Abstract
In recent years, in line with consumer preferences and due to the effects of global climate change, new trends have emerged in wine fermentation and wine technology. Consumers are looking for wines with less ethanol and fruitier aromas, but also with a good balance in terms of acidity and mouthfeel. Nonconventional yeasts contain a wide range of different genera of non-Saccharomyces. If in the past they were considered spoilage yeasts, now they are used to enhance the aroma profile of wine or to modulate wine composition. Recent publications highlight the role of non-Saccharomyces as selected strains for controlling fermentations mostly in cofermentation with Saccharomyces. In this article, I have reviewed the ability of some bacteria and non-Saccharomyces strains to modulate wine acidity.
1. Acids Present in Grapes and Wines and Their Perceived Taste
Organic acids, next to sugars, are the most abundant solids present in grape juice. They are a significant constituent of juice and wine. Responsible for the sour/acid taste, they also influence wine stability, color, and pH. The quality and quantity of organic acids in conjunction with the sugars has a significant effect on the mouthfeel quality of wines [1].
Acid composition and concentration within the grape-must or wine are influenced by many factors, such as grape variety, soil composition, and climatic conditions. Accumulation of grape acids, namely tartaric acid, usually occurs at the beginning of grape berry development and is, to a large extent, completed at the beginning of ripening [2].
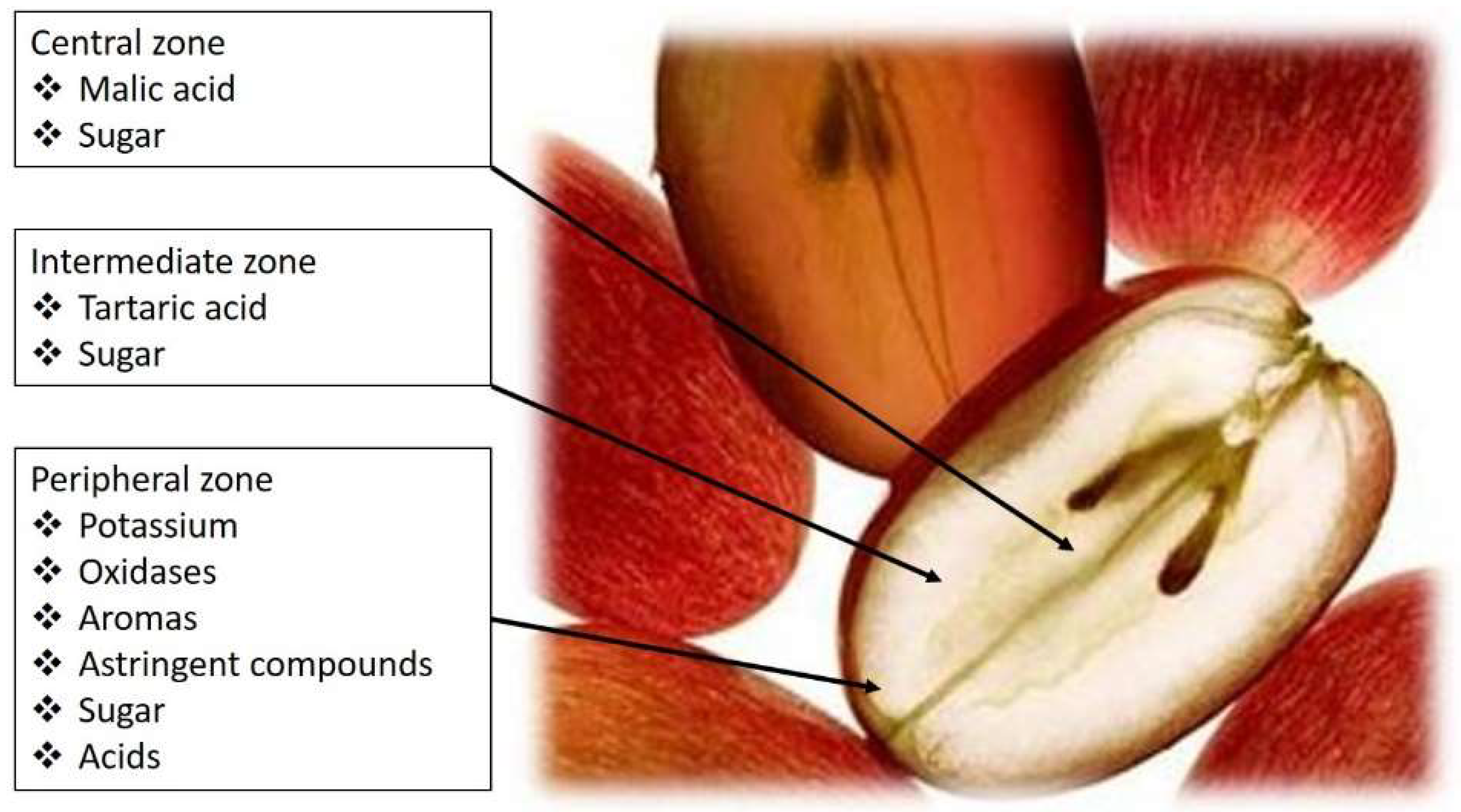
Amerine [3] reported that in berries, tartaric, malic, citric, ascorbic, phosphoric, and tannic acids were present, and soon after, Stafford [4] confirmed the occurrence of all but ascorbic and tannic acids in grapevine leaves, and included oxalic acid, in the form of idioblast crystals of calcium oxalate. Kliewer [5] identified 23 acids in berries, although most of these were found only in trace amounts. Nowadays we know that, by far, the predominant acids are tartaric and malic, which together may account for over 90% of the total acidity in the berry, existing at crudely a 1:1 to 1:3 ratio of tartaric to malic acid [6], both contributing to the pH of the juice, must, and wine during vinification and subsequent aging (Figure 1) [7].
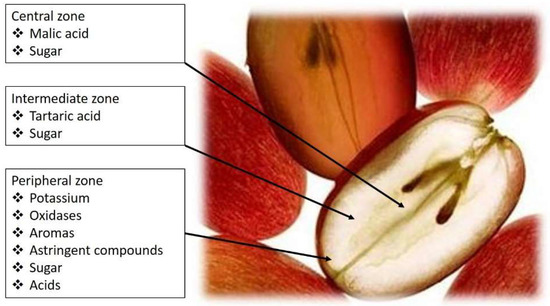
Figure 1.
Grape-berry flavor zones and distribution of tartaric and malic acids.
Tartaric and malic acids are diprotic, with two dissociable protons per molecule. It is the first proton dissociation, with pKa values of around 2.98 (tartaric) and 3.46 (malic) that are meaningful properties in a winemaking context. At a typical wine pH (3.4), tartaric acid will be three times as acidic as malic acid [7].
The bitartrate and bimalate monoanions have important sensory roles to play in wine taste. Malic acid presents a harsh metallic taste (Table 1), sometimes correlated with the taste of green-apples, while the taste attributed to tartaric acid is frequently referred to as being ‘mineral’ or citrus-like. To compensate malic acid ‘lost’ in the late stages of berry ripening, the addition of tartaric acid at crush, or thereafter, can be performed, providing, in this way, control of must/wine pH. However, tartaric acid, unlike malic acid, is not a metabolic substrate for lactic acid bacteria or even yeasts.

Table 1.
Organic acids present in grapes and wines and major acids’ sensory descriptors. Adapted from Boulton et al. [8].
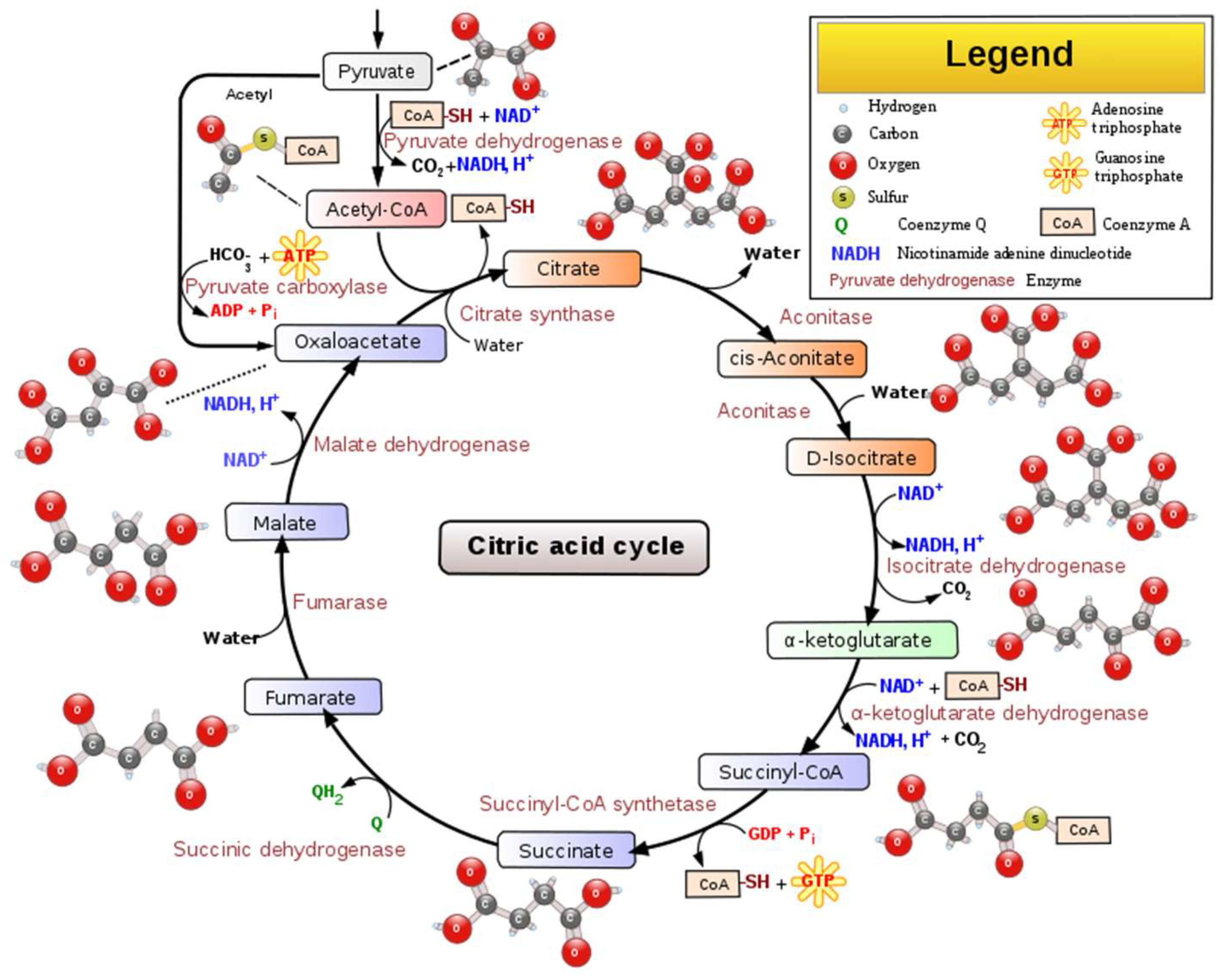
Citric acid, that presents a pleasant citrus-like taste (Table 1), has many uses in wine production. Citric acid is a weak organic acid that presents antimicrobial activity against molds and bacteria. It can create a relationship with antioxidants by chelating metal ions, thus helping in browning prevention. Citric acid occurs in the metabolism of almost every organism because it is an important intermediate in the tricarboxylic acid cycle (TCA cycle) [9], Figure 2.
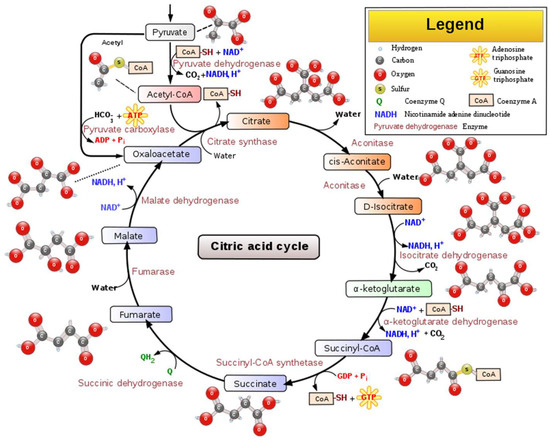
Figure 2.
Schematic representation of the main steps, intermediated compounds, and enzymes of the TCA cycle [10].
During the winemaking process, it is advisable to monitor the concentration of organic acids in order to ensure the quality of the wine, and a distinction is made between acids directly produced in grapes (tartaric, malic, and citric) and those originating during the fermentation—alcoholic and malolactic—succinic, lactic, and acetic acids, among others [11], Table 1.
Acetic acid, in quantities higher than 0.8–0.90 gL−1 [11], is immediately recognizable due to the vinegar smell an acrid taste, causing the wine to be considered spoiled. The maximum acceptable limit for volatile acidity in most wines is 1.2 gL−1 of acetic acid [12]. Acetic acid can appear on the grapes or the grape-must due to the presence of yeasts like Hansenula spp. and Brettanomyces spp., filamentous fungi (Aspergillus niger, Aspergillus tenuis, Cladosporium herbarum, Penicillium spp., and Rhizopus arrhizus), and bacteria (LAB-like indigenous Lactobacilli, and acetic acid bacteria). During alcoholic fermentation, acetic acid usually is formed in small quantities (0.2–0.5 gL−1 acetic acid) as a byproduct of S. cerevisiae metabolism. If the amounts are higher, some contamination spoilage yeasts and bacteria can be present: Candida krusei, Candida stellate, Hansaniaspora uvarum/Kloeckera apiculate, Pichia anomala, Saccharomycodes ludwigii, Acetobacter pasteurianus, and Acetobacter liquefaciens; after malolactic fermentation, heterofermentative species of Oenococcus and Lactobacillus also have the potential to produce acetic acid through the metabolism of residual sugar [13].
Succinic acid, with a sour, salty, and bitter taste, is the major acid produced by yeast during fermentation. This acid is resistant to microbial metabolism under fermentative conditions. During a period from 1991 to 2003, Coulter and coworkers [14] studied 93 red and 45 white Australian wines and found that the concentration of succinic acid in red wines reached from “none” (detection limit of 0.1 gL−1) to 2.6 gL−1, with a mean value of 1.2 gL−1, while the concentration in white wines was between 0.1 gL−1 to 1.6 gL−1, with a mean value of 0.6 gL−1. Thus, succinic acid plays an important role in wine acidity [14].
Lactic acid, that usually is perceived as sour and spicy, is mainly produced by lactic acid bacteria during malolactic fermentation. However, small amounts can also be synthesized by yeast.
Today, the range of wines on the market is huge. On the other hand, wine companies tend to develop a style. Last year’s tendency was to attribute “medals” to balanced flavor profile wines with bordering notes of vegetal-green, chemical, earthy, or sulfur characters, aromas of fruit and oak, hot/full mouthfeel (generally related to the alcohol content), low bitterness, and high sweetness [15]. Consumers and winemakers, giving more importance to flavors and sweetness, tend to “despise” acidity, even if it is one of the most important components of the wine. So, it is important for the wine industry to be able to modulate wine acidity, having in mind the concept of “healthy” and “biological”, without the addition of enological products.
In this article, a review is made of some microorganisms, namely non-Saccharomyces yeasts, that, due to their physiological and genetic traits, are able to modulate wine acidity, either by increasing the wine’s acid content (biological acidification) or by decreasing it—biological deacidification.
2. Wine Biological Acidity Modulation by Bacteria via Malolactic Fermentation
Physicochemical deacidification of wines is time-consuming, requires labor, capital input, and may reduce wine quality [16]. Biological deacidification of wine with malolactic bacteria (MLB), most often strains of Oenococcus oeni, previously known as Leuconostoc oenos [17], is the traditional method used for removing excess wine acidity. However, one important thing must be taken into account; of wine’s total acidity, biological deacidification only affects the malic acid portion, it does not reduce tartaric acid.
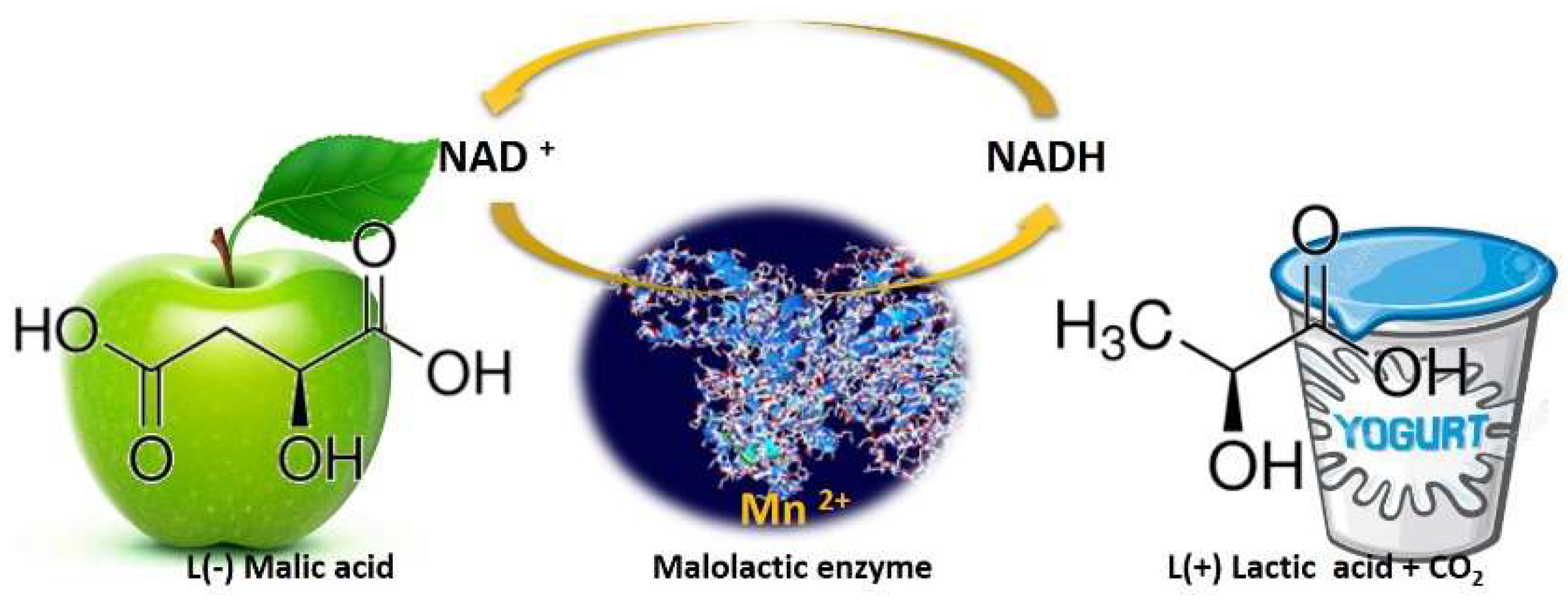
While during alcoholic fermentation, wine yeast strains convert the grape sugars into ethanol and other flavors/mouthfeel compounds, after sugar depletion and the decline of yeasts population, LAB proliferates by utilizing the remaining sugars and thereafter performs malolactic fermentation (MLF). Despite its name “malolactic fermentation”, this biological process is not a fermentation, but an enzymatic reaction in which malic acid (l (−) malic acid) is decarboxylated to lactic acid (l (+) lactic acid) and CO2, Figure 3, [18,19]. This process also reduces the potential carbon source for spoilage microorganisms and leads to wine microbial stabilization [20].
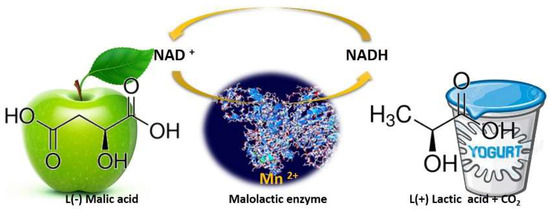
Figure 3.
Schematic representation of malolactic enzyme action. Malolactic bacteria convert sharp green-apple-like malic acid into softer, much less tart lactic acid, releasing CO2 along the way.
During MLF, the metabolism of O. oeni can improve the wine’s sensory characteristics by producing a myriad of secondary metabolites [21]. However, the success of MLF is influenced by oenological parameters, such as temperature, pH, alcohol content, SO2 concentration [22], and yeast inhibitory metabolites, such as medium chain fatty acids [23] or peptic fractions [24].
Several working groups are focused on alternative LAB, such as Lactobacillus plantarum, to perform MLF in wine [25,26]. Lactobacillus plantarum can survive under winemaking stress conditions, and during the fermentation process they are also able to produce a huge number of secondary metabolites important for the wine’s aroma and flavor, including β-glucosidases, esterases, phenolic acid decarboxylases, and citrate lyases [27,28,29] once they contain genes encoding important enzymes that are active under winemaking conditions [30,31]; and can even improve red wine’s color and solve problems associated with wine filtration due to tannase activities [29].
More specific studies have found that depending on the stress conditions in the wine, the gene coding for the malolactic enzyme works differently for O. oeni [32] and L. plantarum [33]. Miller et al. [33] found that the expression of mle (malolactic enzyme) L. plantarum gene presented an increased expression in the middle of MLF and was inducible by the presence of malic acid and low pH wine values, decreasing, nevertheless, in the presence of ethanol. Later, Iorizzo and coworkers [34] reported that some strains of L. plantarum were able to grow at pH values ranging from 3.2 to 3.5 and in the presence of 13% (v/v) ethanol. Several strains of L. plantarum were also found to be able to tolerate the presence of sulfite and in the concentrations used in winemaking [35].
Moreover, L. plantarum strains produce high concentrations of lactic acid, which may contribute to “biological acidification” in low acidity wines, and thus improving wine mouthfeel [36].
3. Wines Biological Acidity Modulation by Nonconventional Yeasts
The malolactic fermentation process is not free from collateral effects (production of off-flavors, wine quality loss, and human health issues due to the production of biogenic amines). Benito et al. [37] developed a new red winemaking methodology by combining the use of two non-Saccharomyces yeast strains as an alternative to the traditional MLF. According to the authors, malic acid is consumed by Schizosaccharomyces pombe, while Lachancea thermotolerans produces lactic acid in order to increase the acidity of wines produced from low acidity musts. The main fermentative properties of interesting non-Saccharomyces yeasts reported as advantageous for fermented beverages and that can modulate wine acidity are described in Table 2.

Table 2.
Percentage of ethanol formed during fermentation, sugars fermented, main volatile compounds formed and effect on wine acidity of seven non-Saccharomyces yeasts.
Schizosaccharomyces pombe, Lachancea thermotolerans, and Torulaspora delbrueckii are presently produced at the industrial level by biotechnological companies [38]. Torulaspora delbrueckii is commercialized in the form of a pure culture, selected for its properties to increase aromatic complexity, mouthfeel, low production of volatile acidity, and high resistance to initial osmotic shock and it is highly recommended for the fermentation of late harvest wines in sequential culture with S. cerevisiae.
3.1. Lachancea thermotolerans: Wine Acidification/Deacetification
L. thermotolerans cells are rather similar in both shape and size to S. cerevisiae and impossible to distinguish by optical microscopy. They also reproduce asexually by multipolar budding. In fermentation conditions, an alcohol degree of 9% (v/v) is the limit of ethanol produced and tolerated [38]. L. thermotolerans can produce lactic acid during fermentation, up to 9.6 gL−1 [58], and glycerol [59].
All these interesting features can be a way to address the problems of increased alcohol content/reduction in the total acidity of wines associated with global climate changes [60]. Since 2013 [59], studies have been made in several wines and wine-regions that elucidate L. thermotolerans wine-making features—Sangiovese and Cabernet-Sauvignon wines where a significant increase in the spicy notes was found [59]; Airén wines, an increased lactic acid concentration up to 3.18 gL−1 and a pH reduction of 0.22 were accomplished [61], Emir wines, where an increase in final total acidity of 5.40–6.28 gL−1 was achieved [60].
However, L. thermotolerans is also capable of another interesting wine-making feature. This yeast can be used to develop a controlled biological deacetification process of wines with high volatile acidity, with the process being oxygen-dependent, which means that its metabolism must shift more towards respiration than fermentation [40,50].
To verify the potential application of L. thermotolerans wine deacetification, the strain was inoculated in two wine-supplemented mineral media, (I) simulating the refermentation of a wine with freshly crushed grapes (130 gL−1 of glucose and 4% ethanol (v/v)) and (II) simulating the refermentation of a wine with the residual marc from a finished wine fermentation (33 gL−1 glucose and 10% ethanol (v/v)). The volatile acidity of both mixtures was 1.13 gL−1 of acetic acid. L. thermotolerans was able to consume 94.6% of the initial acetic acid, in the high-glucose medium, under aerobic conditions and the final “wine” was left with a volatile acidity of 0.06 gL−1 [50].
3.2. Schizosaccharomyces pombe: Biological Acidity Modulation via Maloalcoholic Fermentation
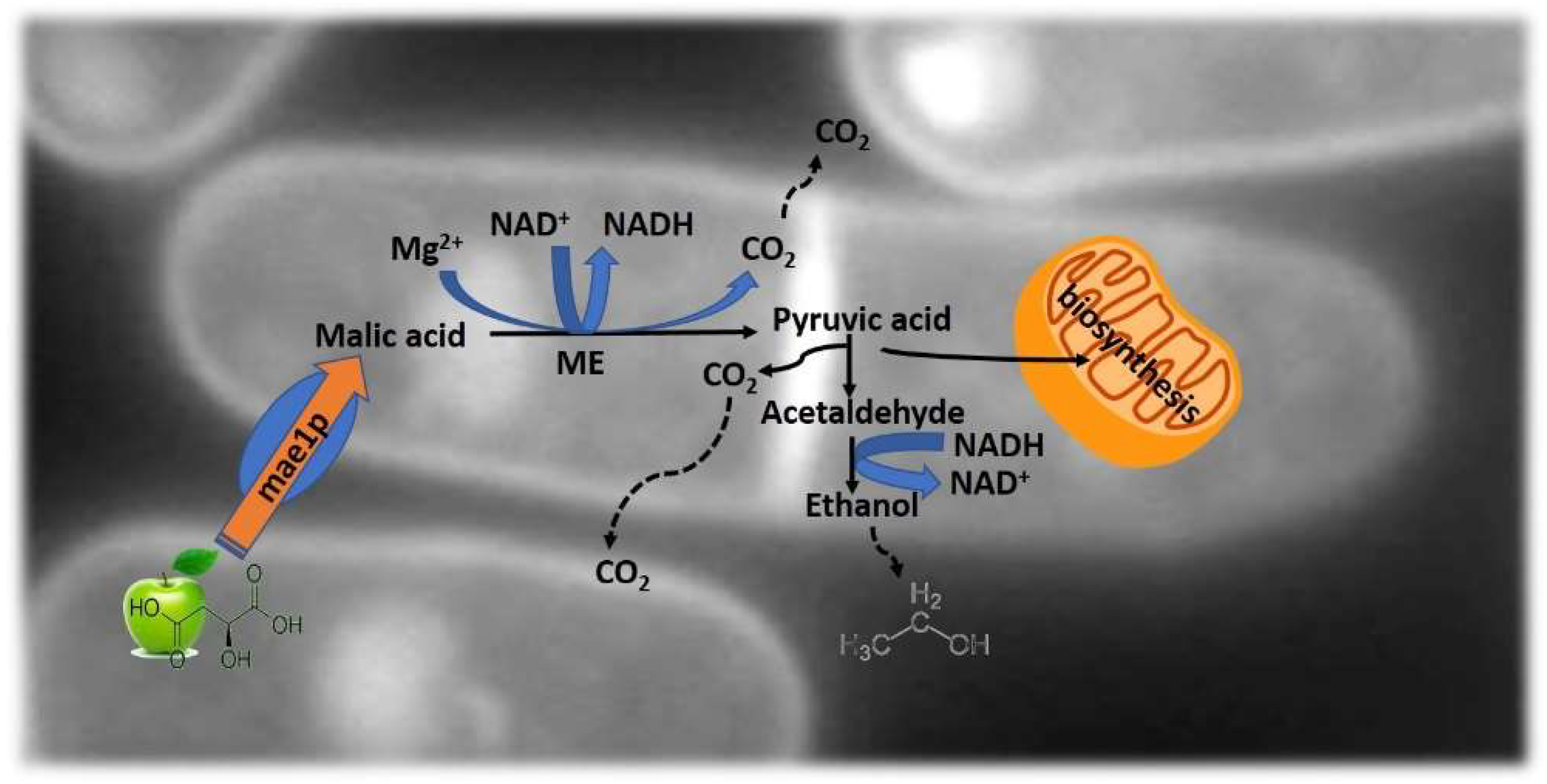
Usually, wine is produced by using Saccharomyces yeast to transform sugars into alcohol, followed by Oenococus bacteria to complete malolactic fermentation and thus rendering the wine with its pleasantness and microbial stability. This methodology has some unsolved problems: (i) the management of highly acidic musts; (ii) the production of potentially toxic products, including biogenic amines and ethyl carbamate [62]. To overcome these issues, the use of non-Saccharomyces yeast strains able to perform alcoholic fermentation and malic acid degradation are being studied. S. cerevisiae has long been known as a poor metabolizer of extracellular malate, due to the lack of a mediated transport system for the acid [63]. One example is the fission yeast from the genus Schizosaccharomyces. These yeasts are able to consume malic acid by converting it to ethanol and CO2 [64], Figure 4.
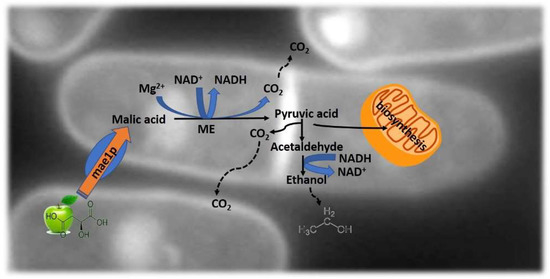
Figure 4.
Schematic representation of the maloalcoholic pathway. Malic acid is transported into the yeast cell by mae1p carboxylic acid transporter. The malic enzyme (ME) converts malate into pyruvate, supplying pyruvic acid to the mitochondria for biosynthesis.
Schizosaccharomyces pombe is highly appreciated in colder regions due to its particular metabolism of maloalcoholic fermentation [64], significantly reducing the levels of ethyl carbamate precursors and biogenic amines without the need for any bacterial MLF [62]. Schizosaccharomyces is also able, during fermentation, to increase the formation of vitisins and vinylphenolic pyranoanthocyanin [65]; these pigments intensify the color of the finished wine [66].
3.3. Candida stellata: Biological Acidity Enrichment
Candida stellata is an imperfect yeast of the genus Candida, order Saccharomycetales, phylum Ascomycota. C. stellata was originally isolated from an overripe grape must in Germany, but we can find it widespread in natural and artificial habitats [67].
During the beginning of alcoholic fermentation, many species can grow simultaneously in the grape must; C. stellata species have been described in this stage of fermentation [68]. C. stellata cells are spherical/ovoid; usually found as single cells. This yeast ferments glucose, sucrose, and raffinose and uses lysine as its sole N source. It is also able to grow at higher pH values and it is not sensitive to ethanol [67]. These features make it a good candidate for co-inoculation with S. cerevisiae. This yeast is also frequently associated with musts proceeding from botrytized grapes [69].
An interesting feature is the ability of C. stellata to form succinic acid. Maurizio and Ferraro [70] found that C. stellata yeast strains produced more succinic acid than a Saccharomyces control strain in synthetic grape juice. The higher production of the acid was also associated with higher glycerol levels. The yeasts also presented high ethanol production and ethanol tolerance [42,71]. Succinic acid could positively influence the sensory/mouthfeel profile of wines with insufficient acidity. Nevertheless, due to its ‘salt-bitter-sour’ taste, excessive levels would negatively influence wine quality.
Moreover, Magyar and Tóth [72] found that C. stellata is similar to C. zemplinina in its strong fructophilic character. However, C. stellata produces more glycerol and more ethanol, which is comparable with that produced by S. uvarum. The latter species is known to produce low acetic acid and low ethanol when compared with S. cerevisiae.
3.4. Torulaspora delbrueckii: Volatile Acidity Modulation in Very Sweet Musts
Torulaspora delbrueckii (formerly Saccharomyces rosei) is reported to have a positive effect on the flavor of alcoholic beverages, and exhibits low production of acetic acid, acetaldehyde, ethyl acetate, and acetoin [71].
T. delbrueckii was studied by Lafon-Lafourcade et al. [73] in order to evaluate the possibility of using it to improve the quality of botrytized wine (wine made with Botrytis cinerea-infected grapes, commonly known as “noble rot”). These wines are made with grapes with sugar concentrations up to 350–450 gL−1 and present a challenge to S. cerevisiae in terms of acetic acid production. It is well known that S. cerevisiae produces acetic acid as a stress response to the high-sugar grape must [13]. The volatile acidity of these wines may be over 1.8 gL−1 (acetic acid), a value above the human nose detection threshold and over the EEC (European Economic Community) legal limit (1.5 gL−1) [12].
Bely et al. [43] used mixed cultures of T. delbrueckii and S. cerevisiae in high-sugar fermentation musts with a higher concentration of T. delbrueckii to promote its growth. This mixed inoculum produced lower levels of acetic acid and acetaldehyde, up to 55% and 68% less, respectively, when compared with the pure cultures of S. cerevisiae. An indication of the positive impact of T. delbrueckii activity on wine quality was also demonstrated by Azzolini et al. [74] in Vino Santo, a sweet wine, due to its low production of acetic acid.
Azzolini et al. [75] studied the impact of T. delbrueckii in the production of Amarone wine (officially named, Amarone della Valpolicella), a high-alcohol dry red wine obtained from withered grapes. Winery trials were inoculated by a selected strain of T. delbrueckii in co-inoculation and/or sequentially with an S. cerevisiae strain. T. delbrueckii was able to promote the formation of alcohols, fermentative esters, fatty acids, and lactones, which are important in the Amarone wine flavor. In terms of aromatic capacity, in white table wines, T. delbrueckii was able to produce dried fruit/pastry aromas, pleasant to the tasters [44,74].
3.5. Zygosaccharomyces Florentinus/Zygotorulaspora Florentina: Volatile Acidity Modulation
The genus Zygosaccharomyces is involved in food and beverage spoilage since it shows high tolerance to osmotic stress, and consequently, the Zygosaccharomyces species can grow in severe environments with high sugar concentration, pH values closer to 2 [76], low ‘water activity’ (aw), and presence of organic acids and preservatives such as SO2 and ethanol [77]. Zygosaccharomyces spp. produces high ethanol and acetoin content in wines and may play an important role as non-Saccharomyces yeasts in differentiated wine products [78]. The odors and flavors produced by Zygosaccharomyces are generally described as being wine-like, so the main problem with this yeast in wines, is the formation of a haze/deposit after bottling.
Kurtzman et al. [79] and Kurtzman & Robnett [80], reported that the genus Zygosaccharomyces comprised 11 species, divided into two groups. After, by means of multigene sequence analysis, Kurtzman [81] proposed a new division: the Zygosaccharomyces clade should be divided into four phylogenetic groups: (1) Zygosaccharomyces, which included Z. bailii, Z. bisporus, Z. kombuchaensis, Z. lentus, Z. mellis, and Z. rouxii; (2) Zygotorulaspora, comprising two species, Zygotorulaspora florentinus (florentina) and Zt. mrakii; (3) Torulaspora, with Torulaspora microellipsoides and (4) the Lachancea clade, in which Lachancea cidri and L. fermentati were included [82].
Zygosaccharomyces are well known for their ability to spoil food and beverages [83]. Nevertheless, some strains are used in industrial production of balsamic vinegar, miso paste, and soy sauce—Z. rouxii [84] and Z. kombuchaensis in the production of kombucha [79].
Domizio and co-workers in 2011 [48] used Zy42 Zygosaccharomyces florentinus strain to perform grape juice fermentation in mixed cultures with a well-known commercial Saccharomyces strain—EC1118 (Lalvin EC-1118 it was originally named “prise de mousse”). It was isolated in Champagne and its use is validated by the Comité Interprofessionnel du Vin de Champagne (CIVC)). The EC1118/Zy42 association increased the production of propanol, higher alcohols, and esters and produced lower levels of acetic acid than the comparative trial with a pure culture of EC1118 (0.3 and 0.4 gL−1, respectively).
Later, Lencioni and coworkers in 2016 [46] evaluated the strain Zygotorulaspora florentina (formerly Zygosaccharomyces florentinus) in mixed culture fermentations with S. cerevisiae, from the laboratory scale to the winery scale. At the lab scale, the resulting mixed fermentations showed a reduction of volatile acidity and an improvement of polysaccharides and 2-phenylethanol (rose-like aroma) concentrations. At the winery scale, they used red grape-must from the Sangiovese grape variety. The resulting wine presented a higher concentration of glycerol and esters; sensorially, the wine astringency was lower and the aromatic character was defined by floral notes.
Recently, the same authors [47], studied the possibility to decrease wine volatile acidity in mixed fermentations with Zygotorulaspora florentina and Starmerella bacillaris (syn., Candida zemplinina). Independent of fermentation temperature, the mixed fermentations with Z. florentina performed best to reduce volatile acidity, thus being a valuable tool for performing fermentation of high-sugar musts.
3.6. Pichia kudriavzevii/Issatchenkia orientalis: Malic Acid Consumption
Since 1966 [85] that there have been several reports on yeast strains degrading extracellular malic acid, including Schizosaccharomyces pombe [64,85]. Among the yeast strains degrading malic acid, S. malidevorans and S. pombe are the strains studied most intensively as a means of reducing wine acidity [64,85,86]. However, it has been reported that Schizosaccharomyces sp. may produce off-flavors/aromas in the wines [87].
Degradation of malic acid by the strain Issatchenkia orientalis KMBL 5774, an acidophilic yeast strain, isolated from Korean grape wine pomace, was investigated by Seo et al. [88]. In the mentioned work, degradation of malic acid by I. orientalis KMBL 5774 was investigated in YNB-malic acid liquid media under various culture conditions (malic acid concentration, pH, temperature, …etc.). The maximal growth was obtained when 3% malic acid was used and maximal malic acid degradation ratio was obtained at 1–2% of the malic acid concentration (94.4 and 94.6%, respectively) [89].
A few years ago, in 2014, Mónaco and coworkers [51] studied fifty-seven Patagonian non-Saccharomyces yeasts of oenological origin, and tested their ability to consume l-malic acid as a carbon source. Only four isolates belonging to Pichia kudriavzevii (also known as Issatchenkia orientalis) species showed this ability, and one was selected and studied further—P. kudriavzevii ÑNI15. This isolate was able to degrade l-malic acid in microvinification assays (38% of l-malic acid reduction when compared with S. cerevisiae (22%)), increasing the pH by 0.2–0.3 units. Furthermore, P. kudriavzevii ÑNI15 produced low levels of ethanol and significant levels of glycerol (10.41 ± 0·48 gL−1). The final wines presented fruity and cooked pears aromas, once Pichia kudriavzevii was unable to synthesize ethyl acetate but it showed good production of ethyl esters from fatty acids when compared with the Saccharomyces yeast [51].
However, under the names Pichia kudriavzevii, Issatchenkia orientalis, and Candida glycerinogenes, the same yeast, including genetically modified strains, is used for industrial-scale production of glycerol and succinate. In 2018, Douglass and coworkers [89] investigated the genomic diversity of a yeast species that is both an opportunistic pathogen and an important industrial yeast. Under the name Candida krusei, it is responsible for about 2% of yeast infections caused by Candida species in humans. Bloodstream infections with C. krusei are problematic because most isolates are fluconazole-resistant. In their work, Douglass et al. [89] sequenced the strains of C. krusei (CBS573T) and P. kudriavzevii (CBS5147T), and the results showed, conclusively, that they are the same species, with collinear genomes 99.6% identical in DNA sequence. Phylogenetic analysis of SNPs does not segregate clinical and environmental isolates into separate clades, suggesting that C. krusei infections are frequently acquired from the environment. More studies are needed to ensure public safety when using these non-Saccharomyces strains in food/wine production.
3.7. Starmerella bacillaris/Candida zemplinina: Wine Biological Acidification and Deacidification
One approach to reducing the ethanol content of wines is by co-inoculation of Saccharomyces and non-Saccharomyces yeast during must fermentation. The selection and use of non-Saccharomyces wine yeasts can potentially lead to a reduction of the overall sugar–ethanol yield [53]. The yeast Starmerella bacillaris (syn. Candida zemplinina) is often isolated from grape and winery environments specifically associated with overripe and botrytized grapes [90]. Its enological use in mixed fermentation with S. cerevisiae has been investigated, and several interesting features, such as low ethanol and high glycerol production, and a fructophilic aptitude, have been found [54]. This yeast also presents acidogenic, psychrotolerant, and osmotolerant properties, and therefore seems to be well adapted to sweet wine fermentations [55].
It was in 2002 that David Mills and coworkers [90] isolated the yeast Starmerella bacillaris for the first time from sweet wines made with botrytized grapes in Napa Valley (California, USA), and due to their enological features, it has been studied since then. Englezos and coworker in 2016 [53] investigated the potential application of Starmerella bacillaris in combination with S. cerevisiae, in co-inoculated and sequential cultures to reduce the ethanol in wines. Lab scale fermentations showed a decrease of up to 0.7% (v/v) of ethanol and an increase of about 4.2 gL−1 of glycerol.
Continuing their work on Starmerella bacillaris and the application of this yeast to the wine industry and winemaking, Englezos and coworker in 2017 [55] wrote an interesting review. In that report, they concluded that among other Starmerella bacillaris enological features, this yeast species could contribute to malic acid degradation due to its ability to produce a wide spectrum of extracellular hydrolytic enzymes [91]. It is also capable of producing pyruvic acid, acting as a natural acidification agent by reducing the wine’s pH [92], which may have an impact on wine color, due to the reaction of pyruvic acid with anthocyanins producing stable colored pigments such as Vitisin A [93].
Moreover, in 2012, Rantsiou and coworkers [94] showed that the use of mixed fermentations with Starmerella bacillaris and S. cerevisiae could be a biological method to reduce acetic acid in sweet wines. According to the mentioned authors, the co-inoculation produced wines with a decrease of 0.3 gL−1 of acetic acid; sequential inoculation produced wines with about half of the acetic acid content, compared to wines produced with pure cultures of S. cerevisiae [94].
Additionally, sequential inoculations of Starmerella bacillaris and S. cerevisiae possess great potential in affecting and modulating the chemical and aromatic profile of white wines, especially those produced from Sauvignon blanc grapes [57]. The volatile profile of Chardonnay, Muscat, Riesling, and Sauvignon blanc white wines, fermented with sequential inoculation of Starmerella bacillaris and S. cerevisiae, were studied by Englezos and coworker in 2018 [57]. Mixed fermentations with both these strains affected the chemical composition of wines by modulating various metabolites of oenological interest. For volatile compounds, mixed fermentations led to a reduction of ethyl acetate, which may be a compound responsible for wine deterioration above certain limits. Interestingly, Sauvignon blanc wines, fermented by mixed cultures, contained significantly higher levels of esters and thiols, both considered positive aromatic attributes [57].
4. Final Remarks
In conclusion, many non-Saccharomyces yeasts present interesting oenological properties in terms of fermentation purity and production of secondary metabolites or even ethanol. When used in single or mixed cultures with S. cerevisiae, these yeasts strains can modulate wine acidity and increase production of some interesting compounds, such as polysaccharides, glycerol, and volatile compounds, such as 2-phenyl ethanol and 2-methyl 1-butanol.
Thus, this review confirms that some non-Saccharomyces yeasts that are most often considered as spoilage yeasts are actually tools of great value for the winemaking industry.
A deeper study of the oenological traits of these yeasts will provide new data for consideration in the control of fermentation, with special regard to warm-climate wine and botrytized sweet wines, where they are commonly found in mixed populations.
Acknowledgments
We appreciate the financial support provided to the Research Unit in Vila Real (PEst-OE/QUI/UI0616/2014) by FCT—Portugal and COMPETE. Additional thanks to the Project NORTE-01-0145-FEDER-000038 (I&D INNOVINE&WINE—Innovation Platform of Vine & Wine).
Conflicts of Interest
The author declares no conflict of interest.
References
- Liu, H.F.; Wu, B.H.; Fan, P.G.; Xu, H.Y.; Li, S.H. Inheritance of sugars and acids in berries of grape (Vitis vinifera L.). Euphytica 2007, 153, 99–107. [Google Scholar] [CrossRef]
- Cosme, F.; Gonçalves, B.; Inês, A.; Jordão, A.M.; Vilela, A. Grape and wine metabolites: Biotechnological approaches to improve wine quality. In Grape and Wine Biotechnology; Morata, A., Loira, I., Eds.; InTechOpen: London, UK, 2016; pp. 187–214. [Google Scholar]
- Amerine, M.A. The Maturation of Wine Grapes. Wines Vines 1956, 37, 53–55. [Google Scholar]
- Stafford, H. Distribution of Tartaric Acid in the Leaves of Certain Angiosperms. Am. J. Bot. 1959, 46, 347–352. [Google Scholar] [CrossRef]
- Kliewer, W. Sugars and Organic Acids of Vitis vinifera. Plant Physiol. 1966, 41, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Lamikanra, O.; Inyang, I.; Leong, S. Distribution and effect of grape maturity on organic acid content of red Muscadine grapes. J. Agric. Food Chem. 1995, 43, 3026–3028. [Google Scholar] [CrossRef]
- Ford, C.M. The Biochemistry of Organic Acids in the Grape. In The Biochemistry of the Grape Berry; e-Book, Gerós, H., Chaves, M., Delrot, S., Eds.; Bentham Science Publishers: Sharjah, UAE, 2012; pp. 67–88. ISBN 978-1-60805-360-5. [Google Scholar]
- Boulton, R.B.; Singleton, V.L.; Bisson, L.F.; Kunkee, R.E. Principles and Practices of Winemaking; Chapman and Hall: New York, NY, USA, 1996; pp. 102–181. [Google Scholar]
- Sharma, R.K. Citric Acid. In Natural Food Antimicrobial Systems; Naidu, A.S., Ed.; CRC Press LLC: New York, NY, USA, 2000. [Google Scholar]
- Citric Acid Cycle. Available online: https://commons.wikimedia.org/wiki/File:Citric_acid_cycle_with_aconitate_2.svg#/media/File:Citric_acid_cycle_with_aconitate_2.svg (accessed on 30 January 2019).
- Ribéreau-Gayon, P.; Glories, Y.; Maujean, A.; Dubourdieu, D. Alcohols, and other volatile compounds. The chemistry of wine stabilization and treatments. In Handbook of Enology, 2nd ed.; John Wiley & Sons Ltd.: Chichester, UK, 2006; Volume 2, pp. 51–64. [Google Scholar]
- Office Internationale de la Vigne et du Vin. International Code of Oenological Practices; OIV: Paris, France, 2010. [Google Scholar]
- Vilela-Moura, A.; Schuller, D.; Mendes-Faia, A.; Silva, R.F.; Chaves, S.R.; Sousa, M.J.; Côrte-Real, M. The impact of acetate metabolism on yeast fermentative performance and wine quality: Reduction of volatile acidity of grape musts and wines—Minireview. Appl. Microbiol. Biotechnol. 2011, 89, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Coulter, A.D.; Godden, P.W.; Pretorius, I.S. Succinic acid-how is it formed, what is its effect on titratable acidity, and what factors influence its concentration in wine? Wine Ind. J. 2004, 19, 16–24. [Google Scholar]
- Hopfer, H.; Heymann, H. Judging wine quality: Do we need experts, consumers or trained panelists? Food Qual. Prefer. 2014, 32, 221–233. [Google Scholar] [CrossRef]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Dicks, L.M.T.; Dellaglio, F.; Collins, M.D. Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int. J. Syst. Bacteriol. 1995, 45, 395–397. [Google Scholar] [CrossRef] [PubMed]
- Henick-Kling, T. Malolactic fermentation. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Reading, UK, 1993; pp. 289–327. [Google Scholar]
- Volschenk, H.; Van Vuuren, H.J.J.; Viljoen-Bloom, M. Malic acid in wine: Origin, function and metabolism during vinification. S. Afr. J. Enol. Vitic. 2006, 27, 123–136. [Google Scholar] [CrossRef]
- Lasik-Kurdyś, M.; Majcher, M.; Nowak, J. Effects of Different Techniques of Malolactic Fermentation Induction on Diacetyl Metabolism and Biosynthesis of Selected Aromatic Esters in Cool-Climate Grape Wines. Molecules 2018, 23, 2549. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.; Borneman, A. Genomic variations of Oenococcus oeni strains and the potential to impact on malolactic fermentation and aroma compounds in wine. Appl. Microbiol. Biotechnol. 2011, 92, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Lerm, E.; Engelbrecht, L.; du Toit, M. Malolactic fermentation: The ABC’s of MLF. S. Afr. J. Enol. Vitic. 2010, 31, 186–212. [Google Scholar] [CrossRef]
- Alexandre, H.; Costello, P.J.; Remize, F.; Guzzo, J.; Guilloux-Benatier, M. Saccharomyces cerevisiae-Oenococcus oeni interactions in wine: Current knowledge and perspectives. Int. J. Food Microbiol. 2004, 93, 141–154. [Google Scholar] [CrossRef]
- Nehme, N.; Mathieu, F.; Taillandier, P. Impact of the co-culture of Saccharomyces cerevisiae-Oenococcus oeni on malolactic fermentation and partial characterization of a yeast-derived inhibitory peptidic fraction. Food Microbiol. 2010, 27, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Berbegal, C.; Peña, N.; Russo, P.; Grieco, F.; Pardo, I.; Ferrer, S.; Spano, G.; Capozzi, V. Technological properties of Lactobacillus plantarum strains isolated from grape must fermentation. Food Microbiol. 2016, 57, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Lucio, O.; Pardo, I.; Krieger-Weber, S.; Heras, J.M.; Ferrer, S. Selection of Lactobacillus strains to induce biological acidification in low acidity wines. LWT Food Sci. Technol. 2016, 73, 334–341. [Google Scholar] [CrossRef]
- Matthews, A.; Grimaldi, A.; Walker, M.; Bartowsky, E.; Grbin, P.; Jiranek, V. Lactic acid bacteria as a potential source of enzymes for use in vinification. Appl. Environ. Microbiol. 2004, 70, 5715–5731. [Google Scholar] [CrossRef]
- Grimaldi, A.; Bartowsky, E.; Jiranek, V. Screening of Lactobacillus spp. and Pediococcus spp. for glycosidase activities that are important in enology. J. Appl. Microbiol. 2005, 99, 1061–1069. [Google Scholar] [CrossRef]
- Brizuela, N.; Tymczyszyn, E.E.; Semorile, L.C.; La Hens, D.V.; Delfederico, L.; Hollmann, A.; Bravo-Ferrada, B. Lactobacillus plantarum as a malolactic starter culture in winemaking: A new (old) player? Electron. J. Biotechnol. 2019, 38, 10–18. [Google Scholar] [CrossRef]
- Du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The next generation of malolactic fermentation starter cultures—An overview. Food Bioprocess Techmol. 2011, 4, 876–906. [Google Scholar] [CrossRef]
- Lerm, E.; Engelbrecht, L.; Toit, M.D. Selection and Characterisation of Oenococcus oeni and Lactobacillus plantarum South African Wine Isolates for Use as Malolactic Fermentation Starter Cultures. S. Afr. J. Enol. Vitic. 2011, 32, 280–295. [Google Scholar] [CrossRef]
- Olguín, N.; Bordons, A.; Reguant, C. Multigenic expression analysis as an approach to understanding the behavior of Oenococcus oeni in wine-like conditions. Int. J. Food Microbiol. 2010, 144, 88–95. [Google Scholar] [CrossRef]
- Miller, B.J.; Franz, C.M.; Cho, G.S.; du Toit, M. Expression of the malolactic enzyme gene (mle) from Lactobacillus plantarum under winemaking conditions. Curr. Microbiol. 2011, 62, 1682–1688. [Google Scholar] [CrossRef]
- Iorizzo, M.; Testa, B.; Lombardi, S.J.; García-Ruiz, A.; Muñoz-González, C.; Bartolomé, B.; Moreno-Arribas, M.V. Selection and technological potential of Lactobacillus plantarum bacteria suitable for wine malolactic fermentation and grape aroma release. LWT Food Sci. Technol. 2016, 73, 557–566. [Google Scholar] [CrossRef]
- Bravo-Ferrada, B.M.; Hollmann, A.; Delfederico, L.; La Hens, D.V.; Caballero, A.; Semorile, L. Patagonian red wines: Selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J. Microbiol. Biotechnol. 2013, 29, 1537–1549. [Google Scholar] [CrossRef]
- Berbegal, C.; Spano, G.; Tristezza, M.; Grieco, F.; Capozzi, V. Microbial Resources and Innovation in the Wine Production Sector. S. Afr. J. Enol. Vitic. 2017, 38, 156–166. [Google Scholar] [CrossRef]
- Benito, A.; Calderón, F.; Palomero, F.; Benito, S. Combined use of selected Schizosaccharomyces pombe and Lachancea thermotolerans yeast strains as an alternative to the traditional malolactic fermentation in red wine production. Molecules 2015, 20, 9510–9523. [Google Scholar] [CrossRef]
- Morata, A.; Suárez-Lepe, J.A. New biotechnologies for wine fermentation and ageing. In Advances in Food Biotechnology, 1st ed.; Ravishankar Rai, V., Ed.; John Wiley & Sons, Ltd.: West Sussex, UK, 2016; pp. 287–301. [Google Scholar]
- Vilela, A. Targeting Demalication and Deacetification Methods: The Role of Carboxylic Acids Transporters. Biochem. Physiol. 2017, 6, 224. [Google Scholar] [CrossRef]
- Vilela, A. Lachancea thermotolerans, the Non-Saccharomyces Yeast that Reduces the Volatile Acidity of Wines. Fermentation 2018, 4, 56. [Google Scholar] [CrossRef]
- Suárez-Lepe, J.A.; Morata, A. New trends in yeast selection for winemaking. Trends Food Sci. Technol. 2012, 23, 39–50. [Google Scholar] [CrossRef]
- Ferraro, L.; Fatichenti, F.; Ciani, M. Pilot scale vinification process using immobilized Candida stellata cells and Saccharomyces cerevisiae. Process. Biochem. 2000, 35, 1125–1129. [Google Scholar] [CrossRef]
- Bely, M.; Stoeckle, P.; Masneuf Pomarède, I.; Dubourdieu, D. Impact of mixed Torulaspora delbrueckii Saccharomyces cerevisiae culture on high sugar fermentation. Int. J. Food Microbiol. 2008, 122, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, R.; Zamora, E.; Álvarez, M.L.; Hernández, L.M.; Ramírez, M. Effects of new Torulaspora delbrueckii killer yeasts on the must fermentation kinetics and aroma compounds of white table wine. Front. Microbiol. 2015, 6, 1222. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Santos, J.; Chaves, S.; Almeida, J.; Leão, C.; Sousa, M.J. The Emerging Role of the Yeast Torulaspora delbrueckii in Bread and Wine Production: Using Genetic Manipulation to Study Molecular Basis of Physiological Responses. In Structure and Function of Food Engineering; Eissa, A.A., Ed.; IntechOpen: London, UK, 2012; Chapter 13; pp. 339–370. [Google Scholar]
- Lencioni, L.; Romani, C.; Gobbi, M.; Comitini, F.; Ciani, M.; Domizio, P. Controlled mixed fermentation at winery scale using Zygotorulaspora florentina and Saccharomyces cerevisiae. Int. J. Food Microbiol. 2016, 234, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Lencioni, L.; Taccari, M.; Ciani, M.; Domizio, P. Zygotorulaspora florentina and Starmerella bacillaris in multistarter fermentation with Saccharomyces cerevisiae to reduce volatile acidity of high sugar musts. Aust. J. Grape Wine Res. 2018, 24, 368–372. [Google Scholar] [CrossRef]
- Domizio, P.; Romani, C.; Lencioni, L.; Comitini, F.; Gobbi, M.; Mannazzu, I.; Ciani, M. Outlining a future for non-Saccharomyces yeasts: Selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 2011, 147, 170–180. [Google Scholar] [CrossRef]
- Pina, C.; Gonçalves, P.; Prista, C.; Loureiro-Dias, M.C. Ffz1, a New Transporter Specific for Fructose from Zygosaccharomyces bailii. Microbiology 2004, 150, 2429–2433. [Google Scholar] [CrossRef]
- Vilela-Moura, A.; Schuller, D.; Mendes-Faia, A.; Côrte-Real, M. Reduction of volatile acidity of wines by selected yeast strains. Appl. Microbiol. Biotechnol. 2008, 80, 881–890. [Google Scholar] [CrossRef]
- Mónaco, S.; Barda, N.; Rubio, N.; Caballero, A. Selection and characterization of a Patagonian Pichia kudriavzevii for wine deacidification. J. Appl. Microbiol. 2014, 117, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Hello, My Name is Pichia kudriavzevii; Eureka Brewing. Available online: https://eurekabrewing.wordpress.com/2014/02/16/hello-my-name-is-pichia-kudriavzevii (accessed on 16 February 2019).
- Englezos, V.; Rantsiou, K.; Cravero, F.; Torchio, F.; Ortiz-Julien, A.; Gerbi, V.; Rolle, L.; Cocolin, L. Starmerella bacillaris and Saccharomyces cerevisiae mixed fermentations to reduce ethanol content in wine. Appl. Microbiol. Biotechnol. 2016, 100, 5515–5526. [Google Scholar] [CrossRef]
- Masneuf-Pomarede, I.; Juquin, E.; Miot-Sertier, C.; Renault, P.; Laizet, Y.; Salin, F.; Alexandre, H.; Capozzi, V.; Cocolin, L.; Colonna-Ceccaldi, B.; et al. The yeast Starmerella bacillaris (synonym Candida zemplinina) shows high genetic diversity in winemaking environments. FEMS Yeast Res. 2015, 5, fov045. [Google Scholar] [CrossRef] [PubMed]
- Englezos, V.; Giacosa, S.; Rantsiou, K.; Rolle, L.; Cocolin, L. Starmerella bacillaris in winemaking: Opportunities and risks. Curr. Opin. Food Sci. 2017, 17, 30–35. [Google Scholar] [CrossRef]
- Sadoudi, M.; Tourdot-Maréchal, R.; Rousseaux, S.; Steyer, D.; Gallardo-Chacón, J.-J.; Ballester, J.; Vichi, S.; Guérin-Schneider, R.; Caixach, J.; Alexandre, H. Yeast–yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 2012, 2, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Englezos, V.; Rantsiou, K.; Cravero, F.; Torchio, F.; Pollon, M.; Fracassetti, D.; Ortiz-Julien, S.; Gerbi, V.; Rolle, L.; Cocolin, L. Volatile profile of white wines fermented with sequential inoculation of Starmerella bacillaris and Saccharomyces cerevisiae. Food Chem. 2018, 257, 350–360. [Google Scholar] [CrossRef]
- Kapsopoulou, K.; Kapaklis, A.; Spyropoulos, H. Growth and fermentation characteristics of a strain of the wine yeast Kluyveromyces thermotolerans isolated in Greece. World J. Microbiol. Biotechnol. 2005, 21, 1599–1602. [Google Scholar] [CrossRef]
- Gobbi, M.; Comitini, F.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: A strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 2013, 33, 271–281. [Google Scholar] [CrossRef]
- Balikci, E.K.; Tanguler, H.; Jolly, N.P.; Erten, H. Influence of Lachancea thermotolerans on cv. Emir wine fermentation. Yeast 2016, 33, 313–321. [Google Scholar] [CrossRef]
- Benito, Á.; Calderón, F.; Palomero, F.; Benito, S. Quality and composition of Airén wines fermented by sequential inoculation of Lachancea thermotolerans and Saccharomyces cerevisiae. Food Technol. Biotechnol. 2016, 54, 135–144. [Google Scholar] [CrossRef]
- Benito, Á.; Jeffares, D.; Palomero, F.; Calderón, F.; Bai, F.Y.; Bähler, J.; Benito, S. Selected Schizosaccharomyces pombe Strains Have Characteristics That Are Beneficial for Winemaking. PLoS ONE 2016, 11, e0151102. [Google Scholar] [CrossRef] [PubMed]
- Salmon, J.M. L-Malic acid permeation in resting cells of anaerobically grown Saccharomyces cerevisiae. Biochim. Biophys. Acta 1987, 901, 30–34. [Google Scholar] [CrossRef]
- Volschenk, H.; Vuuren, H.J.; Viljoen–Bloom, M. Malo-ethanolic fermentation in Saccharomyces and Schizosaccharomyces. Curr. Genet. 2003, 43, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Loira, I.; Morata, A.; Comuzzo, P.; Callejo, M.J.; González, C.; Calderón, F.; Suárez-Lepe, J.A. Use of Schizosaccharomyces pombe and Torulaspora delbrueckii strains in mixed and sequential fermentations to improve red wine sensory quality. Food Res. Int. 2015, 76, 325–333. [Google Scholar] [CrossRef]
- Morata, A.; González, C.; Suárez-Lepe, J.A. Formation of vinylphenolic pyranoanthocyanins by selected yeasts fermenting red grape musts supplemented with hydroxycinnamic acids. Int. J. Food Microbiol. 2007, 116, 144–152. [Google Scholar] [CrossRef] [PubMed]
- García, M.; Esteve-Zarzoso, B.; Cabellos, J.M.; Arroyo, T. Advances in the Study of Candida stellate. Fermentation 2018, 4, 74. [Google Scholar] [CrossRef]
- Mora, J.; Mulet, A. Effects of some treatments of grape juice on the population and growth of yeast species during fermentation. Am. J. Enol. Vitic. 1991, 42, 133–136. [Google Scholar]
- Jackson, R.S. Wine Science-Principles, Practice, Perception, 2nd ed.; Academic Press: San Diego, CA, USA, 2000. [Google Scholar]
- Maurizio, C.; Ferraro, L. Enhanced glycerol content in wines made with immobilized Candida stellata cells. Appl. Environ. Microbiol. 1996, 62, 128–132. [Google Scholar]
- Ciani, M.; Maccarelli, F. Oenological properties of non-Saccharomyces yeasts associated with wine-making. World J. Microbiol. Biotechnol. 1998, 14, 199–203. [Google Scholar] [CrossRef]
- Magyar, I.; Tóth, T. Comparative evaluation of some oenological properties in wine strains of Candida stellata, Candida zemplinina, Saccharomyces uvarum and Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 94–100. [Google Scholar] [CrossRef]
- Lafon-Lafourcade, S.; Lucmaret, V.; Joyeaux, A.; Ribereau-Gayon, P. Utilisation de levains mixtes dans l élaboration’des vins de pourriture noble en vue de réduire lácidité volatile. Comptes Rendues de L’Académie d’Agriculture 1981, 67, 616–622. [Google Scholar]
- Azzolini, M.; Tosi, E.; Lorenzini, M.; Finato, F.; Zapparoli, G. Contribution to the aroma of white wines by controlled Torulaspora delbrueckii cultures in association with Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2015, 31, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Azzolini, M.; Fedrizzi, B.; Tosi, E.; Finato, F.; Vagnoli, P.; Scrinzi, C.; Zapparoli, G. Effects of Torulaspora delbrueckii and Saccharomyces cerevisiae mixed cultures on fermentation and aroma of Amarone wine. Eur. Food Res. Technol. 2012, 235, 303–313. [Google Scholar] [CrossRef]
- Gordon, J.L.; Wolfe, K.H. Recent allopolyploid origin of Zygosaccharomyces rouxii strain ATCC 42981. Yeast 2008, 25, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Steels, H.; James, S.A.; Roberts, I.N.; Stratford, M. Zygosaccharomyces lentus: A significant new osmophilic, preservative-resistant spoilage yeast, capable of growth at low temperature. J. Appl. Microbiol. 1999, 87, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.; Suzzi, G. Higher Alcohol and Acetoin Production by Zygosaccharomyces Wine Yeasts. J. Appl. Bacteriol. 1993, 75, 541–545. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar-Powers, E. Zygosaccharomyces kombuchaensis, a new ascosporogenous yeast from ‘Kombucha tea’. FEMS Yeast Res. 2001, 1, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 2003, 3, 417–432. [Google Scholar] [CrossRef]
- Kurtzman, C.P. Phylogenetic circumscription of Saccharomyces Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003, 4, 233–245. [Google Scholar] [CrossRef]
- Cabral, S.; Prista, C.; Loureiro-Dias, M.; Leandro, M. Occurrence of FFZ genes in yeasts and correlation with fructophilic behavior. Microbiology 2015, 161, 2008–2018. [Google Scholar] [CrossRef]
- Martorell, P.; Stratford, M.; Steels, H.; Fernández-Espinar, M.T.; Querol, A. Physiological characterization of spoilage strains of Zygosaccharomyces bailii, Zygosaccharomyces rouxii isolated from high sugar environments. Int. J. Food Microbiol. 2007, 114, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Sá-Correia, I.; Guerreiro, J.F.; Loureiro-Dias, M.C.; Leão, C.; Côrte-Real, M. Zygosaccharomyces. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.-L., Eds.; Academic Press: New York, NY, USA, 2014; Volume 3, pp. 849–855. [Google Scholar]
- Rankine, B.C. Decomposition of L-malic acid by wine yeasts. J. Sci. Food Agric. 1966, 17, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Snow, P.G.; Gallander, J.F. Deacidification of white table wines through partial fermentation with Schizosaccharomyces pombe. Am. J. Enol. Vitic. 1979, 30, 45–48. [Google Scholar]
- Rodriquez, S.B.; Thornton, R.J. Factors influencing the utilization of L-malate by yeasts. FEMS Microbiol. Lett. 1990, 72, 17–22. [Google Scholar] [CrossRef]
- Seo, S.-H.; Rhee, C.-H.; Park, H.-D. Degradation of Malic Acid by Issatchenkia orientalis KMBL 5774, an Acidophilic Yeast Strain Isolated from Korean Grape Wine Pomace. J. Microbiol. 2007, 45, 521–527. [Google Scholar]
- Douglass, A.P.; Offei, B.; Braun-Galleani, S.; Coughlan, A.Y.; Martos, A.A.; Ortiz-Merino, R.A.; Byrne, K.P.; Wolfe, K.H. Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: One species, four names. PLoS Pathog. 2018, 14, e1007138. [Google Scholar] [CrossRef]
- Mills, D.A.; Johannsen, E.A.; Cocolin, L. Yeast diversity and persistence in botrytis-affected wine fermentations. Appl. Environ. Microbiol. 2002, 68, 4884–4893. [Google Scholar] [CrossRef]
- Englezos, V.; Rantsiou, K.; Torchio, F.; Rolle, L.; Gerbi, V.; Cocolin, L. Exploitation of the non-Saccharomyces yeast Starmerella bacillaris (synonym Candida zemplinina) in wine fermentation: Physiological and molecular characterizations. Int. J. Food Microbiol. 2015, 199, 33–40. [Google Scholar] [CrossRef]
- Magyar, I.; Nyitrai-Sárdy, D.; Leskó, A.; Pomázi, A.; Kállay, M. Anaerobic organic acid metabolism of Candida zemplinina in comparison with Saccharomyces wine yeasts. Int. J. Food Microbiol. 2014, 178, 1–6. [Google Scholar] [CrossRef]
- Romboli, Y.; Mangani, S.; Buscioni, G.; Granchi, L.; Vincenzini, M. Effect of Saccharomyces cerevisiae and Candida zemplinina on quercetin, vitisin A and hydroxytyrosol contents in Sangiovese wines. World J. Microbiol. Biotechnol. 2015, 31, 1137–1145. [Google Scholar] [CrossRef]
- Rantsiou, K.; Dolci, P.; Giacosa, S.; Torchio, F.; Tofalo, R.; Torriani, S.; Suzzi, G.; Rolle, L.; Cocolin, L. Candida zemplinina can reduce acetic acid production by Saccharomyces cerevisiae in sweet wine fermentations. Appl. Environ. Microbiol. 2012, 78, 1987–1994. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).