Changes in the Composition of the Lactic Acid Bacteria Behavior and the Diversity of Oenococcus oeni Isolated from Red Wines Supplemented with Selected Grape Phenolic Compounds
Abstract
:1. Introduction
2. Results
2.1. Effect of Grape Phenolic Compounds on the Growth of LAB
2.2. Influence of the Natural Phenolics Studied on the LAB Metabolism
2.3. Diversity of O. oeni Isolated From Red Wines Treated with Flavonols and Trans-Resveratrol
3. Discussion
4. Materials and Methods
4.1. Malolactic Fermentation Experiments in Red Wine
4.2. Sugars and Organic Acids Determination
4.3. LAB Quantification, Isolation, and Identification
4.4. Commercial LAB Strains
4.5. REA-PFGE
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rammelsberg, M.; Müller, E.; Radler, F. Caseicin 80: Purification and characterization of a new bacteriocin from Lactobacillus casei. Arch. Microbiol. 1990, 154, 249–252. [Google Scholar] [CrossRef]
- Henick-Kling, T.; Stoewsand, G. Lead in wine. Am. J. Enol. Vitic. 1993, 44, 459–463. [Google Scholar]
- Mcdaniel, M.; Henderson, L.; Watson, J.R.; Heatherbell, D. Sensory panel training and screening for descriptive analysis of the aroma of Pinot Noir wine fermented by several strains of malolactic bacteria. Descr. Sens. Anal. Pract. 2008, 351. [Google Scholar] [CrossRef]
- Laurent, M.-H.; Henick-Kling, T.; Acree, T. Changes in the aroma and odor of Chardonnay wine due to malolactic fermentation. Weinwissenschaft 1994, 49, 3–10. [Google Scholar]
- Davis, C.; Wibowo, D.; Lee, T.; Fleet, G. Growth and metabolism of lactic acid bacteria during and after malolactic fermentation of wines at different pH. Appl. Environ. Microbiol. 1986, 51, 539–545. [Google Scholar] [PubMed]
- El Khoury, M.; Campbell-Sills, H.; Salin, F.; Guichoux, E.; Claisse, O.; Lucas, P.M. Biogeography of Oenococcus oeni reveals distinctive but non-specific populations in wine-producing regions. Appl. Environ. Microbiol. 2016. [Google Scholar] [CrossRef]
- Cafaro, C.; Bonomo, M.G.; Guerrieri, A.; Crispo, F.; Ciriello, R.; Salzano, G. Assessment of the genetic polymorphism and physiological characterization of indigenous Oenococcus oeni strains isolated from Aglianico del Vulture red wine. Folia Microbiol. 2016, 61, 1–10. [Google Scholar] [CrossRef]
- Breniaux, M.; Dutilh, L.; Petrel, M.; Gontier, E.; Campbell-Sills, H.; Deleris-Bou, M.; Reguant, C. Adaptation of two groups of Oenococcus oeni strains to red and white wines: The role of acidity and phenolic compounds. J. Appl. Microbiol. 2018. [Google Scholar] [CrossRef]
- Bridier, J.; Claisse, O.; Coton, M.; Coton, E.; Lonvaud-Funel, A. Evidence of distinct populations and specific subpopulations within the species Oenococcus oeni. Appl. Environ. Microbiol. 2010, 76, 7754–7764. [Google Scholar] [CrossRef]
- Campbell-Sills, H.; El Khoury, M.; Favier, M.; Romano, A.; Biasioli, F.; Spano, G.; Coton, M. Phylogenomic analysis of Oenococcus oeni reveals specific domestication of strains to cider and wines. Genome Biol. Evol. 2015, 7, 1506–1518. [Google Scholar] [CrossRef]
- Campbell-Sills, H.; Lorentzen, M.; Lucas, P.M. Genomic Evolution and Adaptation to Wine of Oenococcus oeni. In Biology of Microorganisms on Grapes, in Must and in Wine; Springer: Cham, Switzerland, 2017; pp. 457–468. [Google Scholar]
- del Carmen Portillo, M.; Mas, A. Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. LWT-Food Sci. Technol. 2016, 72, 317–321. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Tabasco, R.; Requena, T.; Claisse, O.; Lonvaud-Funel, A.; Bartolomé, B.; Moreno-Arribas, M.V. Genetic diversity of Oenoccoccus oeni isolated from wines treated with phenolic extracts as antimicrobial agents. Food Microbiol. 2013, 36, 267–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petri, A.; Pfannebecker, J.; Fröhlich, J.; König, H. Fast identification of wine related lactic acid bacteria by multiplex PCR. Food Microbiol. 2013, 33, 48–54. [Google Scholar] [CrossRef] [PubMed]
- López, I.; Torres, C.; Ruiz-Larrea, F. Genetic typification by pulsed-field gel electrophoresis (PFGE) and randomly amplified polymorphic DNA (RAPD) of wild Lactobacillus plantarum and Oenococcus oeni wine strains. Eur. Food Res. Technol. 2008, 227, 547–555. [Google Scholar] [CrossRef]
- Claisse, O.; Lonvaud-Funel, A. Development of a multilocus variable number of tandem repeat typing method for Oenococcus oeni. Food Microbiol. 2012, 30, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Sacchi, K.L.; Bisson, L.F.; Adams, D.O. A review of the effect of winemaking techniques on phenolic extraction in red wines. Am. J. Enol. Vitic. 2005, 56, 197–206. [Google Scholar]
- Del Llaudy, M.C.; Canals, R.; Canals, J.M.; Zamora, F. Influence of ripening stage and maceration length on the contribution of grape skins, seeds and stems to phenolic composition and astringency in wine-simulated macerations. Eur. Food Res. Technol. 2008, 226, 337–344. [Google Scholar] [CrossRef]
- Setford, P.C.; Jeffery, D.W.; Grbin, P.R.; Muhlack, R.A. Factors affecting extraction and evolution of phenolic compounds during red wine maceration and the role of process modelling. Trends Food Sci. Technol. 2017, 69, 106–117. [Google Scholar] [CrossRef]
- Olejar, K.J.; Fedrizzi, B.; Kilmartin, P.A. Influence of harvesting technique and maceration process on aroma and phenolic attributes of Sauvignon blanc wine. Food Chem. 2015, 183, 181–189. [Google Scholar] [CrossRef]
- Caridi, A.; Sidari, R.; Giuffrè, A.M.; Pellicanò, T.M.; Sicari, V.; Zappia, C.; Poiana, M. Test of four generations of Saccharomyces cerevisiae concerning their effect on antioxidant phenolic compounds in wine. Eur. Food Res. Technol. 2017, 243, 1287–1294. [Google Scholar] [CrossRef]
- Monagas, M.; Bartolomé, B.; Gómez-Cordovés, C. Updated knowledge about the presence of phenolic compounds in wine. Crit. Rev. Food Sci. Nutr. 2005, 45, 85–118. [Google Scholar] [CrossRef] [PubMed]
- Hernández, T.; Estrella, I.; Carlavilla, D.; Martín-Álvarez, P.J.; Moreno-Arribas, M.V. Phenolic compounds in red wine subjected to industrial malolactic fermentation and ageing on lees. Anal. Chim. Acta 2006, 563, 116–125. [Google Scholar] [CrossRef]
- Bouzanquet, Q.; Barril, C.; Clark, A.C.; Dias, D.A.; Scollary, G.R. A novel glutathione-hydroxycinnamic acid product generated in oxidative wine conditions. J. Agric. Food Chem. 2012, 60, 12186–12195. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.; Oliveira, C.; Santos, C.; Campos, F.M.; Couto, J.A. Phenolic composition of monovarietal red wines regarding volatile phenols and its precursors. Eur. Food Res. Technol. 2018, 244, 1985–1994. [Google Scholar] [CrossRef]
- Burns, J.; Gardner, P.T.; O’Neil, J.; Crawford, S.; Morecroft, I.; McPhail, D.B.; Lean, M.E. Relationship among antioxidant activity, vasodilation capacity, and phenolic content of red wines. J. Agric. Food Chem. 2000, 48, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Bae, H. An overview of stress-induced resveratrol synthesis in grapes: Perspectives for resveratrol-enriched grape products. Molecules 2017, 22, 294. [Google Scholar] [CrossRef] [PubMed]
- Rozès, N.; Arola, L.; Bordons, A. Effect of phenolic compounds on the co-metabolism of citric acid and sugars by Oenococcus oeni from wine. Lett. Appl. Microbiol. 2003, 36, 337–341. [Google Scholar] [CrossRef] [Green Version]
- Campos, F.M.; Couto, J.A.; Figueiredo, A.R.; Tóth, I.V.; Rangel, A.O.; Hogg, T.A. Cell membrane damage induced by phenolic acids on wine lactic acid bacteria. Int. J. Food Microbiol. 2009, 135, 144–151. [Google Scholar] [CrossRef] [Green Version]
- Stead, D. The effect of hydroxycinnamic acids on the growth of wine-spoilage lactic acid bacteria. J. Appl. Bacteriol. 1993, 75, 135–141. [Google Scholar] [CrossRef]
- Campos, F.; Couto, J.; Hogg, T. Influence of phenolic acids on growth and inactivation of Oenococcus oeni and Lactobacillus hilgardii. J. Appl. Microbiol. 2003, 94, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Bloem, A.; Bertrand, A.; Lonvaud-Funel, A.; De Revel, G. Vanillin production from simple phenols by wine-associated lactic acid bacteria. Lett. Appl. Microbiol. 2007, 44, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Harris, V.; Jiranek, V.; Ford, C.M.; Grbin, P.R. Inhibitory effect of hydroxycinnamic acids on Dekkera spp. Appl. Microbiol. Biotechnol. 2010, 86, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Campos, F.M.; Figueiredo, A.R.; Hogg, T.A.; Couto, J.A. Effect of phenolic acids on glucose and organic acid metabolism by lactic acid bacteria from wine. Food Microbiol. 2009, 26, 409–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devi, A.; Anu-Appaiah, K. Diverse physiological and metabolic adaptations by Lactobacillus plantarum and Oenococcus oeni in response to the phenolic stress during wine fermentation. Food Chem. 2018, 268, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, M.R.; Alberto, M.; de Nadra, M.M. Antibacterial effect of phenolic compounds from different wines. Food Control 2007, 18, 93–101. [Google Scholar] [CrossRef]
- Harborne, J.B.; Williams, C.A. Flavone and flavonol glycosides. In The Flavonoids; Springer: Berlin/Heidelberg, Germany, 1988; pp. 303–328. [Google Scholar]
- Treutter, D. Significance of flavonoids in plant resistance: A review. Environ. Chem. Lett. 2006, 4, 147. [Google Scholar] [CrossRef]
- Alberto, M.R.; Farías, M.E.; de Nadra, M.C.M. Effect of gallic acid and catechin on Lactobacillus hilgardii 5w growth and metabolism of organic compounds. J. Agric. Food Chem. 2001, 49, 4359–4363. [Google Scholar] [CrossRef]
- Reguant, C.; Bordons, A.; Arola, L.; Rozes, N. Influence of phenolic compounds on the physiology of Oenococcus oeni from wine. J. Appl. Microbiol. 2000, 88, 1065–1071. [Google Scholar] [CrossRef]
- de Llano, D.G.; Gil-Sánchez, I.; Esteban-Fernández, A.; Ramos, A.M.; Cueva, C.; Moreno-Arribas, M.; Bartolomé, B. Some Contributions to the Study of Oenological Lactic Acid Bacteria through Their Interaction with Polyphenols. Beverages 2016, 2, 27. [Google Scholar] [CrossRef]
- Pastorkova, E.; Zakova, T.; Landa, P.; Novakova, J.; Vadlejch, J.; Kokoska, L. Growth inhibitory effect of grape phenolics against wine spoilage yeasts and acetic acid bacteria. Int. J. Food Microbiol. 2013, 161, 209–213. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Moreno-Arribas, M.V.; Martín-Álvarez, P.J.; Bartolomé, B. Comparative study of the inhibitory effects of wine polyphenols on the growth of enological lactic acid bacteria. Int. J. Food Microbiol. 2011, 145, 426–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Ruiz, A.; Bartolomé, B.; Cueva, C.; Martín-Álvarez, P.; Moreno-Arribas, M. Inactivation of oenological lactic acid bacteria (Lactobacillus hilgardii and Pediococcus pentosaceus) by wine phenolic compounds. J. Appl. Microbiol. 2009, 107, 1042–1053. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, T.; Estrella, I.; Pérez-Gordo, M.; Alegría, E.; Tenorio, C.; Ruiz-Larrrea, F.; Moreno-Arribas, M. Contribution of malolactic fermentation by Oenococcus oeni and Lactobacillus plantarum to the changes in the nonanthocyanin polyphenolic composition of red wine. J. Agric. Food Chem. 2007, 55, 5260–5266. [Google Scholar] [CrossRef] [PubMed]
- Zoechling, A.; Reiter, E.; Eder, R.; Wendelin, S.; Liebner, F.; Jungbauer, A. The flavonoid kaempferol is responsible for the majority of estrogenic activity in red wine. Am. J. Enol. Vitic. 2009, 60, 223–232. [Google Scholar]
- Komes, D.; Ulrich, D.; Ganic, K.K.; Lovric, T. Study of phenolic and volatile composition of white wine during fermentation and a short time of storage. VITIS-J. Grapevine Res. 2007, 46, 77–84. [Google Scholar]
- Rossouw, M.; Marais, J. The phenolic composition of south African Pinotage, Shiraz and Cabernet Sauvignon wines. S. Afr. J. Enol. Vitic. 2017, 25, 94–104. [Google Scholar] [CrossRef]
- Miles, A.; Misra, S.; Irwin, J. The estimation of the bactericidal power of the blood. J. Hyg. 1938, 38, 732–749. [Google Scholar] [CrossRef] [PubMed]
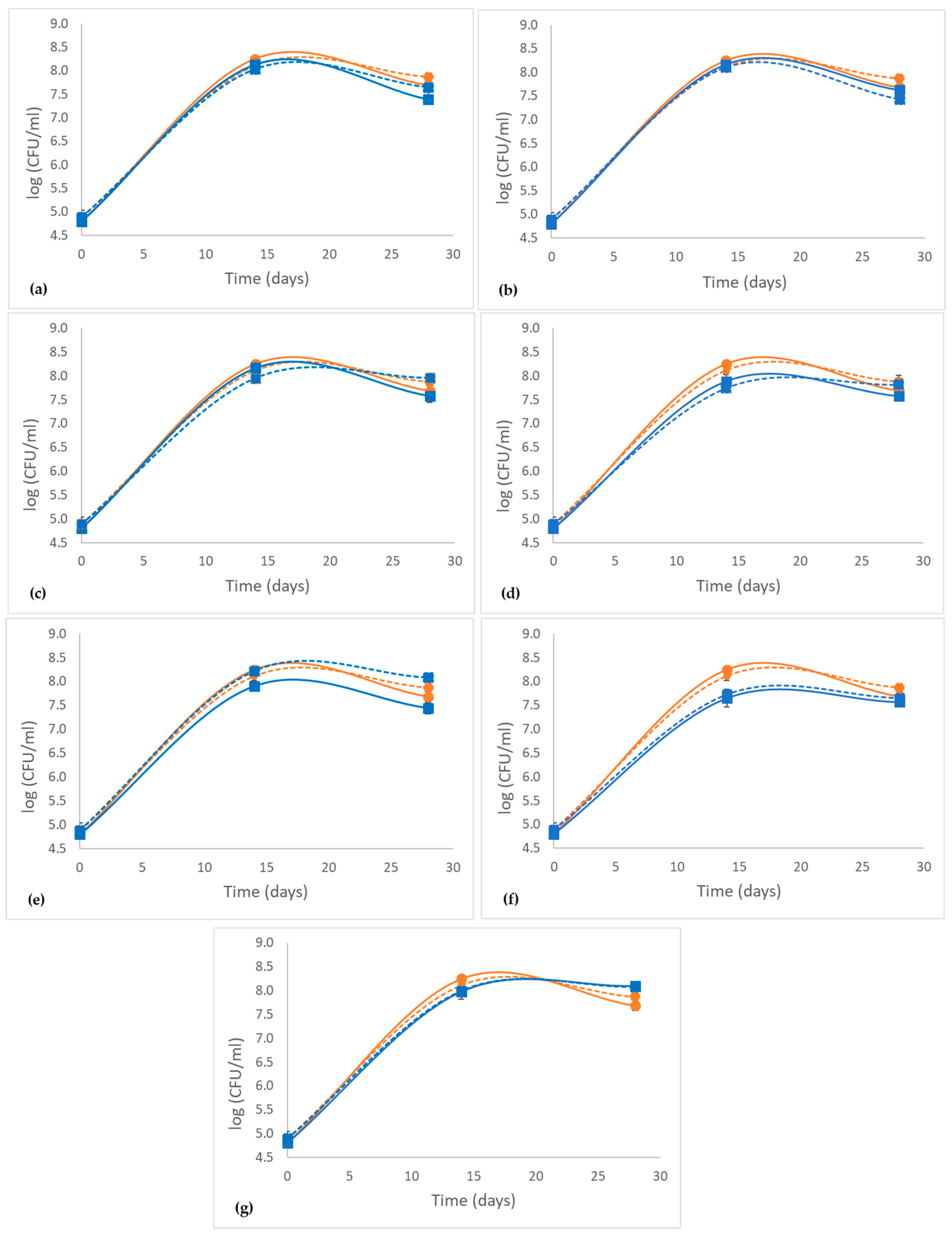
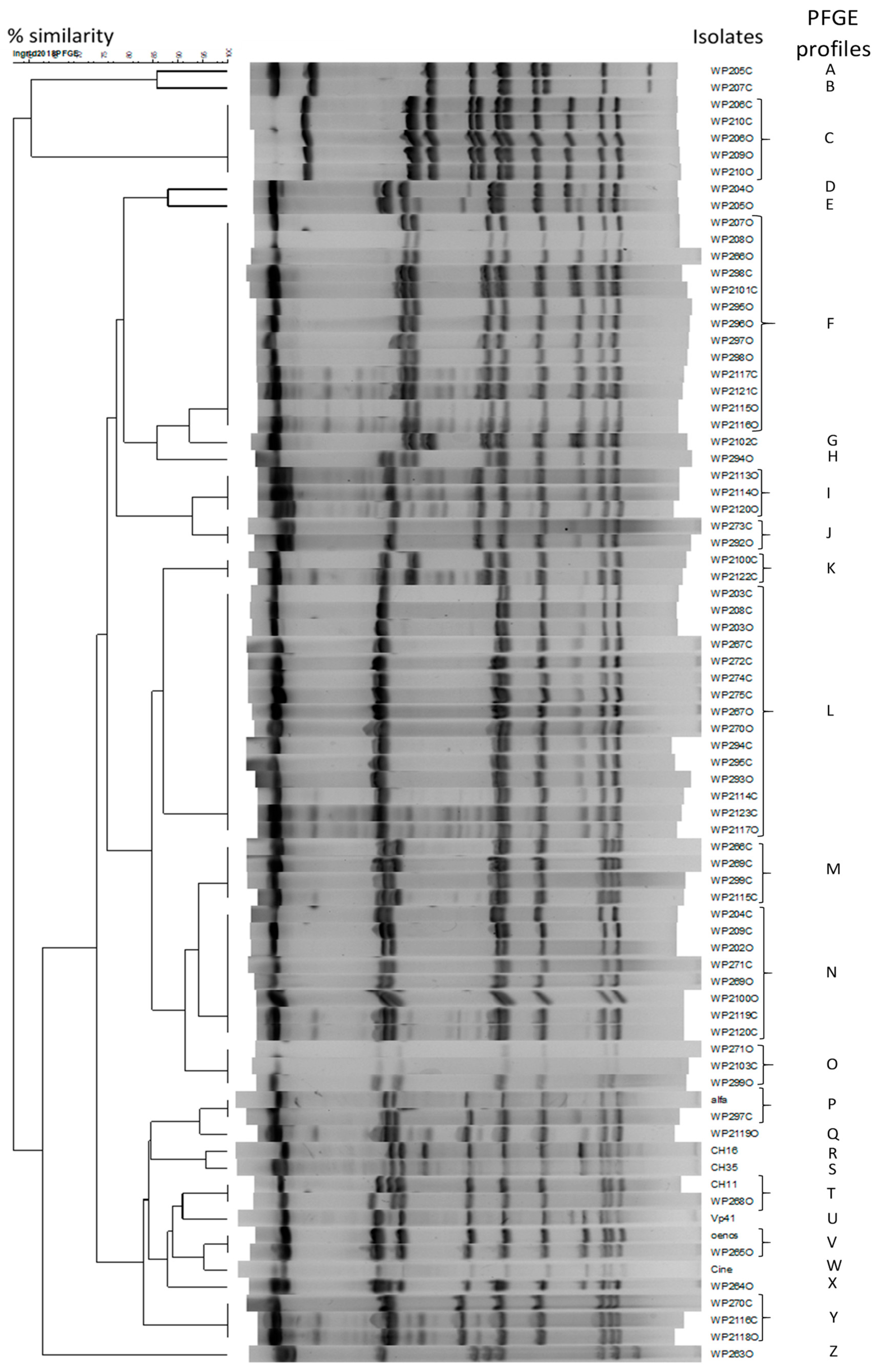
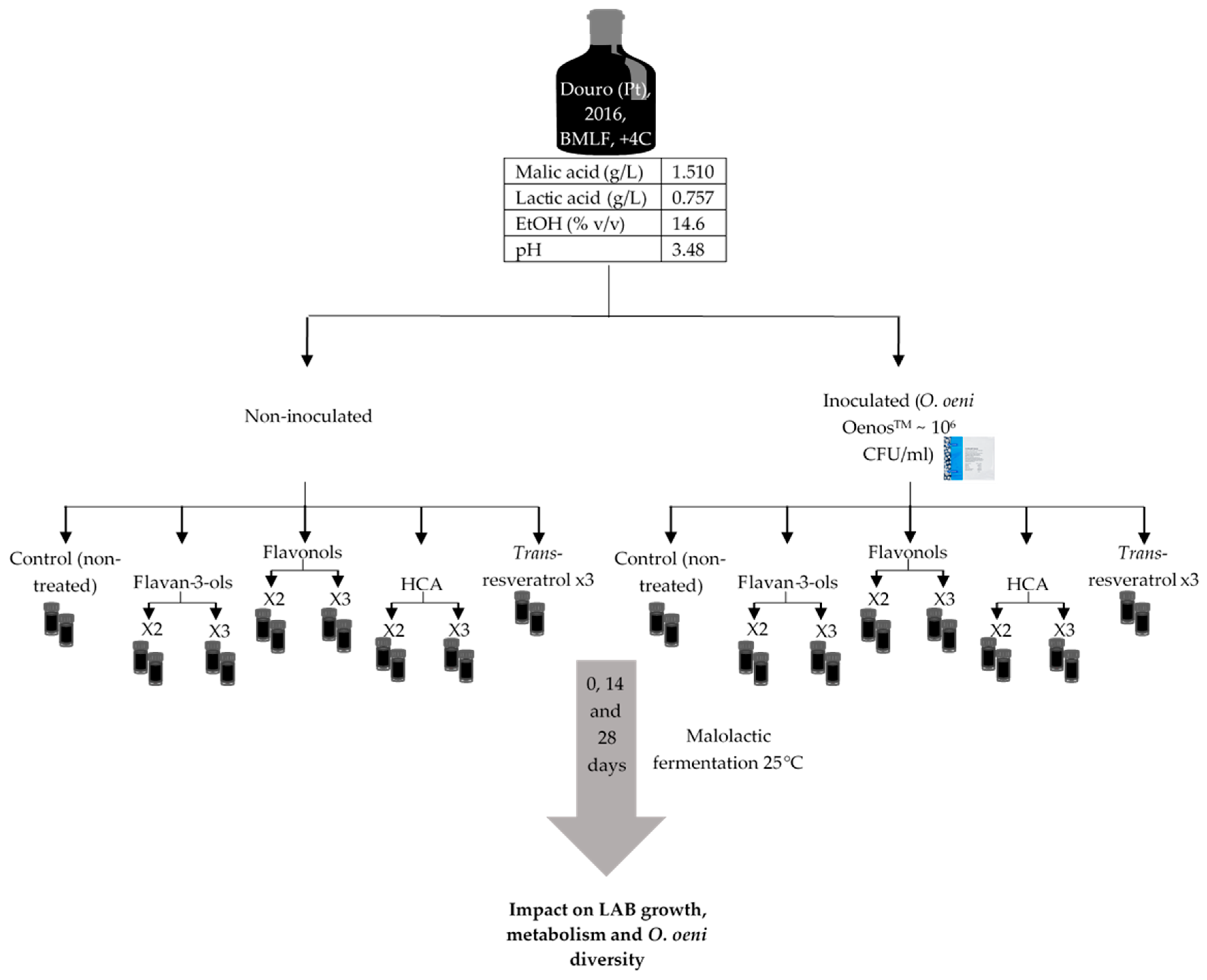
| Treatments | Fructose (g/L) | Lactic Acid (g/L) | Acetic Acid (g/L) | |||
|---|---|---|---|---|---|---|
| Before | After | Before | After | Before | After | |
| Control | 0.84 ± 0.04 y | 0.62 ± 0.06 | 0.78 ± 0.05 | 2.77 ± 0.03 | 0.26 ± 0.00 | 0.59 ± 0.02 |
| 2× Flavan-3-ols | 0.51 ± 0.06 | 2.71 ± 0.00 | 0.69 ± 0.02 | |||
| 3× Flavan-3-ols | 0.47 ± 0.02 | 2.76 ± 0.00 | 0.59 ± 0.02 | |||
| 2× Flavonols | 0.49 ± 0.09 | 2.38 ± 0.12 | 0.59 ± 0.00 | |||
| 3× Flavonols | 0.51 ± 0.01 | 2.39 ± 0.05 | 0.59 ± 0.00 | |||
| 2× HCA | 0.46 ± 0.02 | 2.41 ± 0.17 | 0.59 ± 0.00 | |||
| 3× HCA | 0.49 ± 0.02 | 2.29 ± 0.10 | 0.65 ± 0.02 | |||
| 3× Trans-resveratrol | 0.41 ± 0.01 | 2.38 ± 0.07 | 0.69 ± 0.02 | |||
| Treatments | Citric Acid (g/L) | Fructose (g/L) | Lactic Acid (g/L) | Acetic Acid (g/L) | ||||
|---|---|---|---|---|---|---|---|---|
| Before | After h | Before | After | Before | After | Before | After | |
| Control | 1.46 ± 0.07 y | nd | 0.87 ± 0.02 | 0.41 ± 0.01 | 0.78 ± 0.03 | 2.66 ± 0.02 | 0.26 ± 0.01 | 0.72 ± 0.02 |
| 2× Flavan-3-ols | 0.63 ± 0.08 | 0.47 ± 0.03 | 2.65 ± 0.00 | 0.71 ± 0.00 | ||||
| 3× Flavan-3-ols | 0.80 ± 0.21 | 0.50 ± 0.01 | 2.46 ± 0.10 | 0.71 ± 0.00 | ||||
| 2× Flavonols | 0.80 ± 0.37 | 0.52 ± 0.02 | 2.65 ± 0.00 | 0.71 ± 0.00 | ||||
| 3× Flavonols | 0.61 ± 0.10 | 0.49 ± 0.04 | 2.65 ± 0.00 | 0.71 ± 0.00 | ||||
| 2× HCA | 0.69 ± 0.05 | 0.42 ± 0.01 | 2.65 ± 0.00 | 0.71 ± 0.00 | ||||
| HCA ×3 | 0.93 ± 0.29 | 0.51 ± 0.02 | 2.65 ± 0.00 | 0.71 ± 0.00 | ||||
| Trans-resveratrol ×3 | 0.76 ± 0.26 | 0.42 ± 0.05 | 2.65 ± 0.00 | 0.71 ± 0.00 | ||||
| Red Wine | Treatment | Sampling Time (days) | O. oeni Isolated | PFGE Profile | Red Wine | Treatment | Sampling Time (days) | O. oeni Isolated | PFGE Profile | Commercialized Strains | PFGE Profile |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-inoculated | Control | 0 | WP201C | ND | Inoculated | Control | 0 | WP201O | ND | Oenos™ | V |
| WP202C | ND | WP202O | N | CH11™ | T | ||||||
| WP203C | L | WP203O | L | CH16™ | R | ||||||
| WP204C | N | WP204O | D | CH35™ | S | ||||||
| WP205C | A | WP205O | E | CiNe™ | W | ||||||
| WP206C | C | WP206O | C | Alpha™ | P | ||||||
| WP207C | B | WP207O | F | VP41™ | U | ||||||
| WP208C | L | WP208O | F | ||||||||
| WP209C | N | WP209O | C | ||||||||
| WP210C | C | WP210O | C | ||||||||
| 28 | WP266C | M | 28 | WP263O | Z | ||||||
| WP267C | L | WP264O | X | ||||||||
| WP268C | ND | WP265O | V | ||||||||
| WP269C | M | WP266O | F | ||||||||
| WP270C | Y | WP267O | L | ||||||||
| WP271C | N | WP268O | T | ||||||||
| WP272C | L | WP269O | N | ||||||||
| WP273C | J | WP270O | L | ||||||||
| WP274C | L | WP271O | O | ||||||||
| WP275C | L | WP272O | ND | ||||||||
| Flavonols | 28 | WP294C | L | Flavonols | 28 | WP291O | ND | ||||
| WP295C | L | WP292O | J | ||||||||
| WP296C | ND | WP293O | L | ||||||||
| WP297C | P | WP294O | H | ||||||||
| WP298C | F | WP295O | F | ||||||||
| WP299C | M | WP296O | F | ||||||||
| WP2100C | K | WP297O | F | ||||||||
| WP2101C | F | WP298O | F | ||||||||
| WP2102C | G | WP299O | O | ||||||||
| WP2103C | O | WP2100O | N | ||||||||
| Stilbene | 28 | WP2114C | L | Stilbene | 28 | WP2111O | ND | ||||
| WP2115C | M | WP2112O | ND | ||||||||
| WP2116C | Y | WP2113O | I | ||||||||
| WP2117C | F | WP2114O | I | ||||||||
| WP2118C | ND | WP2115O | F | ||||||||
| WP2119C | N | WP2116O | F | ||||||||
| WP2120C | N | WP2117O | L | ||||||||
| WP2121C | F | WP2118O | Y | ||||||||
| WP2122C | K | WP2119O | Q | ||||||||
| WP2123C | L | WP2120O | I |
| Phenolic Compounds | Concentrations (mg/L) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bottled red wines (Zoechling et al. (2009)) [46] | White wine during fermentation and short storage (Komes et al. (2007)) [47] | Red wines during MLF (Hernandez et al. (2006)) [23] | Red wines during ageing (Hernandez et al. (2006)) [23] | Red wine inoculated with different O. oeni strains (Hernandez et al. (2007)) [45] | Red wine (Rossouw et al. (2017)) [48] | |||||||
| Min | Max | Min | Max | Min | Max | Min | Max | Min | Max | Min | Max | |
| Flavan-3-ols | ||||||||||||
| (+)-catechin | 22.00 | 141.70 | 1.84 | 3.03 | 16.54 | 19.82 | 7.11 | 22.44 | 11.50 | 23.50 | 41.82 | 50.40 |
| (−)-epicatechin | 7.50 | 94.50 | 0.67 | 4.68 | 9.89 | 11.35 | 3.18 | 15.84 | 3.69 | 7.53 | 29.94 | 40.58 |
| Flavonols | ||||||||||||
| Quercetin | nd | 7.40 | - | - | 14.09 | 14.69 | 3.56 | 8.73 | 0.98 | 4.72 | 11.19 | 18.27 |
| Kaempferol | nd | 5.40 | - | - | - | - | - | - | - | - | 2.58 | 3.60 |
| HCA | ||||||||||||
| Trans-p-coumaric acid | 1.30 | 6.60 | 0.34 | 2.63 | 0.70 | 21.34 | 10.01 | 26.30 | 2.06 | 16.01 | 7.36 | 11.47 |
| Trans-ferulic acid | 0.30 | 1.00 | 2.02 | 3.20 | - | - | 0.73 | 1.41 | nd | 0.84 | - | - |
| Stilbene | ||||||||||||
| Trans-resveratrol | 0.07 | 3.95 | - | - | 0.59 | 1.01 | nd | 0.79 | 0.63 | 5.25 | - | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Collombel, I.; Campos, F.M.; Hogg, T. Changes in the Composition of the Lactic Acid Bacteria Behavior and the Diversity of Oenococcus oeni Isolated from Red Wines Supplemented with Selected Grape Phenolic Compounds. Fermentation 2019, 5, 1. https://doi.org/10.3390/fermentation5010001
Collombel I, Campos FM, Hogg T. Changes in the Composition of the Lactic Acid Bacteria Behavior and the Diversity of Oenococcus oeni Isolated from Red Wines Supplemented with Selected Grape Phenolic Compounds. Fermentation. 2019; 5(1):1. https://doi.org/10.3390/fermentation5010001
Chicago/Turabian StyleCollombel, Ingrid, Francisco M. Campos, and Tim Hogg. 2019. "Changes in the Composition of the Lactic Acid Bacteria Behavior and the Diversity of Oenococcus oeni Isolated from Red Wines Supplemented with Selected Grape Phenolic Compounds" Fermentation 5, no. 1: 1. https://doi.org/10.3390/fermentation5010001
APA StyleCollombel, I., Campos, F. M., & Hogg, T. (2019). Changes in the Composition of the Lactic Acid Bacteria Behavior and the Diversity of Oenococcus oeni Isolated from Red Wines Supplemented with Selected Grape Phenolic Compounds. Fermentation, 5(1), 1. https://doi.org/10.3390/fermentation5010001





