with H. uvarum. M: 100 bp DNA ladder, lane1: H. uvarum strain isolated from shiraz juice, lane 2: H. uvarum strain isolated from shiraz juice, lane 3: H. uvarum reference used for implantations, lane 4 to 12: dominant non-Saccharomyces yeasts isolated from wines inoculated with H. uvarum and S. cerevisiae.
3.3. LAB Development and Progression of MLF
The growth and development of the naturally occurring and inoculated LAB are shown in Figure 3. The naturally occurring LAB were present at ~3.5 × 104 CFU/mL in the Shiraz grape must and decreased during alcoholic fermentation in most of the treatments, with the increase in numbers at the end (day 5). This is also the typical winemaking scenario [4,31]. Individually, the numbers of naturally occurring LAB varied notably in wines, fermented with the selected yeast combinations. Based on the LAB counts from day 2 to 5, Sc1 had a greater inhibitory effect on LAB growth (decreased from 3.5 × 104 to 8.8 × 102 CFU/mL) than Sc2 or H. uvarum in combination with Sc1 or Sc2 (decreased from 3.5 × 104 to 1.8 × 103 CFU/mL). This is in agreement with findings of Du Plessis et al. [24].
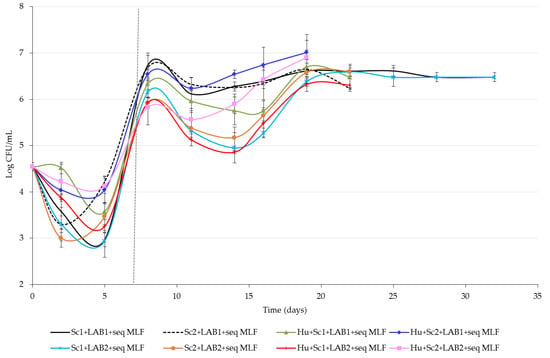
Figure 3.
Cell counts (colony forming units per milliliters, CFU/mL) of the naturally occurring and sequentially inoculated lactic acid bacteria (LAB) in shiraz juice and wine produced with Saccharomyces cerevisiae (Sc1 or Sc2) on its own or in combination with Hanseniaspora uvarum (Hu) and two LAB species (LAB1 or LAB2). The dashed vertical line at day 7 indicates inoculation of the commercial LAB for sequential malolactic fermentation (seq MLF). Values are means of three replicate fermentations and error bars indicate standard deviation.
The alcoholic fermentation was completed after five days and the commercial LAB were inoculated on day 7 to induce sequential MLF in the selected treatments. The addition of commercial LAB resulted in an expected increase of LAB numbers from ~1 × 103–104 to >7 × 105 CFU/mL (Figure 3). No notable delays in MLF was observed in sequentially inoculated wines (Table 2), despite inoculated LAB1 (O. oeni) and LAB2 (L. plantarum) counts decreasing from 5.0 × 106 to 4.5 × 105 CFU/mL, and 6.8 to 1.9 × 105 CFU/mL, respectively (Figure 3). Wines produced with Hu + Sc1 + LAB1 and Hu + Sc2 + LAB2 completed MLF in the shortest time (18 days), while wines produced with Sc1 + LAB1 and Sc1 + LAB2 took the longest to complete MLF (34 days). The delay in MLF of the Sc1+LAB2 wines can be correlated to lower LAB numbers (<1 × 106 CFU/mL), but the trend was not observed for Sc1 + LAB1 wines, which contained high LAB numbers (>1 × 106 CFU/mL) throughout MLF (Figure 3).
The development of LAB that were inoculated at the same time as the yeasts are shown in Figure 4. LAB1 (O. oeni) numbers remained above 1 × 106 CFU/mL and completed MLF within 10 days (Table 2), while LAB2 (L. plantarum) numbers decreased to below 1 × 106 CFU/mL, before increasing again, which resulted in the MLF taking longer to complete. For the wines inoculated with LAB2, the Hu + Sc2 treatment completed MLF within 15 days, while the Sc1 + LAB2 treatment took 34 days to complete MLF. There was a negative interaction between Sc1 and LAB2. The inhibition of LAB2 growth might be due to the depletion of essential nutrients needed for LAB growth or the production of toxic metabolites.
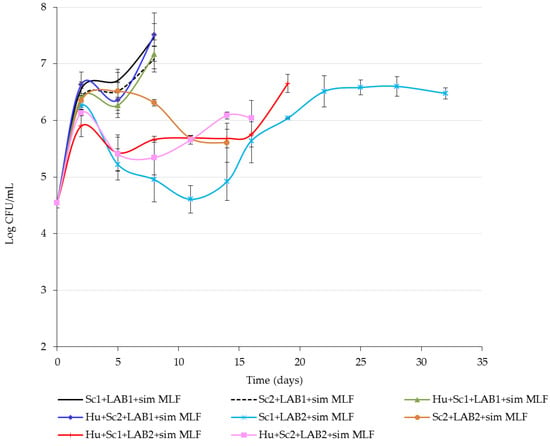
Figure 4.
Cell counts (colony forming units per milliliters, CFU/mL) of the naturally occurring and inoculated lactic acid bacteria (LAB) in shiraz juice and wine produced with Saccharomyces cerevisiae (Sc1 or Sc2) on its own or in combination with Hanseniaspora uvarum and two LAB species (LAB1 or LAB2). Malolactic fermentation induced as a simultaneous inoculation (sim MLF). Values are means of three replicate fermentations and error bars indicate standard deviation.
In general, O. oeni is known to be better suited to harsh conditions found in wine than L. plantarum, which explains why LAB1 performed better than LAB2. Overall, simultaneous MLF completed in a shorter time than sequential MLF. This trend is in agreement with findings of other researchers [20,32].
3.4. Standard Oenological Parameters
The interaction between yeast combinations, LAB strains and MLF strategies had a significant effect (p ≤ 0.05) on pH, VA, malic acid, and lactic acid concentrations of wines (Table S1). In addition, yeast combination also had a significant effect on alcohol and glycerol concentrations of wines, while the interaction between LAB strain and MLF strategy had a significant impact on TA, and glycerol concentrations.
3.4.1. Wines without MLF
All wines were fermented to dryness and contained residual sugar levels of less than 4 g/L (Table 2). Alcohol concentrations of wines produced with H. uvarum in combination with Sc1 and Sc2 were slightly lower than those produced with only Sc1 or Sc2. This trend is in agreement with findings of Mendoza et al. [13,33]. Wines produced with only S. cerevisiae yeasts contained significantly higher glycerol concentrations than wines produced with the H. uvarum and S. cerevisiae combinations. Mendoza et al. [13] reported similar findings, but Liu et al. [34] reported the contrary, which indicates that this is not a species trait, but rather strain dependent.
None of the treatments produced excessively high concentrations of VA (>0.7 g/L). However, VA concentrations in wines produced with H. uvarum in combination with Sc1 (0.29 g/L) and Sc2 (0.43 g/L) were slightly higher than wines produced with Sc1 (0.24 g/L) and Sc2 (0.35 g/L) on their own. This is in agreement with the findings of Mendoza et al. [13] and also confirmed reports that some H. uvarum (K. apiculata) strains can produce lower VA levels comparable to those of S. cerevisiae [11,35,36]. Malic acid concentrations in wines produced with H. uvarum in combination with Sc1 (1.82 g/L) and Sc2 (1.69 g/L) were significantly lower than wines produced with Sc1 (2.81 g/L) and Sc2 (2.11 g/L) on their own. The ability of this H. uvarum strain to degrade malic acid has been reported by Du Plessis et al. [24,37].
3.4.2. Wines That Underwent MLF
In most cases, non-MLF wines contained lower alcohol levels than MLF wines (Table 2). These findings are contrary to those of Mendoza et al. [13] and Abrahamse and Bartowsky [20], but in agreement with results of Izquierdo-Cañas et al. [37] and Du Plessis et al. [24]. The differences reported might be due to the LAB strain used or LAB and yeast interactions. In general, the alcohol levels were lower for simultaneous MLF wines than for sequential MLF wines, which are in agreement with the findings of Mendoza et al. [13] and Abrahamse and Bartowsky [20], but contrary to the findings of Izquierdo-Cañas et al. [32] and Tristezza et al. [22]. MLF wines had significantly higher glycerol levels than non-MLF wines. In most cases, simultaneous MLF wines contained slightly lower glycerol levels than sequential MLF wines.
Overall, MLF wines contained significantly higher VA values (0.38 to 0.58 g/L) than non-MLF wines (0.24 to 0.43 g/L). Similar results have been reported by Mendoza et al. [13] and Izquierdo-Cañas et al. [37]. Most of the simultaneous MLF wines had slightly lower VA levels than sequential MLF wines, which is similar to reports of Tristezza et al. [22].
The conversion of malic acid to lactic acid resulted in a significant decrease in the total acidity levels of the MLF wines, with the expected increase in the pH of those wines. In most cases, simultaneous MLF wines had slightly higher total acidity levels than sequential MLF wines, which is similar to the findings of Mendoza et al. [13].
3.5. Multivariate Data Analysis of Wines
Principal component analysis (PCA) was used to investigate the association among yeast combinations, LAB strain, MLF strategy, and volatile composition of shiraz wines (Figure 5). The first two principal components explain 65% of the variance in the data (PC1 = 38.78% and PC2 = 26.27%). Four distinct clusters (indicated by different colors) can be observed, i.e., Hu + Sc1 non-MLF and MLF wines (top right quadrant), Hu + Sc2 non-MLF and MLF wines (top left quadrant), Sc2 non-MLF and MLF wines (bottom left quadrant), and Sc1 non-MLF and MLF wines (bottom right quadrant). Results clearly show that the yeast combinations had a significant effect on volatile chemical composition of the wines (Table S2 and Figure 5). The distribution of the data points within the aforementioned four clusters shows that there was some within-group variation. This within-group variation is due to the LAB strain or MLF strategy that was applied. These results indicate that yeast combination has the greatest impact on the chemical composition, but LAB strain and MLF strategy also have a significant effect (p ≤ 0.05) on the chemical composition of the wines (Table S2).
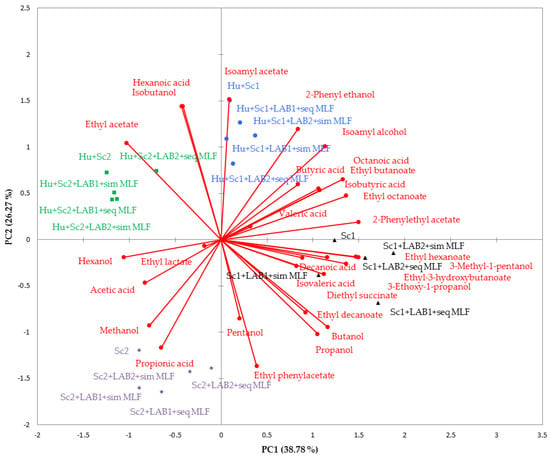
Figure 5.
Principal component biplot of volatile compounds of shiraz wines produced with Saccharomyces cerevisiae (Sc1 and Sc2) in combination with Hanseniaspora uvarum, two lactic bacteria strains (LAB1 and LAB2), and two malolactic fermentation (MLF) strategies (simultaneous or sequential inoculation). Mean values of three replicate fermentations. Abbreviations: LAB1 = Oenococcus oeni, LAB2 = Lactobacillus plantarum, sim = simultaneous MLF, and seq = sequential MLF.
Based on the contribution and squared cosines of the variables, the main compounds responsible for differentiating among wines produced with the selected yeast combinations, LAB strain and MLF strategies were, isoamyl acetate, ethyl hexanoate, ethyl octanoate, ethyl-3-hydroxybutanoate, ethyl phenylacetate, 2-phenyl acetate, isobutanol, 3-methyl-1-pentanol, hexanoic acid, and octanoic acid (Figure 5).
All wines produced with Sc1 show a positive correlation with 2-phenylethyl acetate, 3-methyl-pentanol, ethyl hexanoate, decanoic acid, ethyl-3-hydroxybutanoate, 3-ethoxy-1-propanol, isovaleric acid, diethyl succinate, ethyl decanoate, butanol, and propanol. The aforementioned wines were negatively correlated with ethyl acetate.
The Sc2 wines show a positive correlation with methanol, propionic acid, pentanol, and ethyl phenylacetate, and a negative correlation with isoamyl acetate, 2-phenyl ethanol, isoamyl alcohol, octanoic acid, isobutyric acid, and ethyl butanoate.
All wines produced with Hu + Sc1 show a positive correlation with isoamyl acetate, 2-phenyl ethanol, isoamyl alcohol, and butyric acid, and a negative correlation with propionic acid, methanol, and acetic acid. Octanoic acid, ethyl butanoate, isobutyric acid, valeric acid, and ethyl octanoate show a positive correlation with wines produced with Sc1 only and those produced with H. uvarum in combination with Sc1.
The Hu + Sc2 wines show a positive correlation with ethyl acetate and are negatively correlated with ethyl decanoate, butanol, propanol, diethyl succinate, isovaleric acid, decanoic acid, and ethyl-3-hydroxybutanoate. Isobutanol and hexanoic acid show a positive correlation with wines produced with H. uvarum in combination with Sc1 and Sc2. This indicates that these compounds are linked to the growth and metabolism of the H. uvarum strain.
3.6. Sensory Evaluation
Sensory evaluation results indicate how yeast selection, LAB combination, and MLF strategy can impact the volatile composition and sensory profiles of wines. ANOVA of the sensory data show that the interactions among the selected yeast combinations, LAB strains and even MLF strategies had a significant (p ≤ 0.05) impact on fresh vegetative, cooked vegetative, spicy, and floral aromas (Table 3). Yeast treatment had a significant effect on fresh vegetative and spicy aroma, as well as body and astringency of the wines. Wines produced with the selected LAB strains and MLF strategies were significantly different with regard to berry, fruity, sweet associated, and spicy aroma, as well as acidity and body. Only the sensory attributes that showed significant differences for at least two of the treatment interactions will be discussed in detail (Table 3).

Table 3.
Probability (p) values 1 of shiraz wines produced with different yeast treatments and malolactic fermentation (MLF) strategies. Probability (p) values 1 of the sensory descriptors of shiraz wines produced with Saccharomyces cerevisiae (Sc1 or Sc2) only, or in combination with Hanseniaspora uvarum (Hu), two lactic acid bacteria strains (LAB 1 or LAB2), and two MLF strategies (simultaneous or sequential inoculation).
3.6.1. Fresh Vegetative Aroma
Non-MLF wines produced with S. cerevisiae only (Sc1 and Sc2) scored lower for fresh vegetative aroma than non-MLF wines produced with H. uvarum in combination with the two S. cerevisiae strains (Figure 6). Sequential MLF wines scored higher for fresh vegetative aroma than simultaneous MLF and non-MLF wines. Of all the different treatments, Hu + Sc1 + LAB2 seq MLF wines scored the highest (35.27%) for fresh vegetative aroma (Table S3). The Hu + Sc1 combination consistently produced wines with high fresh vegetative aroma scores and this was observed for non-MLF and MLF. The opposite trend was found for wines produced with Sc2. These results indicate that this Hu + Sc1 combination can be used to enhance the fresh vegetative character in wines where this attribute is lacking or to produce a wine style with a predominant fresh vegetative flavor profile. On the other hand, if a wine with low fresh vegetative character is preferred, the use of a yeast strain, such as Sc2 is recommended.
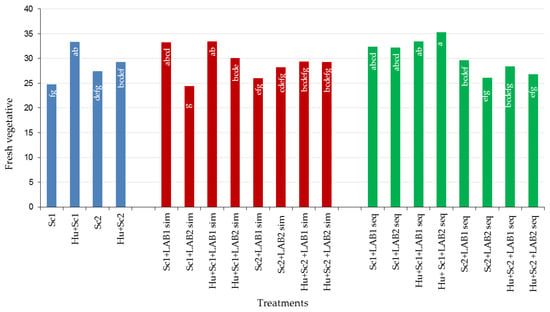
Figure 6.
Percentage (%) of fresh vegetative aroma of shiraz wines produced with Saccharomyces cerevisiae (Sc1 and Sc2) in combination with Hanseniaspora uvarum, two lactic bacteria strains (LAB1 and LAB2), and two malolactic fermentation (MLF) strategies (simultaneous or sequential inoculation). Abbreviations: LAB1 = Oenococcus oeni, LAB2 = Lactobacillus plantarum, sim = simultaneous MLF, and seq = sequential MLF. The letters inside the bars refer to differences among the treatments and treatments that have the same letter/s do not differ significantly (p ≤ 0.05).
Differences in fresh vegetative aroma scores were observed for wines produced with the two LAB strains, and were also affected by MLF strategy applied. In most cases, wines inoculated with LAB1 scored higher for vegetative aroma than wines inoculated with LAB2. Therefore to increase fresh vegetative flavor in Shiraz wines O. oeni should be used to induce MLF, but to reduce the fresh vegetative flavor, L. plantarum is recommended.
3.6.2. Spicy
Non-MLF wines produced with Sc2 scored the highest for spicy aroma (32.71%; Figure 7 and Table S3). Overall, sequential MLF wines scored higher for spicy aroma than simultaneous MLF and non-MLF wines (Figure 7). Of all the various treatments, sequential MLF wines produced with Hu + Sc1 + LAB2 scored the highest for spicy aroma. Differences in spicy aroma scores were found for wines produced with the two LAB strains, and were affected by yeast combination as well as the MLF strategy. Therefore to increase spicy flavor in wine MLF should be induced as a sequential inoculation.
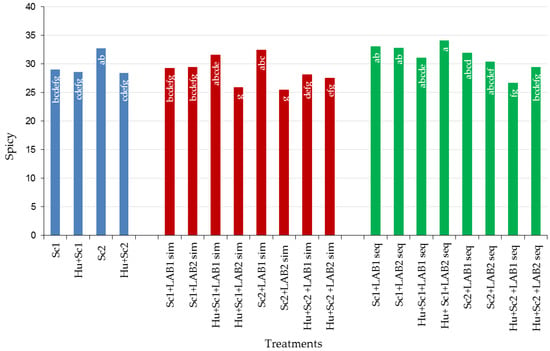
Figure 7.
Percentage (%) of spicy aroma of shiraz wines produced with Saccharomyces cerevisiae (Sc1 and Sc2) in combination with Hanseniaspora uvarum, two lactic bacteria strains (LAB1 and LAB2), and two malolactic fermentation (MLF) strategies (simultaneous or sequential inoculation). Abbreviations: LAB1 = Oenococcus oeni, LAB2 = Lactobacillus plantarum, sim = simultaneous MLF, and seq = sequential MLF. The letters inside the bars refer to differences among the treatments and treatments that have the same letter/s do not differ significantly (p ≤ 0.05).
3.6.3. Body
The non-MLF wines produced with Sc1 and Sc2 scored lower for the taste descriptor, body (mouth-feel) than those where H. uvarum was used (Figure 8). MLF wines scored higher for body than non-MLF wines. MLF wines produced with Sc1 scored slightly higher for body than those inoculated with Sc2. MLF wines produced with LAB2 scored higher for body than those inoculated LAB1. It is noteworthy that the relative scores for body varied according to the yeast combination used. Winemakers can manipulate the body (mouth-feel) of wines by applying the aforementioned combinations to achieve the wine style they prefer. To increase the body of a wine, H. uvarum in combination with S. cerevisiae should be used and MLF should be induced using LAB2 (L. plantarum).
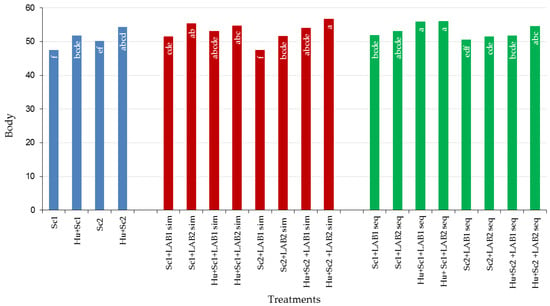
Figure 8.
Percentage (%) of body of shiraz wines produced with Saccharomyces cerevisiae (Sc1 and Sc2) in combination with Hanseniaspora uvarum, two lactic bacteria strains (LAB1 and LAB2), and two malolactic fermentation (MLF) strategies (simultaneous or sequential inoculation). Abbreviations: LAB1 = Oenococcus oeni, LAB2 = Lactobacillus plantarum, sim = simultaneous MLF, and seq = sequential MLF. The letters inside the bars refer to differences among the treatments and treatments that have the same letter/s do not differ significantly (p ≤ 0.05).
3.7. Overall Effects
Chemical and sensory results support our opinion that the selected H. uvarum strain contributed positively to wine flavor. None of the treatment combinations produced off-flavors. Wines produced with H. uvarum in combination with Sc1 and Sc2 were different to wines produced with the Sc1 or Sc2 on their own. These results show that H. uvarum can be used to reduce the duration of MLF and to change the style or flavor profile of a wine. Wines where yeast and LAB were added as a simultaneous inoculation, reduced MLF duration and the flavor profiles differed from those that were sequentially inoculated. Notable differences were also observed between wines inoculated with LAB1 and LAB2 with regard to their flavor profiles, which supports the concept of L. plantarum playing a greater role in the future of MLF as envisaged by Du Toit et al. [38]. The yeast treatments, LAB strains and MLF strategies had a significant effect on the standard chemical parameters and volatile composition of the wines, and these differences in chemical composition translated to perceivable sensory differences.
4. Conclusions
H. uvarum had a positive effect on the growth of inoculated and naturally occurring LAB, which resulted in shorter MLF periods for wines. Allowing the naturally occurring yeast population to develop for at least 24 hours may be beneficial to winemakers that want MLF to proceed quickly and successfully. Wines produced with the selected yeasts, LAB strains and MLF strategies differed with regard to fermentation kinetics, chemical composition, and sensory properties. Yeast treatment had a greater effect on the volatile chemical composition of the wines than LAB strain or MLF strategy, but LAB strain and MLF strategy also had a significant impact. The sensory differences between non-MLF and MLF wines were as significant as wines produced with different yeast strains. H. uvarum in combination with O. oeni as a sequential inoculation can be used to increase vegetative aroma of shiraz wines. The spicy flavor can be increased by inducing MLF as a sequential inoculation and increased body can be achieved by using H. uvarum in combination with S. cerevisiae to conduct the alcoholic fermentation and L. plantarum to induce MLF. The flavor profile of shiraz wines can be enhanced by using different yeasts, LAB strains, MLF strategies, or a combination of the aforementioned options.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2311-5637/5/3/64/s1.
Author Contributions
H.d.P., M.d.T. and N.J. conceived and designed the experiments; H.d.P. and J.H. performed the experiments; H.N. assisted with FTIR and GC analyses and interpretation of data; M.v.d.R. contributed to the statistical analysis and interpretation of the data; H.d.P., M.d.T., H.N. and N.J. contributed to writing the paper.
Acknowledgments
The authors thank the ARC, Winetech and the National Research Foundation of South Africa (THRIP programme; grant numbers UID 71526 and 90103) for funding. The opinions, findings and conclusions expressed in this publication are those of the authors. The National Research Foundation accepts no liability in this regard. Mses P. Adonis, C. du Plessis, D. Blaauw, R. Louw and L. Isaacs, as well as Messrs H. Jumat, M. Mewa Ngongang and J. Boonzaier, are thanked for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Jolly, N.P.; Valera, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G.H. Yeast interactions and wine flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donéche, B.; Lonvaud, A. Handbook of Enology, Volume 1: The Microbiology of Wine and Vinifications, 2nd ed.; John Wiley & Sons Ltd.: London, UK, 2006. [Google Scholar]
- Hranilovic, A.; Li, S.; Boss, P.K.; Bindon, K.; Ristic, R.; Grbin, P.R.; Van der Westhuizen, T.; Jiranek, V. Chemical and sensory profiling of Shiraz wines co-fermented with commercial non-Saccharomyces inocula. Aust. J. Grape Wine Res. 2018, 24, 166–180. [Google Scholar] [CrossRef]
- Romano, P.; Capece, A.; Jespersen, L. Taxonomic and ecological diversity of food and beverage yeasts. In Yeasts in Food and Beverages; Querol, A., Fleet, G., Eds.; Springer: Berlin, Germany, 2006; pp. 13–54. [Google Scholar]
- Capozzi, V.; Garofalo, C.; Chiriatti, M.A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2015, 181, 75–83. [Google Scholar] [CrossRef]
- Ye, D.Q.; Sun, Y.; Song, Y.Y.; Ma, X.L.; Ren, X.N.; Gong, X.; Liu, Y.L. Diversity and identification of yeasts isolated from tumultuous stage of spontaneous table grape fermentations in central China. S. Afr. J. Enol. Vitic. 2019, 40, 1–7. [Google Scholar] [CrossRef]
- Romano, P.; Fiore, C.; Paraggio, M.; Caruso, M.; Capece, A. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef]
- De Benedictis, M.; Bleve, G.; Grieco, F.; Tristezza, M.; Tufariello, M. An optimized procedure for the enological selection of non-Saccharomyces starter cultures. Antonie Leeuwenhoek 2011, 99, 189–200. [Google Scholar] [CrossRef]
- Tristezza, M.; Tufariello, M.; Capozzi, V.; Spano, G.; Mita, G.; Grieco, F. The oenological potential of Hanseniaspora uvarum in simultaneous and sequential co-fermentation with Saccharomyces cerevisiae for industrial wine production. Front. Microbiol. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Moreira, N.; Mendes, F.; de Pinho, R.G.; Hogg, T.; Vasconcelos, I. Heavy sulphur compounds, higher alcohols and esters production profile of Hanseniaspora uvarum and Hanseniaspora guilliermondii grown as pure and mixed cultures in grape must. Int. J. Food Microbiol. 2008, 124, 231–238. [Google Scholar] [CrossRef]
- Mendoza, L.M.; Merín, M.G.; Morata, V.I.; Farías, M.E. Characterization of wines produced by mixed culture of autochthonous yeasts and Oenococcus oeni from the northwest region of Argentina. J. Ind. Microbiol. Biotechnol. 2011, 38, 1777–1785. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J.; Costello, P.; Henschke, P.A. Management of malolactic fermentation—Wine flavour manipulation. Aust. N. Z. Grapegrow. Winemak. 2002, 461, 7–8 and 10–12. [Google Scholar]
- Lerm, E.; Engelbrecht, L.; Du Toit, M. Malolactic fermentation: The ABC’s of MLF. S. Afr. J. Enol. Vitic. 2010, 31, 186–212. [Google Scholar] [CrossRef]
- Dimopoulou, M.; Raffenne, J.; Claisse, O.; Miot-Sertier, C.; Iturmendi, N.; Moine, V.; Coulon, J.; Dols-Lafargue, M. Oenococcus oeni exopolysaccharide biosynthesis, a tool to improve malolactic starter performance. Front. Microbiol. 2018, 9, 1276. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.; Bartowsky, E.; Jiranek, V. Screening of Lactobacillus spp. and Pediococcus spp. for glycosidase activities that are important in oenology. J. Appl. Microbiol. 2005, 99, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Mtshali, P.S.; Divol, B.; Van Rensburg, P.; du Toit, M. Genetic screening of wine-related enzymes in Lactobacillus species isolated from South African wines. J. Appl. Microbiol. 2010, 108, 1389–1397. [Google Scholar] [CrossRef]
- Brizuela, N.; Tymczyszyn, E.E.; Semorile, L.C.; La Hens, D.V.; Delfederico, L.; Hollmann, A.; Bravo-Ferrada, B. Lactobacillus plantarum as a malolactic starter culture in winemaking: A new (old) player? Electron. J. Biotechnol. 2019, 38, 10–19. [Google Scholar] [CrossRef]
- Abrahamse, C.; Bartowsky, E. Timing of malolactic fermentation inoculation in Shiraz grape must and wine: influence on chemical composition. World J. Microbiol. Biot. 2012, 28, 255–265. [Google Scholar] [CrossRef]
- Bartowsky, E.J.; Costello, P.J.; Chambers, P.J. Emerging trends in the application of malolactic fermentation. Aust. J. Grape Wine Res. 2015, 21, 663–669. [Google Scholar] [CrossRef]
- Tristezza, M.; di Feo, L.; Tufariello, M.; Grieco, F.; Capozzi, V.; Spano, G.; Mita, G. Simultaneous inoculation of yeasts and lactic acid bacteria: Effects on fermentation dynamics and chemical composition of Negroamaro wine. LWT-Food Sci. Technol. 2016, 66, 406–412. [Google Scholar] [CrossRef]
- Lesschaeve, I. Sensory evaluation of wine and commercial realities: Review of current practices and perspectives. Am. J. Enol. Vitic. 2007, 58, 252–258. [Google Scholar]
- Du Plessis, H.; du Toit, M.; Nieuwoudt, H.; van der Rijst, M.; Kidd, M.; Jolly, N. Effect of Saccharomyces, non-Saccharomyces yeasts and malolactic fermentation strategies on fermentation kinetics and flavor of Shiraz wines. Fermentation 2017, 3, 64. [Google Scholar] [CrossRef]
- Minnaar, P.P.; Ntushelo, N.; Ngqumba, Z.; Van Breda, V.; Jolly, N.P. Effect of Torulaspora delbrueckii yeast on the anthocyanin and flavanol concentrations of Cabernet franc and Pinotage wines. S. Afr. J. Enol. Vitic. 2015, 36, 50–58. [Google Scholar] [CrossRef]
- Anonymous. Methods of analyses for wine laboratories. Compiled by the South African Wine Laboratories Association. South African Society for Viticulture and Oenology, P.O. Box 2092, Dennesig, Stellenbosch 7601, South Africa. 2003. Available online: http://www.sasev.org (accessed on 8 July 2019).
- Louw, L.; Roux, K.; Tredoux, A.; Tomic, O.; Naes, T.; Nieuwoudt, H.H.; Van Rensburg, P. Characterisation of selected South African cultivar wines using FTMIR spectroscopy, gas chromatography and multivariate data analysis. J. Agric. Food. Chem. 2009, 57, 2623–2632. [Google Scholar] [CrossRef] [PubMed]
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques 2011, 50, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 1999, 49, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Pfliegler, W.P.; Horváth, E.; Kállai, Z.; Sipiczki, M. Diversity of Candida zemplinina isolates inferred from RAPD, micro/minisatellite and physiological analysis. Microbiol. Res. 2014, 169, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Costantini, A.; García-Moruno, E.; Moreno-Arribas, M.V. Biochemical transformations produced by malolactic fermentation. In Wine Chemistry and Biochemistry; Moreno-Arribas, M.V., Polo, M.C., Eds.; Springer: Berlin, Germany, 2009; pp. 27–57. [Google Scholar]
- Izquierdo-Cañas, P.M.; Romero, E.G.; Pérez-Martín, F.; Seseña, S.; Palop, M.L. Sequential inoculation versus co-inoculation in Cabernet Franc wine fermentation. Food Sci. Technol. Int. 2015, 21, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, L.M.; Manca de Nadra, M.C.; Farías, M.E. Kinetics and metabolic behaviour of a composite culture of Kloeckera apiculata and Saccharomyces cerevisiae wine related strains. Biotechnol. Lett. 2007, 29, 1057–1063. [Google Scholar] [CrossRef]
- Liu, P.T.; Lu, L.; Duan, C.Q.; Yan, G.L. The contribution of indigenous non-Saccharomyces wine yeast to improved aromatic quality of Cabernet Sauvignon wines by spontaneous fermentation. LWT-Food Sci. Technol. 2016, 71, 356–363. [Google Scholar] [CrossRef]
- Ciani, M.; Beco, L.; Comitini, F. Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 2006, 108, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Whitener, M.E.B.; Stanstrup, J.; Carlin, S.; Divol, B.; Du Toit, M.; Vrhovsek, U. Effect of non-Saccharomyces yeasts on the volatile chemical profile of Shiraz wine. Aust. J. Grape Wine Res. 2017, 23, 179–192. [Google Scholar] [CrossRef]
- Izquierdo-Cañas, P.M.; Mena-Morales, A.; García-Romero, E. Malolactic fermentation before or during wine aging in barrels. LWT-Food Sci. Technol. 2016, 66, 468–474. [Google Scholar] [CrossRef]
- Du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The next generation of malolactic fermentation starter cultures—an overview. Food Bioproc. Tech. 2010, 4, 876–906. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).