Effects of the Starch Molecular Structures in Barley Malts and Rice Adjuncts on Brewing Performance
Abstract
:1. Introduction
2. Results
2.1. Chemical Compostions and Enzymatic Activities of the Malts and Rice Adjuncts
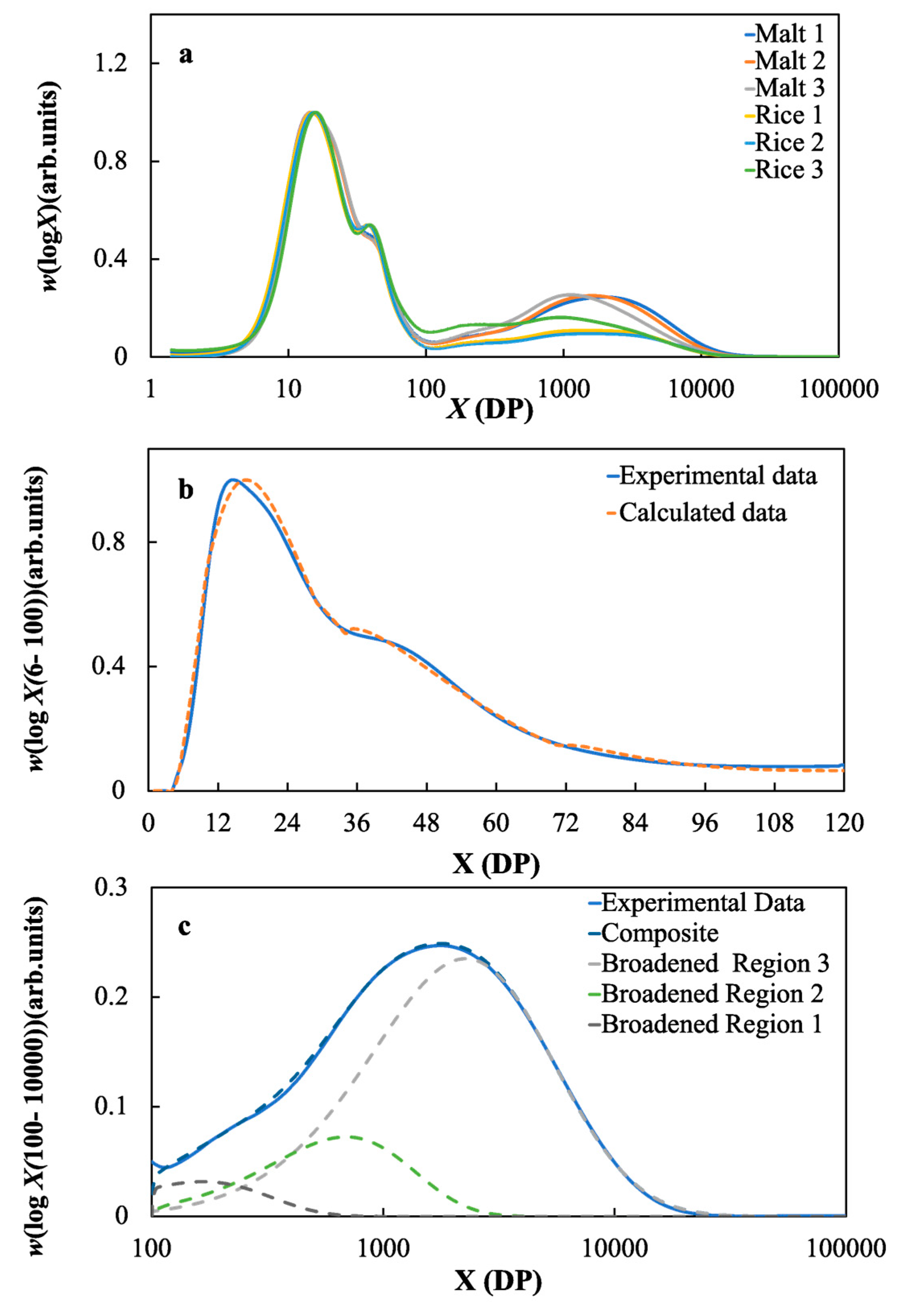
2.2. Starch Structural Parameters of the Malts and Rice Samples
2.3. Thermal Properties of the Malts and Rice adjuncts
2.4. Fermentable Sugars in the Wort
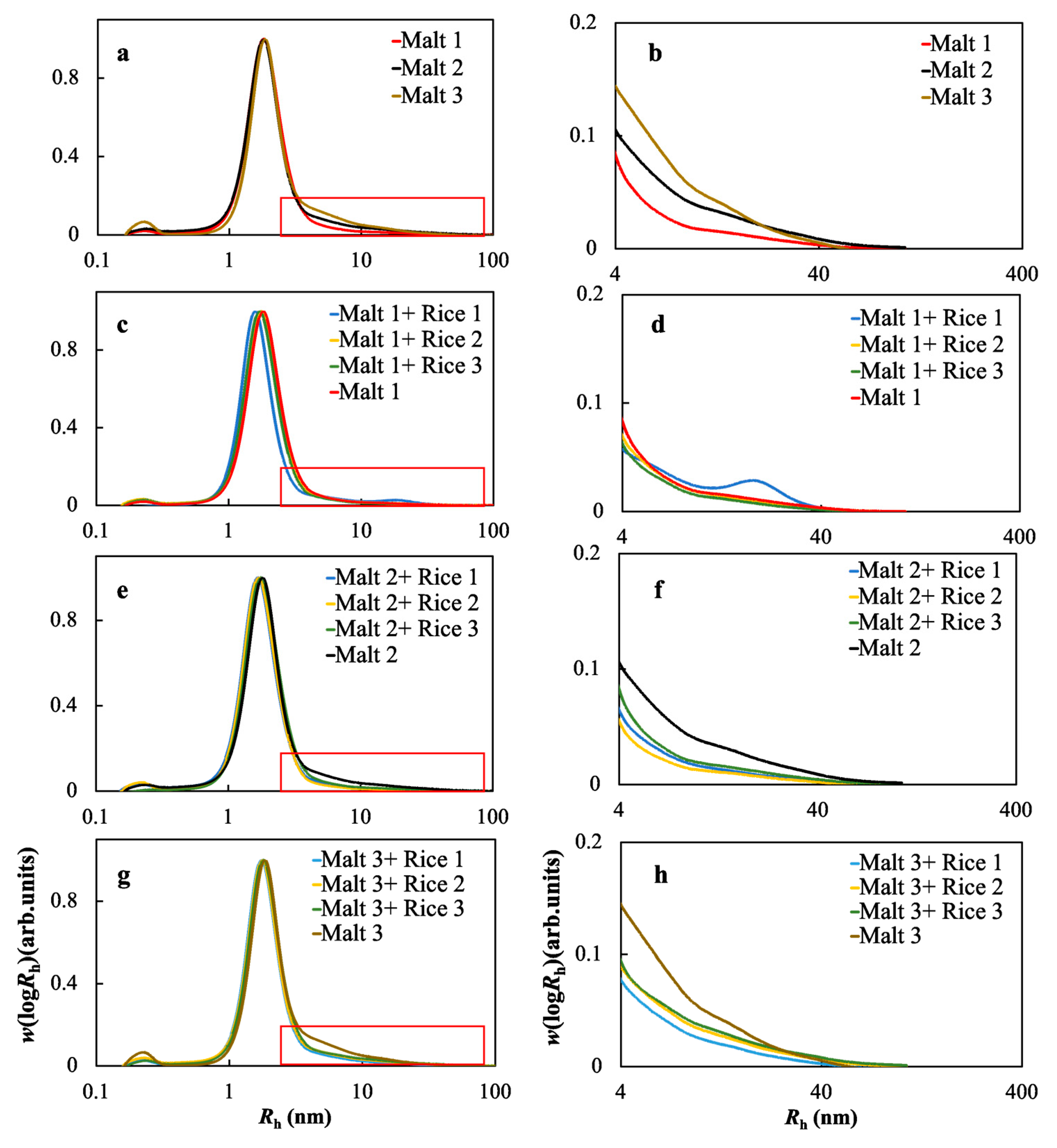
2.5. Molecular Size Distributions of the Soluble Starches in the Wort Samples
2.6. Contents of Soluble Starch and Nitrogen in Wort After Mashing
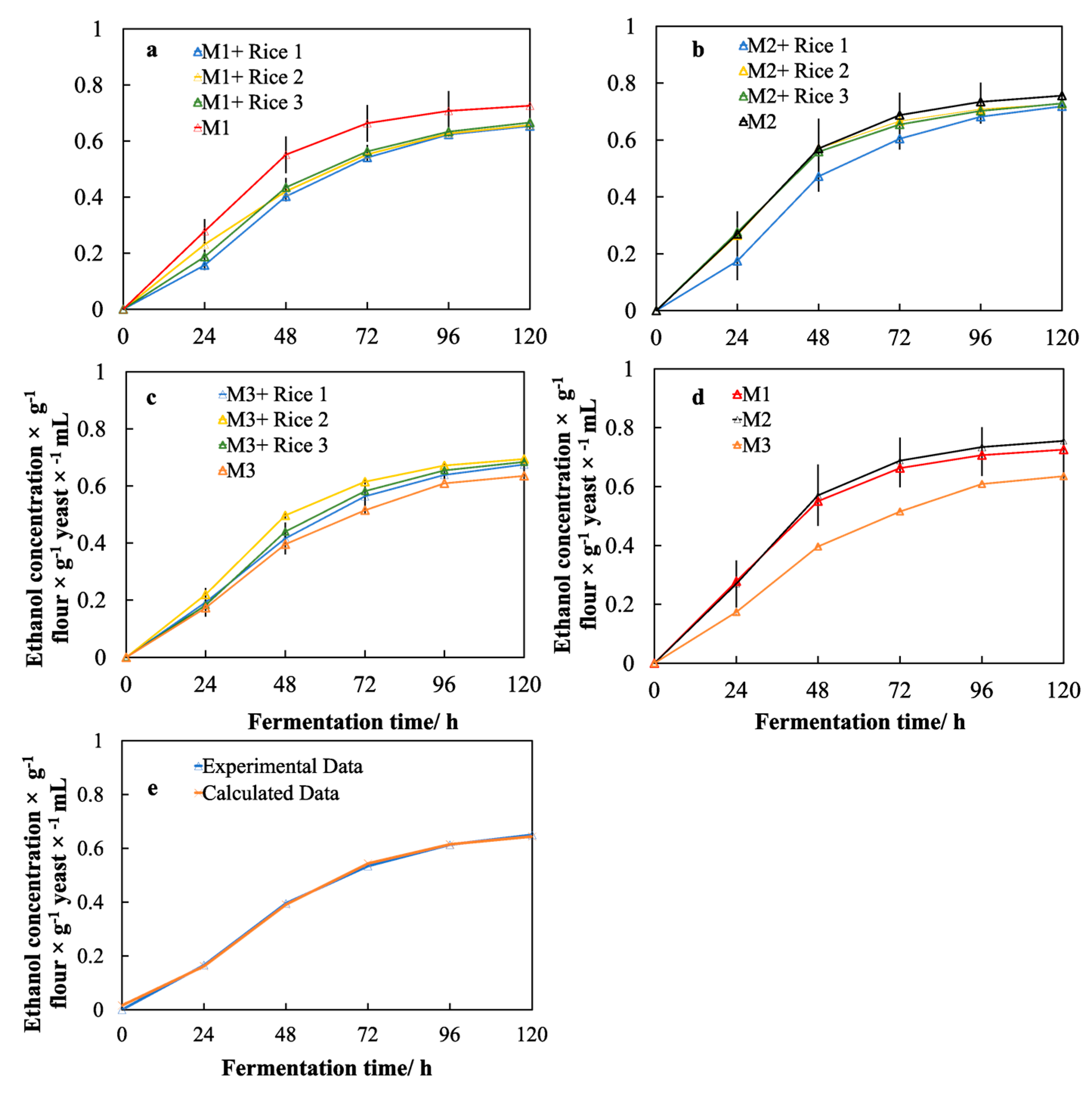
2.7. Ethanol Production and Model Fitting
2.8. Correlations Among Protein Contents, Starch Structural Parameters and Fermentable Sugar Contents in Wort Samples
2.9. Correlations among Fermentable Sugars, Starch Structural Parameters, Wort Components and Fermentation Parameters
3. Discussion
3.1. Effects of Different Malts on the Fermentable Sugar Contents in Wort Samples
3.2. Effects of Rice Adjuncts on Fermentable Sugar Production and Soluble Starch Content
- The increase of maltose content by the addition of rice adjuncts: During the pre- gelatinisation of rice starch, starch molecules would have leached out from inside granules, leading to the production of more rapidly degraded materials. These have starch chains exposed to the solution and so are readily available for attack by amylase [33,34]. Based on this, during mashing, the increase of those rapidly degraded materials would be easily degraded by both α- and β-amylase, leading to an increase of maltose content following the addition of rice adjunct.
- The decrease of glucose content by the addition of rice adjuncts: During mashing, significant β-glucanolysis will take place, and glucose would be further hydrolysed by various enzymes, to disaccharides and glucose [35]. The addition of rice adjuncts means less activity of amylase and amyloglucosidase, and β-glucanase will also release glucose by hydrolysing barley β-glucan. Accordingly, less glucose would be formed, as seen here.
3.3. Effects of Individual Rice Adjuncts on the Fermentable Sugar Contents in the Wort Samples
3.4. The Effects of the Molecular Structure of Starch on the Fermentable Sugar Content in the Wort Samples
3.5. Effects of Different Malts on Ethanol Production
3.6. Effects of the Rice Adjuncts on the Performance of Yeast Fermentation
3.7. Effects of the Molecular Structure of Rice Starch on the Performance of Yeast Fermentation
4. Materials and methods
4.1. Malts
4.2. Milling
4.3. Chemical Composition of the Malted Barley and Rice
4.4. Differential Scanning Calorimetry
4.5. Enzyme Activity of Amylases and Limit Dextrinase in the Malted Barley
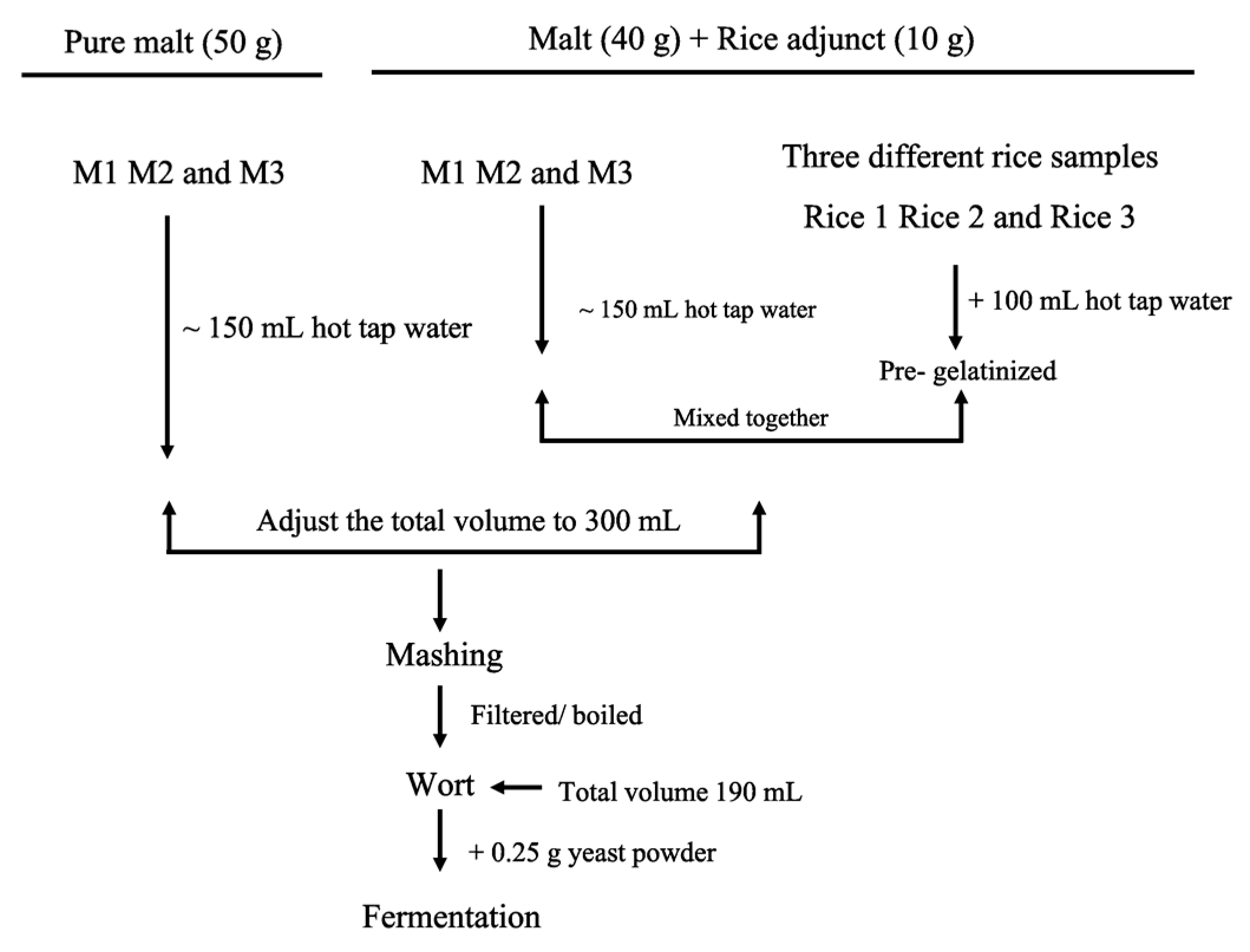
4.6. Pre-Gelatinisation of Rice
4.7. Mashing
4.8. Wort Filtration
4.9. Sugar Analysis of Wort Samples
4.10. Soluble Nitrogen Content in Wort
4.11. Fermentation
4.12. Original Gravity
4.13. Model to Predict Ethanol Production
4.14. Starch Extraction
4.15. Starch Structural Analysis
4.16. Amylose and Amylopectin Contents
4.17. Fitting Amylose and Amylopectin CLDs with Two Mathematical Models
4.18. Molecular Size Distributions of Soluble Starches in the Wort Samples
4.19. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Walker, G.M.; Stewart, G.G. Saccharomyces cerevisiae in the production of fermented beverages. Beverages 2016, 2, 30. [Google Scholar] [CrossRef]
- Yu, W.; Zou, W.; Dhital, S.; Wu, P.; Gidley, M.J.; Fox, G.P.; Gilbert, R.G. The adsorption of α-amylase on barley proteins affects the in vitro digestion of starch in barley flour. Food Chem. 2018, 241, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Agu, R.C. Some relationships between malted barleys of different nitrogen levels and the wort properties. J. Inst. Brew. 2003, 109, 106–109. [Google Scholar] [CrossRef]
- Evans, D.E.; Goldsmith, M.; Redd, K.S.; Nischwitz, R.; Lentini, A. Impact of mashing conditions on extract, its fermentability, and the levels of wort free amino nitrogen (FAN), beta-glucan, and lipids. J. Am. Soc. Brew. Chem. 2012, 70, 39–49. [Google Scholar]
- Gorinstein, S.; Zemser, M.; Vargas-Albores, F.; Ochoa, J.L.; Paredes-Lopez, O.; Scheler, C.; Salnikow, J.; Martin-Belloso, O.; Trakhtenberg, S. Proteins and amino acids in beers, their contents and relationships with other analytical data. Food Chem. 1999, 67, 71–78. [Google Scholar] [CrossRef]
- Procopio, S.; Krause, D.; Hofmann, T.; Becker, T. Significant amino acids in aroma compound profiling during yeast fermentation analyzed by PLS regression. LWT Food Sci. Technol. 2013, 51, 423–432. [Google Scholar] [CrossRef]
- Evans, D.E.; Dambergs, R.; Ratkowsky, D.; Li, C.; Harasymow, S.; Roumeliotis, S.; Eglinton, J.K. Refining the Prediction of Potential Malt Fermentability by Including an Assessment of Limit Dextrinase Thermostability and Additional Measures of Malt Modification, Using Two Different Methods for Multivariate Model Development. J. Inst. Brew. 2010, 116, 86–96. [Google Scholar] [CrossRef] [Green Version]
- Espinosa-Ramirez, J.; Perez-Carrillo, E.; Serna-Saldivar, S.O. Maltose and glucose utilization during fermentation of barley and sorghum lager beers as affected by beta-amylase or amyloglucosidase addition. J. Cereal Sci. 2014, 60, 602–609. [Google Scholar] [CrossRef]
- MacGregor, A.W.; Bazin, S.L.; Macri, L.J.; Babb, J.C. Modelling the contribution of alpha-amylase, beta-amylase and limit dextrinase to starch degradation during mashing. J. Cereal Sci. 1999, 29, 161–169. [Google Scholar] [CrossRef]
- Evans, D.E.; Collins, H.; Eglinton, J.; Wilhelmson, A. Assessing the impact of the level of diastatic power enzymes and their thermostability on the hydrolysis of starch during wort production to predict malt fermentability. J. Am. Soc. Brew. Chem. 2005, 63, 185–198. [Google Scholar] [CrossRef]
- Duke, S.H.; Henson, C.A. A Comparison of Barley Malt Amylolytic Enzyme Activities as Indicators of Malt Sugar Concentrations. J. Am. Soc. Brew. Chem. 2009, 67, 99–111. [Google Scholar] [CrossRef]
- Evans, D.E.; Fox, G.P. Comparison of Diastatic Power Enzyme Release and Persistence During Modified Institute of Brewing 65 degrees C and Congress Programmed Mashes. J. Am. Soc. Brew. Chem. 2017, 75, 302–311. [Google Scholar]
- Fox, G.; Yu, W.; Nischwitz, R.; Harasymow, S. Variation in maltose in sweet wort from barley malt and rice adjuncts with differences in amylose structure. J. Inst. Brew. 2018. [Google Scholar] [CrossRef]
- Gous, P.W.; Fox, G.P. Review: Amylopectin synthesis and hydrolysis - Understanding isoamylase and limit dextrinase and their impact on starch structure on barley (Hordeum vulgare) quality. Trends Food Sci. Technol. 2017, 62, 23–32. [Google Scholar] [CrossRef]
- Gous, P.W.; Warren, F.; Mo, O.W.; Gilbert, R.G.; Fox, G.P. The effects of variable nitrogen application on barley starch structure under drought stress. J. Inst. Brew. 2015, 121, 502–509. [Google Scholar] [CrossRef] [Green Version]
- Tester, R.F. Influence of growth conditions on barley starch properties. Int. J. Biol. Macromol. 1997, 21, 37–45. [Google Scholar] [CrossRef]
- Swanston, J.S.; Ellis, R.P.; Stark, J.R. Effects on grain and malting quality of genes altering barley starch composition. J. Cereal Sci. 1995, 22, 265–273. [Google Scholar] [CrossRef]
- Bamforth, C.W. Brewing and brewing research: Past, present and future. J. Sci. Food Agric. 2000, 80, 1371–1378. [Google Scholar] [CrossRef]
- Bamforth, C.W. Current perspectives on the role of enzymes in brewing. J. Cereal Sci. 2009, 50, 353–357. [Google Scholar] [CrossRef]
- De Rouck, G.; Jaskula, B.; De Causmaecker, B.; Malfliet, S.; Van Opstaele, F.; De Clippeleer, J.; De Brabanter, J.; De Cooman, L.; Aerts, G. The Influence of Very Thick and Fast Mashing Conditions on Wort Composition. J. Am. Soc. Brew. Chem. 2013, 71, 1–14. [Google Scholar] [CrossRef]
- Agu, R.C.; Palmer, G.H. alpha-Glucosidase activity of sorghum and barley malts. J. Inst. Brew. 1997, 103, 25–29. [Google Scholar] [CrossRef]
- You, S.G.; Izydorczyk, M.S. Molecular characteristics of barley starches with variable amylose content. Carbohydr. Polym. 2002, 49, 33–42. [Google Scholar] [CrossRef]
- Yu, W.; Tan, X.; Zou, W.; Hu, Z.; Fox, G.P.; Gidley, M.J.; Gilbert, R.G. Relationships between protein content, starch molecular structure and grain size in barley. Carbohydr. Polym. 2017, 155, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Li, C.; Yu, W.; Gilbert, R.G.; Li, E. How amylose molecular fine structure of rice starch affects functional properties. Carbohydr. Polym. 2009, 204, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Li, H.; Zou, W.; Tao, K.; Zhu, J.; Gilbert, R.G. Using starch molecular fine structure to understand biosynthesis-structure-property relations. Trends Food Sci. Technol. 2018. [Google Scholar] [CrossRef]
- Yu, W.; Tao, K.; Gilbert, R.G. Improved methodology for analyzing relations between starch digestion kinetics and molecular structure. Food Chem. 2018, 264, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.C. A comparison of maize, sorghum and barley as brewing adjuncts. J. Inst. Brew. 2002, 108, 19–22. [Google Scholar]
- Rubsam, H.; Gastl, M.; Becker, T. Influence of the range of molecular weight distribution of beer components on the intensity of palate fullness. Eur. Food Res. Technol. 2013, 236, 65–75. [Google Scholar] [CrossRef]
- Rubsam, H.; Gastl, M.; Becker, T. Determination of the influence of starch sources and mashing procedures on the range of the molecular weight distribution of beer using field-flow fractionation. J. Inst. Brew. 2013, 119, 139–148. [Google Scholar] [CrossRef]
- Yu, W.; Tao, K.; Gidley, M.J.; Fox, G.P.; Gilbert, R.G. Molecular brewing: Molecular structural effects involved in barley malting and mashing. Carbohydr. Polym. 2019, 206, 583–592. [Google Scholar]
- Zou, W.; Sissons, M.; Gidley, M.J.; Gilbert, R.G.; Warren, F.J. Combined techniques for characterising pasta structure reveals how the gluten network slows enzymic digestion rate. Food Chem. 2015, 188, 559–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, R.S.; Manners, D.J. The action of malted-barley alpha-amylase on amylopectin. Carbohydr. Res. 1978, 66, 295–297. [Google Scholar] [CrossRef]
- Baldwin, A.J.; Egan, D.L.; Warren, F.J.; Barker, P.D.; Dobson, C.M.; Butterworth, P.J.; Ellis, P.R. Investigating the Mechanisms of Amylolysis of Starch Granules by Solution-State NMR. Biomacromolecules 2015, 16, 1614–1621. [Google Scholar] [CrossRef] [PubMed]
- Warren, F.J.; Royall, P.G.; Gaisford, S.; Butterworth, P.J.; Ellis, P.R. Binding interactions of α-amylase with starch granules: The influence of supramolecular structure and surface area. Carbohydr. Polym. 2011, 86, 1038–1047. [Google Scholar] [CrossRef]
- Bamforth, C.; Martin, H.L. The degradation of β-glucan during malting and mashing: The role of β-glucanase. J. Inst. Brew. 1983, 89, 303–307. [Google Scholar] [CrossRef]
- Tester, R.F.; Morrison, W.R. Swelling and gelatinization of cereal starches. I. Effects of amylopectin, amylose, and lipids. Cereal Chem. 1990, 67, 551–557. [Google Scholar]
- Noda, T.; Takahata, Y.; Sato, T.; Suda, I.; Morishita, T.; Ishiguro, K.; Yamakawa, O. Relationships between chain length distribution of amylopectin and gelatinization properties within the same botanical origin for sweet potato and buckwheat. Carbohydr. Polym. 1998, 37, 153–158. [Google Scholar] [CrossRef]
- O’Connor-Cox, E.S.C.; Ingledew, W.M. Wort Nitrogenous Sources—Their Use by Brewing Yeasts: A Review. J. Am. Soc. Brew. Chem. 1989, 47, 102–108. [Google Scholar] [CrossRef]
- Mallawarachchi, K.S.; Bandara, L.R.L.M.; Dilshan, S.K.D.H.S.; Ariyadasa, T.U.; Gunawardena, S.H.P. Optimization of mashing process in beer production using rice as an adjunct. In Proceedings of the 2016 Moratuwa Engineering Research Conference (MERCon), Moratuwa, Sri Lanka, 5–6 April 2016; pp. 289–292. [Google Scholar]
- Zwietering, M.; Jongenburger, I.; Rombouts, F.; Van’t Riet, K. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar]
- Vilaplana, F.; Gilbert, R.G. Characterization of branched polysaccharides using multiple-detection size separation techniques. J. Sep. Sci. 2010, 33, 3537–3554. [Google Scholar] [CrossRef]
- Castro, J.V.; Ward, R.M.; Gilbert, R.G.; Fitzgerald, M.A. Measurement of the molecular weight distribution of debranched starch. Biomacromolecules 2005, 6, 2260–2270. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M.A.; Bergman, C.J.; Resurreccion, A.P.; Möller, J.; Jimenez, R.; Reinke, R.F.; Martin, M.; Blanco, P.; Molina, F.; Chen, M.-H.; et al. Addressing the Dilemmas of Measuring Amylose in Rice. Cereal Chem. 2009, 86, 492–498. [Google Scholar] [CrossRef]
- Vilaplana, F.; Hasjim, J.; Gilbert, R.G. Amylose content in starches: Toward optimal definition and validating experimental methods. Carbohydr. Polym. 2012, 88, 103–111. [Google Scholar] [CrossRef]
- Feng, G.; Flanagan, B.M.; Williams, B.A.; Mikkelsen, D.; Yu, W.; Gidley, M.J. Extracellular depolymerisation triggers fermentation of tamarind xyloglucan and wheat arabinoxylan by a porcine faecal inoculum. Carbohydr. Polym. 2018, 201, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.C.; Gilbert, R.G. Molecular weight distributions of starch branches reveal genetic constraints on biosynthesis. Biomacromolecules 2010, 11, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.C.; Morell, M.K.; Gilbert, R.G. A parameterized model of amylopectin synthesis provides key insights into the synthesis of granular starch. PLoS ONE 2013, 8, e65768. [Google Scholar] [CrossRef] [PubMed]
- Nada, S.S.; Zou, W.; Li, C.; Gilbert, R.G. Parameterizing amylose chain-length distributions for biosynthesis-structure-property relations. Anal. Bioanal. Chem. 2017, 409, 6813–6819. [Google Scholar] [CrossRef]
| Sample ID | Industrial Utilisation | Amylose Content (%) | Moisture Content (%) | Protein Content 2 (%) | Starch Content 2 (%) | Enzyme Activity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α-amylase 2 | β-amylase 2 | Limit Dextrinase 2 | ||||||||||
| Malt 1 | Pale | (32.5 ± 3.2)a | (2.3 ± 0.5)a | (8.8 ± 0.1)b | (58.3 ±0.6)a | (239.3 ±1.0)b | (99.3 ± 2.9)a | (2.0 ± 0.2)a | ||||
| Malt 2 | Amber | (32.3 ± 1.1)a | (3.1 ± 0.4)a | (8.4 ± 0.1)a | (56.6 ±1.3)a | (202.6 ±10.2)a | (147.2 ± 3.8)b | (9.7 ± 0.3)c | ||||
| Malt 3 | Ale | (32.0 ± 0.8)a | (2.3 ± 0.3)a | (9.0 ± 0.0)b | (62.0 ±0.4)b | (433.6 ±5.1)c | (98.8 ± 1.0)a | (3.2 ± 0.3)b | ||||
| Rice 1 | Food | (19.6 ± 0.0)a | (11.8 ± 0.7)a | (12.5 ± 0.1)b | (78.2 ±2.0)a | ND | ND | ND | ||||
| Rice 2 | Food | (18.2 ±0.4)a | (12.4 ± 0.3)a | (13.9 ± 0.1)a | (79.6 ±2.9)a | ND | ND | ND | ||||
| Rice 3 | Food | (27.6 ± 1.1)b | (11.8 ± 0.4)a | (11.7 ± 0.0)c | (81.3 ±1.2)a | ND | ND | ND | ||||
| Amylose and amylopectin fitting parameters | ||||||||||||
| hAm, 3 × 102 | hAm, 2 × 102 | hAm, 1 × 102 | βAm, 3 × 104 | βAm, 2 × 104 | βAm, 1 × 103 | hAp, 1 | hAp, 2 × 102 | hAp, 3 ×104 | βAp, 1 × 102 | βAp, 2 × 102 | βAp, 3 × 102 | |
| Malt 1 | (15.9 ± 1.9)a | (10.4 ± 1.8)a | (3.5 ± 2.1)a | (5.8 ± 0.1)a | (23.1 ± 0.4)a | (12.4 ± 0.15)a | (1.0 ± 0.0)b | (2.4 ± 0.0)b | (8.2 ± 0.1)a | (10.4 ± 0.1)a | (5.5 ± 0.2)a | (2.7 ± 0.0)a |
| Malt 2 | (14.5 ± 0.8)a | (11.9 ± 1.1)a | (3.4 ± 0.4)a | (6.2 ± 0.1)b | (22.2 ± 0.2)a | (14.0 ± 1.4)a | (1.0 ± 0.0)b | (2.3 ± 0.0)ab | (7.1 ± 0.0)a | (10.5 ± 0.1)a | (5.5 ± 0.1)a | (2.8 ± 0.0)a |
| Malt 3 | (12.3 ± 8.8)a | (13.3 ± 10.5)a | (3.7 ± 0.7)a | (7.5 ± 0.6)c | (26.0 ± 4.0)a | (13.0 ± 1.36)a | (1.1 ± 0.0)b | (2.3 ± 0.0)a | (6.9 ± 0.0)a | (10.4 ± 0.0)a | (5.4 ± 0.1)a | (2.8 ± 0.2)a |
| Rice 1 | (8.1 ± 0.2)b | (4.7 ± 0.10)a | (3.0 ± 0.2)a | (7.2 ± 0.0)a | (28.7 ± 0.7)a | (12.9 ± 0.5)a | (0.9 ± 0.0)a | (3.4 ± 0.0)c | (9.4 ± 1.3)a | (12.0 ± 0.1)c | (6.9 ± 0.2)c | (4.5 ± 0.1)d |
| Rice 2 | (7.3 ± 0.3)a | (4.0 ± 0.1)a | (2.6 ± 0.2)a | (7.0 ± 0.1)a | (29.9 ± 0.9)a | (12.9 ± 0.5)a | (0.9 ± 0.0)a | (3.5 ± 0.0)c | (8.7 ± 0.6)a | (11.7 ± 0.0)b | (6.6 ± 0.1)c | (4.1 ± 0.0)c |
| Rice 3 | (9.1 ± 0.3)c | (8.4 ± 1.4)b | (7.2 ± 0.3)b | (7.8 ± 0.9)a | (31.5 ± 1.5)a | (13.5 ± 0.6)a | (1.0 ± 0.0)b | (3.9 ± 0.1)d | (19.1 ± 0.1)b | (11.7 ± 0.0)b | (6.0 ± 0.0)b | (3.2 ± 0.1)b |
| Gelatinisation characteristics of malt and rice samples | ||||||||||||
| To | Tp | Tc | Ethalpy (J/g) | |||||||||
| Malt 1 | (57.3 ± 0.2)a | (63.3 ± 0.1)a | (68.7 ± 0.0)a | (6.7 ± 0.2)a | ||||||||
| Malt 2 | (57.0 ± 0.3)a | (63.2 ± 0.5)a | (69.7 ± 0.2)b | (6.5 ± 0.1)a | ||||||||
| Malt 3 | (57.9 ± 0.5)a | (62.9 ± 0.0)a | (68.1 ± 0.3)a | (7.2 ± 0.9)a | ||||||||
| Rice 1 | (60.0 ± 0.1)b | (67.8 ± 0.9)b | (72.8 ± 0.2)b | (9.8 ± 0.2)a | ||||||||
| Rice 2 | (58.1 ± 0.1)a | (64.9 ± 0.4)a | (71.5 ± 0.7)a | (9.5 ± 1.3)a | ||||||||
| Rice 3 | (69.8 ± 0.2)c | (74.2 ± 0.2)c | (79.9 ± 0.4)c | (9.9 ± 0.7)a | ||||||||
| Sample ID | Glucose (mg/g) | Sucrose (mg/g) | Maltose (mg/g) | Maltotriose (mg/g) | Total Sugar (mg/g) |
|---|---|---|---|---|---|
| Pure Malt 1 | (44.4 ± 5.2)ab | (32.3 ± 5.2)bc | (463.8 ± 48.3)a | (122.4 ± 13.7)ab | (663.8 ± 72.4)a |
| Malt 1 + Rice 1 | (42.0 ± 1.8)ab | (31.1 ± 2.5)abc | (541.6 ± 34.6)bc | (135.9 ± 1.9)bc | (750.6 ± 40.7)abc |
| Malt 1 + Rice 2 | (38.8 ± 2.7)a | (28.1 ± 0.7)a | (480.3 ± 16.2)ab | (118.2 ± 10.7)ab | (662.3 ± 30.2)a |
| Malt 1 + Rice 3 | (38.8 ± 4.1)a | (27.0 ± 3.4)ab | (506.1 ± 28.5)abc | (131.6 ± 13.7)ab | (703.5 ± 49.6)abc |
| Pure Malt 2 | (65.3 ± 5.3)f | (36.4 ± 4.0)c | (498.2 ± 37.0)abc | (157.7± 8.0)d | (757.6 ± 54.2)abc |
| Malt 2 + Rice 1 | (57.5 ± 0.6)e | (31.8 ± 0.7)abc | (552.6 ± 15.2)c | (163.9 ± 7.1)d | (805.8 ± 8.3)c |
| Malt 2 + Rice 2 | (56.3 ± 4.0)de | (31.0 ± 3.4)abc | (532.4 ± 44.4)abc | (154.1 ± 10.1)cd | (773.8 ± 61.9)bc |
| Malt 2 + Rice 3 | (57.0 ± 1.4)e | (30.9 ± 4.7)abc | (531.96 ± 22.0)abc | (157.2 ± 7.5)d | (777.0 ± 35.6)bc |
| Pure Malt 3 | (53.1 ± 3.3)cde | (31.4 ± 3.9)abc | (475.1 ± 21.9)ab | (111.3 ± 6.0)a | (670.8 ± 35.1)a |
| Malt 3 + Rice 1 | (49.2 ± 2.7)abc | (28.4 ± 2.7)abc | (540.6 ± 18.9)bc | (119.3 ± 4.0)ab | (737.5 ± 28.3)abc |
| Malt 3 + Rice 2 | (47.7 ± 0.7)bc | (27.9 ± 0.6)ab | (530.6 ± 5.3)abc | (115.8 ± 0.8)ab | (721.9 ± 4.4)abc |
| Malt 3 + Rice 3 | (44.5 ± 3.1)ab | (24.9 ± 3.2)a | (513.9 ± 12.3)abc | (112.1 ± 8.1)a | (695.4 ± 26.8)ab |
| Soluble Starch Content (mg/mL) 1 | Soluble Nitrogen Content (mg/ mL) 1 | |
|---|---|---|
| Pure Malt 1 | (1.5 ± 0.1)d | (182.1 ± 46.7)bc |
| Malt 1 + Rice 1 | (1.9 ± 0.0)f | (147.0 ± 10.1)abc |
| Malt 1 + Rice 2 | (1.7 ± 0.0)ef | (147.8 ± 0.4)abc |
| Malt 1 + Rice 3 | (1.7 ± 0.0)ef | (145.6 ± 9.6)ab |
| Pure Malt 2 | (0.7 ± 0.1)a | (185.6 ± 24.4)c |
| Malt 2 + Rice 1 | (1.1 ± 0.1)b | (147.9 ± 6.6)abc |
| Malt 2 + Rice 2 | (1.1 ± 0.1)b | (153.7 ± 3.2)abc |
| Malt 2 + Rice 3 | (1.0 ± 0.0)b | (145.4 ± 1.9)ab |
| Pure Malt 3 | (1.3 + 0.1)c | (147.4 ± 1.2)abc |
| Malt 3 + Rice 1 | (1.6 ± 0.0)e | (142.6 ± 7.8)ab |
| Malt 3 + Rice 2 | (1.7 ± 0.0)e | (146.5 ± 3.8)abc |
| Malt 3 + Rice 3 | (1.7 ± 0.1)ef | (137.6 ± 5.6)a |
| Sample ID | Experimental Parameter | Model Fitting Parameters | ||
|---|---|---|---|---|
| CExp × 102 (%) | Cfinal × 102 (%) | k × 10 (% h-1) | T (h) | |
| Pure Malt 1 | (72.5 ± 14.5)a | (72.0 ± 11.4)a | (2.0 ± 0.1)abc | (5.8 ± 1.1)a |
| M1 + Rice 1 | (65.3 ± 0.0)a | (66.4 ± 0.3)a | (1.6 ± 0.1)ab | (9.5 ± 2.1)a |
| M1 + Rice 2 | (65.7 ± 0.4)a | (66.4 ± 1.1)a | (1.7 ± 0.1)abc | (8.7 ± 3.7)a |
| M1 + Rice 3 | (66.6 ± 2.3)a | 66.8 ± 0.9)a | (1.8 ± 0.3)abc | (8.3 ± 1.4)a |
| Pure Malt 2 | (72.5 ± 12.9)a | (75.3 ± 9.5)a | (2.2 ± 0.5)bc | (8.7 ± 2.9)a |
| M2 + Rice 1 | (71.8 ± 3.4)a | (71.3 ± 2.3)a | (1.9 ± 0.0)abc | (11.9 ± 7.4)a |
| M2 + Rice 2 | (72.8 ± 1.2)a | (72.0 ± 0.6)a | (2.3 ± 0.2)c | (7.9 ± 2.1)a |
| M2 + Rice 3 | (72.8 ± 1.1)a | (72.0 ± 1.7)a | (1.1 ± 0.2)abc | (5.3 ± 0.9)a |
| Pure Malt 3 | (63.5 ± 1.8)a | (64.9 ± 1.1)a | (1.5 ± 0.1)a | (6.7 ± 1.6)a |
| M3 + Rice 1 | (67.4 ± 1.7)a | (69.0 ± 0.6)a | (1.6 ± 0.3)ab | (8.2 ± 4.6)a |
| M3 + Rice 2 | (69.5 ± 1.9)a | (72.0 ± 4.1)a | (1.7 ± 0.2)abc | (7.0 ± 1.0)a |
| M3 + Rice 3 | (68.4 ± 0.5)a | (69.0 ± 0.3)a | (1.7 ± 0.0)abc | (8.7 ± 3.4)a |
| Experimental Parameter | Model Fitting Parameters | Fermentable Sugars | TotalProtein | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CExp | Cfinal | k | T | Glucose | Sucrose | Maltose | Maltotriose | TotalSugar | |||
| Fermentable sugars | Sucrose | 0.59 * | 0.55 | 0.53 | −0.07 | 0.72 ** | |||||
| Maltose | 0.15 | 0.19 | −0.01 | 0.45 | 0.24 | -0.06 | |||||
| Maltotriose | 0.70 * | 0.58 * | 0.70 * | 0.30 | 0.67 * | 0.59 * | 0.46 | 1.00 | |||
| Total sugar | 0.54 | 0.51 | 0.42 | 0.40 | 0.65 * | 0.40 | 0.84 ** | 0.85 ** | 1.00 | ||
| Total protein | −0.62 * | −0.51 | −0.60 * | 0.10 | −0.67 * | −0.84 ** | 0.25 | −0.62 * | −0.28 | 1.00 | |
| Total starch | −0.65 * | −0.52 | −0.75 ** | −0.04 | −0.52 | −0.80 ** | 0.20 | −0.73 ** | −0.32 | 0.83 ** | |
| Soluble nitrogen content | 0.62 * | 0.60 * | 0.63 * | −0.19 | 0.38 | 0.76 ** | −0.49 | 0.28 | −0.06 | −0.72 ** | |
| Soluble starch content | −0.73 ** | −0.66 * | −0.70 * | 0.05 | −0.93 ** | −0.71 ** | −0.02 | −0.73 ** | −0.53 | 0.81 ** | |
| Amylose content | −0.36 | −0.33 | −0.48 | −0.43 | −0.25 | −0.27 | −0.43 | −0.64 * | −0.59 * | 0.03 | |
| Amylose fitting parameters | hAm, 3 | 0.07 | −0.06 | 0.22 | −0.21 | −0.29 | 0.31 | −0.65 * | 0.03 | −0.41 | −0.49 |
| hAm, 2 | −0.14 | −0.04 | −0.34 | −0.34 | 0.26 | −0.02 | −0.21 | −0.45 | −0.26 | −0.05 | |
| hAm, 1 | −0.28 | −0.29 | −0.37 | −0.35 | −0.30 | −0.45 | −0.16 | −0.42 | −0.36 | 0.09 | |
| Amylopectin fitting parameters | hAp, 3 | −0.60 * | −0.47 | −0.74 ** | −0.16 | −0.36 | −0.65 * | 0.02 | −0.75 ** | −0.41 | 0.59 * |
| hAp, 2 | −0.40 | −0.39 | −0.40 | 0.10 | −0.55 | −0.87 ** | 0.46 | −0.27 | 0.01 | 0.74 ** | |
| hAp, 1 | −0.23 | −0.31 | −0.21 | −0.11 | −0.54 | −0.62 * | 0.09 | −0.16 | −0.15 | 0.24 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, W.; Quek, W.P.; Li, C.; Gilbert, R.G.; Fox, G.P. Effects of the Starch Molecular Structures in Barley Malts and Rice Adjuncts on Brewing Performance. Fermentation 2018, 4, 103. https://doi.org/10.3390/fermentation4040103
Yu W, Quek WP, Li C, Gilbert RG, Fox GP. Effects of the Starch Molecular Structures in Barley Malts and Rice Adjuncts on Brewing Performance. Fermentation. 2018; 4(4):103. https://doi.org/10.3390/fermentation4040103
Chicago/Turabian StyleYu, Wenwen, Wei Ping Quek, Cheng Li, Robert G. Gilbert, and Glen P. Fox. 2018. "Effects of the Starch Molecular Structures in Barley Malts and Rice Adjuncts on Brewing Performance" Fermentation 4, no. 4: 103. https://doi.org/10.3390/fermentation4040103
APA StyleYu, W., Quek, W. P., Li, C., Gilbert, R. G., & Fox, G. P. (2018). Effects of the Starch Molecular Structures in Barley Malts and Rice Adjuncts on Brewing Performance. Fermentation, 4(4), 103. https://doi.org/10.3390/fermentation4040103






