Changes in Ruminal Dynamics and Microbial Populations Derived from Supplementation with a Protein Concentrate for Cattle with the Inclusion of Non-Conventional Feeding Sources
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. General Description of the Experiment
2.3. Pelleted Concentrate Formulation and Supplementation Period
2.4. In Vitro Fermentation of Grazed Forage
2.5. pH and N-NH3 Measurement
2.6. In Situ Degradability of Grazed Forage
2.7. Microbial DNA Extraction and Sequencing
2.8. Bioinformatic Analysis
2.9. Statistical Analysis
3. Results
3.1. In Vitro Gas Production, pH, and Ammoniacal Nitrogen Concentration
3.2. In Situ Degradability Parameters
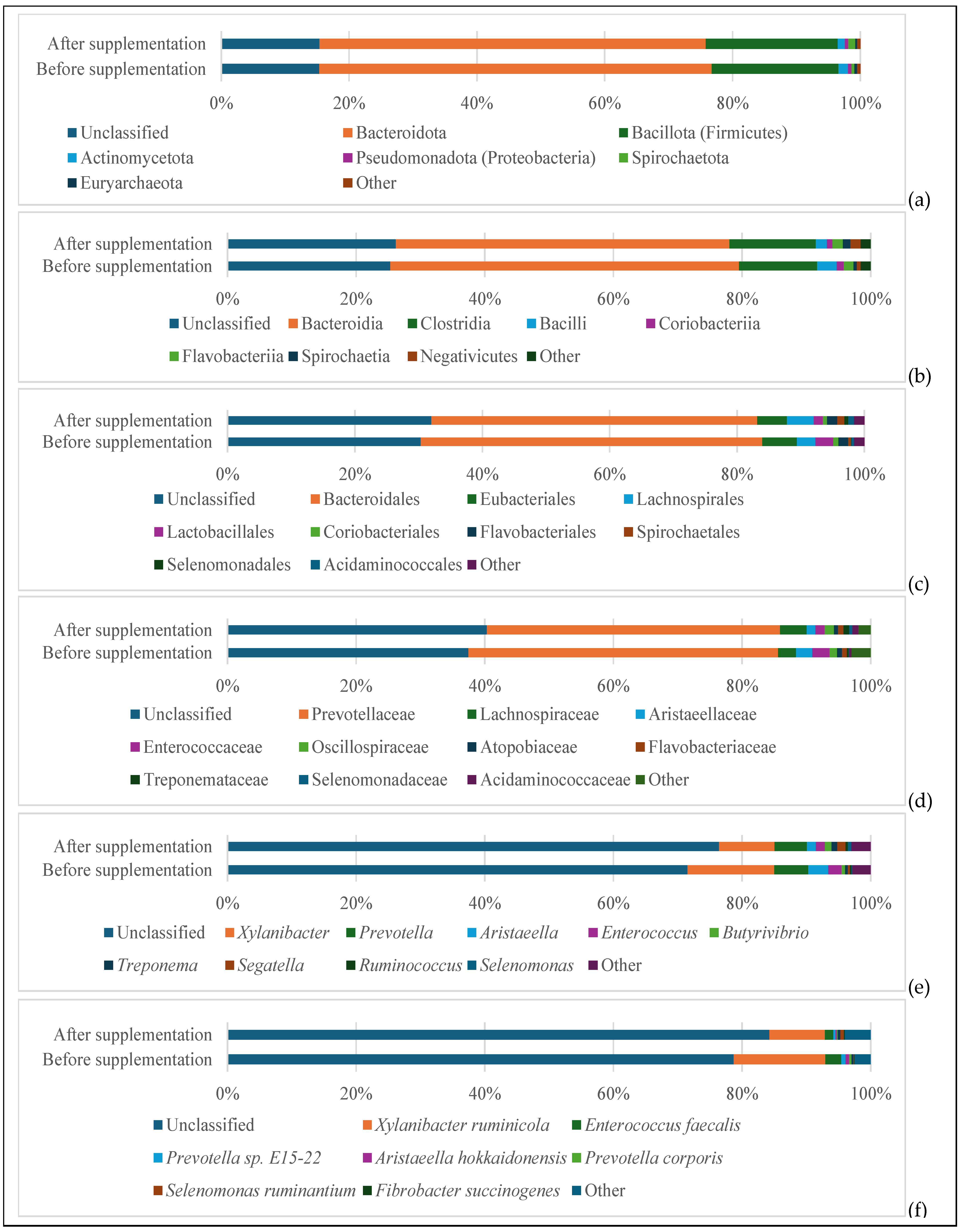
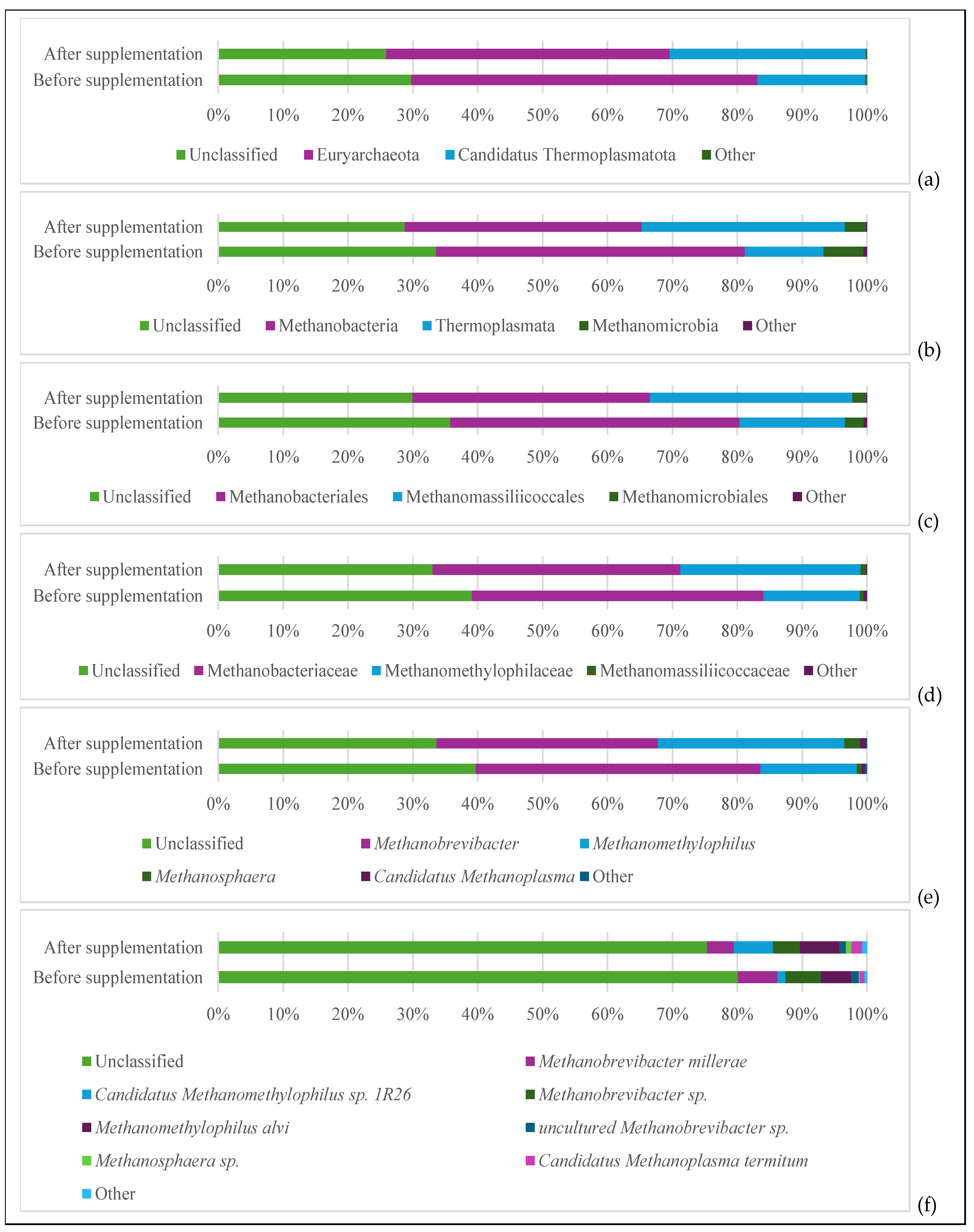
3.3. Microbial Population Analysis
3.4. Correlation Analysis
4. Discussion
4.1. Effects on In Vitro Gas Production, pH, and Ammoniacal Nitrogen Concentration
4.2. Effects on In Situ Degradability Parameters
4.3. Effects on Microbial Populations
4.3.1. Bacterial Community Shifts Based on 16S rRNA Analysis
4.3.2. Methanogenic Archaea Shifts Based on mcrA Analysis
4.4. Limitations and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A | The water-soluble fraction (%, in situ degradability parameter) |
| ADF | Acid Detergent fiber |
| AS | After the supplementation period |
| B | The potentially degradable water-insoluble fraction (%, in situ degradability parameter) |
| BS | Before the supplementation period |
| CEL | Cellulose |
| CP | Crude protein |
| D | Amount of feed that disappears over time (in situ degradability analisys) |
| DM | Dry matter |
| EE | Ethereal extract |
| HEM | Hemicellulose |
| Kd | Degradation rate (%/h, in situ degradability parameter) |
| LIG | Lignin |
| mcrA | α subunit of the methyl coenzyme M reductase |
| NDF | Neutral Detergent fiber |
| N-NH3 | Ammoniacal nitrogen concentration in ruminal liquid |
| NSC | Non-structural carbohydrates (Nonfibrous carbohydrates) |
| OM | Organic matter |
| SC | Spearman correlation |
| SCFA | Short chain fatty acid |
| SEM | Standard error of the means |
| T | Time in hours of incubation (in situ degradability analysis) |
| TC | Total carbohydrates |
| VFA | Volatile fatty acid |
Appendix A
| % | Acacia farnesiana | Acacia schaffneri | Agave duranguensis bagasse |
|---|---|---|---|
| DM | 92.1 ± 0.09 | 85.1 ± 0.55 | 81.3 ± 1.78 |
| OM | 90.1 ± 0.46 | 91.7 ± 0.001 | 86.4 ± 0.74 |
| CP | 19.2 ± 0.37 | 22.4 ± 0.03 | 4.42 ± 0.056 |
| EE | 1.7 ± 0.09 | 3.9 ± 0.20 | 2.47 ± 0.09 |
| NDF | 55.3 ± 0.81 | 52.7 ± 1.47 | 43.82 ± 1.005 |
| ADF | 41.9 ± 0.40 | 32.3 ± 0.40 | 36.22 ± 0340 |
| HEM | 13.5 ± 0.40 | 20.3 ± 1.88 | 7.6 ± 1.4 |
| CEL | 29.5 ± 0.46 | 23.2 ± 0.37 | 30.82 ± 0.49 |
| LIG | 11.8 ± 0.26 | 8.2 ± 0.23 | 0.80 ± 0.52 |
| TC | 71.2 ± 0.27 | 58.8 ± 0.32 | 78.49 ± 0.6 |
| NSC | 15.8 ± 0.94 | 6.1 ± 1.79 | 34.67 ± 0.50 |
| TF | 2.3 ± 0.25 | 2.71 ± 0.10 | ND |
| CT | 1.56 ± 0.02 | 1.74 ± 0.08 | ND |
| Amplicons | Primers | Sequence (“Overhand” Adapter Included) | Reference | PCR Conditions | Reaction |
|---|---|---|---|---|---|
| Hypervariable region V3/V4 of the ribosomal gen 16S | 341F | TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG | [17] | 3 min at 95 °C, 25 cycles (30 s at 95 °C,30 s at 55 °C and 30 s at 72 °C), 5 min at 72 °C | 1.5 mM MgCl2; 0.2 mM dATP, dCTP, dGTP, y dTTP; 0.2 M primers; 1.25 IU of Taq DNA polymerase (Promega Corp., Madison, WI, USA) |
| 805R | GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C | ||||
| α region of the functional gen of Methyl Coenzime M reductase | mlas | TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GGT GGT GTM GGD TTC ACM CAR TA | [21] | 10 min at 95 °C, 30 cycles (30 s at 95 °C,45 s at 60 °C and 45 s at 72 °C), 10 min at 72 °C | 4 mM MgCl2; 0.5 mM dATP, dCTP, dGTP, y dTTP; 0.25 M primers; 1.25 IU of Taq DNA polymerase (Promega Corp., Madison, WI, USA) |
| mrcA-rev | GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA G CGT TCA TBG CGT AGT TVG GRT AGT |
Appendix B
References
- Chatellier, V. International trade in animal products and the place of the European Union: Main trends over the last 20 years. Animal 2021, 15, 100289. [Google Scholar] [CrossRef]
- Da Silva, V.P.; Pereira, O.G.; Da Silva, L.D.; Agarussi, M.C.N.; de Campos Valadares Filho, S.; Ribeiro, K.G. Stylosanthes silage as an alternative to reduce the protein concentrate in diets for finishing beef cattle. Livest. Sci. 2022, 258, 104873. [Google Scholar] [CrossRef]
- Nguyen, D.V.; Vu, C.C.; Nguyen, T.V. The Current Utilisation and Possible Treatments of Rice Straw as Ruminant Feed in Vietnam: A Review. Pak. J. Nutr. 2020, 19, 91–104. [Google Scholar] [CrossRef]
- Moran, D.; Blair, K. Review: Sustainable livestock systems: Anticipating demand-side challenges. Animal 2021, 15, 100288. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). Livestock Solutions for Climate Change; FAO: Rome, Italy, 2017; pp. 1–8. [Google Scholar]
- Wang, K.; Xiong, B.; Zhao, X. Could propionate formation be used to reduce enteric methane emission in ruminants? Sci. Total Environ. 2023, 855, 158867. [Google Scholar] [CrossRef]
- Beriso, Y. Base Line Information on Nutritional Profiling of Non-Conventional Livestock Feed Resources. Greener J. Agric. Sci. 2022, 12, 205–218. [Google Scholar] [CrossRef]
- García-Piña, E.Y.; Herrera-Torres, E.; Murillo-Ortíz, M.; Reyes-Jáquez, D.; Carrete-Carreón, F.O.; Pámanes-Carrasco, G. Dietary alternatives in livestock production for mitigation of greenhouse gas emissions in Mexico: GHG mitigation in Mexico. Agro Product. 2020, 13, 75–81. [Google Scholar] [CrossRef]
- Pulina, G.; Acciaro, M.; Atzori, A.; Battacone, G.; Crovetto, G.; Mele, M.; Pirlo, G.; Rassu, S. Animal board invited review–Beef for future: Technologies for a sustainable and profitable beef industry. Animal 2021, 15, 100358. [Google Scholar] [CrossRef]
- Pedro, S.I.; Gonçalves, J.; Horta, C.; Gonçalves, J.C.; Gominho, J.; Gallardo, E.; Anjos, O. A Systematic Analysis of Nutritional and Mineral Composition and Toxicity in Acacia Species Leaves. Appl. Sci. 2024, 14, 9437. [Google Scholar] [CrossRef]
- Araiza-Rosales, E.E.; Pámanes-Carrasco, G.A.; Sánchez-Arroyo, J.F.; Herrera-Torres, E.; Rosales-Castro, M.; Carrete-Carreón, F.O. Caracterización nutricional y producción de gas de especies vegetales con potencial alimenticio para la alimentación de rumiantes. Rev. MVZ Córdoba 2022, 27, e2142. [Google Scholar] [CrossRef]
- Pérez-Cruz, Y.M.; Noveron-Neri, I.A.; Hernandez-Rodríguez, D.; Hernández-Mendo, O. El huizache (Vachellia farnesiana): Una alternativa no convencional en la alimentación ovina. AgroDivulgación 2022, 2, 4. [Google Scholar]
- Torres-Velázquez, D.S.; Murillo-Ortiz, M.; Cervantes-Guerrero, M.; Páez-Lerma, J.B.; Reyes-Jáquez, D.; Soto-Cruz, N.O.; Araiza-Ponce, K.A. Huizache Leaves and Agave Bagasse Incorporated into Granulated and Pelletized Concentrates and Their Effects on Methane Production and in vitro Fermentation Patterns in Ruminant Diets. Indian J. Anim. Res. 2023, BF-1699, 1–7. [Google Scholar] [CrossRef]
- Dias, M.R.; Moraes, K.A.K.d.; Oliveira, A.S.d.; Batista, E.D.; Salomão, A.M.R.; Zambenedetti, A.; Petrenko, N.B.; Sousa, J.N.; Ortelam, J.C.; Ickert, A.; et al. Nutritional Performance of Grazing Beef Cattle Supplemented with High-Protein Distillers’ Dried Grain. Animals 2024, 14, 1209. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Global Rumen Census Collaborators; Janssen, P.H. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geo graphical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- Huws, S.A.; Oyama, L.B.; Creevey, C.J. The Sustainable Use and Conservation of Microorganisms of Relevance to Ruminant Digestion (Draft Study on the Sustainable Use and Conservation of Microorganisms of Relevance to Ruminant Digestion-FAO Intergovernmental Technical Working Group on Animal Genetic Resources for Food and Agriculture); Food and Agriculture Organization of the United Nations: Rome, Italy, 2022; pp. 4–25. [Google Scholar]
- Márquez Mota, C.C.; Piña-González, L.; Sánchez-Tapia, M.; Torres, N.; Tovar, A.R.; Loor, J.J.; Alharthi, A.; Corona, L. Changes in the bacterial diversity of the ruminal liquid fraction of dairy cows on a corn stover based diet. Vet. Mex. OA 2023, 10, 1–15. [Google Scholar] [CrossRef]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The rumen microbiome: A crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef]
- Andrade, B.G.; Bressani, F.A.; Cuadrat, R.R.; Tizioto, P.C.; de Oliveira, P.S.; Mourão, G.B.; Coutinho, L.L.; Reecy, J.M.; Koltes, J.E.; Walsh, P.; et al. The structure of microbial populations in Nelore GIT reveals inter dependency of methanogens in feces and rumen. J. Anim. Sci. Biotechnol. 2020, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Newbold, C.J.; Ramos-Morales, E. Review: Ruminal microbiome and microbial metabolome: Effects of diet and ruminant host. Animal 2020, 14, s78–s86. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, L.M.; Regan, J.M. Phylogenetic Comparison of the Methanogenic Communities from an Acidic, Oligotrophic Fen and an Anaerobic Digester Treating Municipal Wastewater Sludge. Appl. Environ. Microbiol. 2008, 74, 6663–6671. [Google Scholar] [CrossRef]
- Steinberg, L.M.; Regan, J.M. mcrA-Targeted Real-Time Quantitative PCR Method to Examine Methanogen Communities. Appl. Environ. Microbiol. 2009, 75, 4435–4442. [Google Scholar] [CrossRef]
- Luton, P.E.; Wayne, J.M.; Sharp, R.J.; Riley, P.W. The mcrA gene as an alternative to 16s rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 2002, 148, 3521–3530. [Google Scholar] [CrossRef]
- NOM-062-ZOO-1999; Especificaciones Técnicas para la Producción, Cuidado y Uso de los Animales de Laboratorio. Norma Oficial Mexicana; Diario Oficial de la Federación: Mexico City, Mexico, 2001; pp. 128–134.
- Theodorou, M.K.; Williams, B.A.; Dhanoa, M.S.; McAllan, A.B.; France, J. A simple gas production method a pressure transducer todetermine the fermentation kinetics ofruminant feeds. Anim. Feed Sci. Technol. 1994, 48, 185–197. [Google Scholar] [CrossRef]
- Fievez, V.; Babayemi, O.J.; Demeyer, D. Estimation of direct and indirect gas production in syringes: A tool to estimate short chain fatty acid production that requires minimal laboratory facilities. Anim. Feed Sci. Technol. 2005, 123, 197–210. [Google Scholar] [CrossRef]
- Galyean, M. Laboratory Procedure in Animal Nutrition Research; Department of Animal and Life Science, New Mexico State University: Las Cruces, NM, USA, 2010; p. 98. [Google Scholar]
- AOAC International. Official Methods of Analysis of AOAC International, 21st ed.; AOAC: Gaithersburg, MD, USA, 2019. [Google Scholar]
- Salazar, J.A.E.; Rojas, C.R.M. Fistulación en bovinos y uso de la técnica de degradabilidad ruminal para análisis de alimentos. Nutr. Anim. Trop. 2020, 14, 209–229. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome 2019, 1, 257. [Google Scholar] [CrossRef]
- Cabezas-Garcia, E.H.; Danielsson, R.; Ramin, M.; Huhtanen, P. In Vitro Incubations Do Not Reflect In Vivo Differences Based on Ranking of Low and High Methane Emitters in Dairy Cows. Animals 2021, 11, 3112. [Google Scholar] [CrossRef]
- Sandoval-Pelcastre, A.; Ramírez-Mella, M.; Rodríguez-Avila, N.; Martínez, B. Tropical trees and shrubs with potential to reduce the production of methane in ruminants. Trop. Subtrop. Agroecosyst. 2020, 23, 1–16. [Google Scholar] [CrossRef]
- Galindo-Blanco, J.; Rodríguez-García, I.; González-Ibarra, N.; García-López, R.; Herrera-Villafranca, M. Silvopastoral system with Tithonia diversifolia (Hemsl.) A. Gray: Effect on the rumen microbial population of cows. Pastos y Forrajes 2018, 41, 273–280. [Google Scholar]
- Salami, S.A.; Luciano, G.; O’Grady, M.N.; Biondi, L.; Newbold, C.J.; Kerry, J.P. Sustainability of feeding plant by-products: A review of the implications for ruminant meat production. Anim. Feed Sci. Technol. 2019, 251, 37–55. [Google Scholar] [CrossRef]
- Moorby, J.; Fraser, M. Review: New feeds and new feeding systems in intensive and semi-intensive forage-fed ruminant livestock systems. Animal 2021, 15, 100297. [Google Scholar] [CrossRef]
- Ogata, T.; Kim, Y.H.; Masaki, T.; Iwamoto, E.; Ohtani, Y.; Orihashi, T.; Ichijo, T.; Sato, S. Effects of an increased concentrate diet on rumen pH and the bacterial community in Japanese Black beef cattle at different fattening stages. J. Vet. Med. Sci. 2019, 81, 968–974. [Google Scholar] [CrossRef] [PubMed]
- Aikman, P.C.; Henning, P.H.; Humphries, D.J.; Horn, C.H. Rumen pH and fermentation characteristics in dairy cows supplemented with Megasphaera elsdenii NCIMB 41125 in early lactation. J. Dairy Sci. 2011, 94, 2840–2849. [Google Scholar] [CrossRef]
- Pérez, E.; Vera, J.K.; Ramírez, L.; Martínez, S. Suplementación con G. sepium: Su efecto en la digestión ruminal y el comportamiento de bovinos en pastoreo intensivo en la época de lluvias. Pastos y Forrajes 2002, 25, 4. [Google Scholar]
- Frutos, P.; Mantecón, Á.R.; Hervás, G.; Giráldez, F.J. Tannins and ruminant nutrition. Span. J. Agric. Res. 2004, 2, 191–202. [Google Scholar] [CrossRef]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; The, K.H.; Lambie, S.C.; Cookson, A.L.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Hitch, T.C.; Bisdorf, K.; Afrizal, A.; Riedel, T.; Overmann, J.; Strowig, T.; Clavel, T. A taxonomic note on the genus Prevotella: Description of four novel genera and emended description of the genera Hallella and Xylanibacter. Syst. Appl. Microbiol. 2022, 45, 126354. [Google Scholar] [CrossRef]
- Mahoney-Kurpe, S.C.; Palevich, N.; Noel, S.J.; Gagic, D.; Biggs, P.J.; Soni, P.; Reid, P.M.; Koike, S.; Kobayashi, Y.; Janssen, P.H.; et al. Aristaeella hokkaidonensis gen. nov. sp. nov. and Aristaeella lactis sp. nov., two rumen bacterial species of a novel proposed family, Aristaeellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2023, 73, 5. [Google Scholar] [CrossRef]
- Bi, Y.; Zeng, S.; Zhang, R.; Diao, Q.; Tu, Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018, 18, 69. [Google Scholar] [CrossRef]
- Maake, T.W.; Aiyegoro, O.A.; Adeleke, M.A. Effects of Lactobacillus rhamnosus and Enterococcus faecalis Supplementation as Direct-Fed Microbials on Rumen Microbiota of Boer and Speckled Goat Breeds. Vet. Sci. 2021, 8, 103. [Google Scholar] [CrossRef]
- Mamuad, L.L.; Kim, S.H.; Biswas, A.A.; Yu, Z.; Cho, K.K.; Kim, S.B.; Lee, K.; Lee, S.S. Rumen fermentation and microbial community composition influenced by live Enterococcus faecium supplementation. AMB Express 2019, 9, 123. [Google Scholar] [CrossRef]
- Ueki, A.; Akasaka, H.; Suzuki, D.; Hattori, S.; Ueki, K. Xylanibacter oryzae gen. nov., sp. nov., a novel strictly anaerobic, Gram-negative, xylanolytic bacterium isolated from rice-plant residue in flooded rice-field soil in Japan. Int. J. Syst. Evol. Microbiol. 2006, 56, 2215–2221. [Google Scholar] [CrossRef][Green Version]
- Murovec, U.; Accetto, T. Transcriptomic analysis of polysaccharide utilization loci reveals substrate preferences in ruminal generalists Segatella bryantii TF1-3 and Xylanibacter ruminicola KHP1. BMC Genom. 2024, 25, 495. [Google Scholar] [CrossRef]
- Malik, P.K.; Trivedi, S.; Kolte, A.P.; Sejian, V.; Bhatta, R.; Rahman, H. Diversity of rumen microbiota using metagenome sequencing and methane yield in Indian sheep fed on straw and concentrate diet. Saudi J. Biol. Sci. 2022, 29, 103345. [Google Scholar] [CrossRef]
- Xie, X.; Yang, C.; Guan, L.L.; Wang, J.; Xue, M.; Liu, J.X. Persistence of Cellulolytic Bacteria Fibrobacter and Treponema After Short-Term Corn Stover-Based Dietary Intervention Reveals the Potential to Improve Rumen Fibrolytic Function. Front. Microbiol. 2018, 9, 1363. [Google Scholar] [CrossRef]
- Orzuna-Orzuna, J.F.; Dorantes-Iturbide, G.; Lara-Bueno, A.; Mendoza-Martinez, G.D.; Miranda-Romero, L.A.; Hernandez-Garcia, P.A. Effects of dietary tannins’ supplementation on growth performance, rumen fermentation, and enteric methane emissions in beef cattle: A meta-analysis. Sustainability 2021, 13, 7410. [Google Scholar] [CrossRef]
- Bekele, A.Z.; Koike, S.; Kobayashi, Y. Phylogenetic diversity and dietary association of rumen Treponema revealed using group-specific 16Sr RNA gene-based analysis. FEMS Microbiol. Lett. 2011, 316, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xu, F.; Thomas, S.C.; Zhang, Y.; Paul, B.; Sakilam, S.; Chae, S.; Li, P.; Almeter, C.; Kamer, A.R.; et al. Targeting the succinate receptor effectively inhibits periodontitis. Cell Rep. 2022, 40, 12. [Google Scholar] [CrossRef] [PubMed]
- Marchandin, H.; Teyssier, C.; Campos, J.; Jean-Pierre, H.; Roger, F.; Gay, B.; Carlier, J.P.; Jumas-Bilak, E. Negativicoccus succinicivorans gen. nov., sp. nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. nov. and Acidaminococcaceae fam. nov. in the bacterial phylum Firmicutes. Int. J. Syst. Evol. Microbiol. 2010, 60, 1271–1279. [Google Scholar] [CrossRef]
- Van Gylswyk, N.O. Suciniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Bacteriol. 1995, 45, 297–300. [Google Scholar] [CrossRef]
- Latham, E.A.; Weldon, K.K.; Wickersham, T.A.; Coverdale, J.A.; Pinchak, W.E. Responses in the rumen microbiome of Bos taurus and indicus steers fed a low-quality rice straw diet and supplemented protein. J. Anim. Sci. 2018, 96, 1032–1044. [Google Scholar] [CrossRef]
- Lin, M.; Dai, X.; Weimer, P.J. Shifts in fermentation end products and bacterial community composition in long-term, sequentially transferred in vitro ruminal enrichment cultures fed switchgrass with and without ethanol as a co-substrate. Bioresour. Technol. 2019, 285, 121324. [Google Scholar] [CrossRef]
- Karri, S.; Vadela, M.B.; Gundi, V.A. Fiber degradation strategies of bacteria in rumen ecosystem. In Recent Developments in Applied Microbiology and Biochemistry; Viswanath, B., Ed.; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2021; Volume 2, Chapter 25; pp. 153–159. [Google Scholar] [CrossRef]
- Sasikumar, S.; Dharumadurai, D. Symbiotic functions of rumen microbial community in dairy cows. In Developments in Applied Microbiology and Biotechnology, Microbial Symbionts; Dharumadurai, D., Ed.; Academic Press: Cambridge, MA, USA, 2023; Chapter 26; pp. 479–491. [Google Scholar] [CrossRef]
- Arjun, S.; Neha, P.; Sai, S.M.; Ravi, L. Microbial symbionts in ruminants. In Developments in Applied Microbiology and Biotechnology, Microbial Symbionts; Dharumadurai, D., Ed.; Academic Press: Cambridge, MA, USA, 2023; Chapter 27; pp. 493–509. [Google Scholar] [CrossRef]
- Wang, K.; Nan, X.M.; Zhao, Y.G.; Tong, J.J.; Jiang, L.S.; Xiong, B.H. Effects of propylene glycol on in vitro ruminal fermentation, methanogenesis, and microbial community structure. J. Dairy Sci. 2021, 104, 2924–2934. [Google Scholar] [CrossRef]
- Huang, J.; Li, Y. Rumen methanogen and protozoal communities of Tibetan sheep and Gansu Alpine Finewool sheep grazing on the Qinghai–Tibetan Plateau, China. BMC Microbiol. 2018, 18, 212. [Google Scholar] [CrossRef]
- Solís-García, A.; Rivas-García, P.; Escamilla-Alvarado, C.; Rico-Martínez, R.; Bravo-Sánchez, M.G.; Botello-Álvarez, J.E. Methanol production kinetics during agave cooking for mezcal industry. Revista Mexicana de Ingeniería Química 2017, 16, 827–834. [Google Scholar]
- Borrel, G.; Fadhlaoui, K.; Ben-Hania, W.; Gaci, N.; Pehau-Arnaudet, G.; Chaudhary, P.P.; Vandekerckove, P.; Ballet, N.; Alric, M.; O’Toole, P.W.; et al. Methanomethylophilus alvi gen. nov., sp. nov., a Novel Hydrogenotrophic Methyl-Reducing Methanogenic Archaea of the Order Methanomassiliicoccales Isolated from the Human Gut and Proposal of the Novel Family Methanomethylophilaceae fam. nov. Microorganisms 2023, 11, 2794. [Google Scholar] [CrossRef]
- McCabe, M.S.; Cormican, P.; Keogh, K.; O’Connor, A.; O’Hara, E.; Palladino, R.A.; Kenny, D.A.; Waters, S.M. Illumina MiSeq Phylogenetic Amplicon Sequencing Shows a Large Reduction of an Uncharacterised Succinivibrionaceae and an Increase of the Methanobrevibacter gottschalkii Clade in Feed Restricted Cattle. PLoS ONE 2015, 10, e0133234. [Google Scholar] [CrossRef]
- Feldewert, C.; Lang, K.; Brune, A. The hydrogen threshold of obligately methyl-reducing methanogens. FEMS Microbiol. Lett. 2020, 367, fnaa137. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, L.J.; Zundt, M.; Tsujiguchi, M.P.; Giotto, F.M.; Barbosa, M.J.P.T.; Grandis, F.A.; Silva, I.G.; Pereira, M.C.S.; Ribeiro, E.L.A. The use of condensed tannin in lambs’ diet alters the rumen protozoa population without affecting growth performance. Small Rumin. Res. 2023, 229, 107122. [Google Scholar] [CrossRef]
- Patra, A.K.; Saxena, J. Exploitation of dietary tannins to improve rumen metabolism and ruminant nutrition. J. Sci. Food Agric. 2011, 91, 24–37. [Google Scholar] [CrossRef]
- Battelli, M.; Colombini, S.; Parma, P.; Galassi, G.; Crovetto, G.M.; Spanghero, M.; Pravettoni, D.; Zanzani, S.A.; Manfredi, M.T.; Rapetti, L. In vitro effects of different levels of quebracho and chestnut tannins on rumen methane production, fermentation parameters, and microbiota. Front. Vet. Sci. 2023, 10, 1178288. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, P.; Liu, S.; Miao, B.; Zeng, B.; Jiang, Y.; Li, L.; Wang, L.; Chen, Y.; Zhang, H. Characterization of the rumen microbiota and volatile fatty acid profiles of weaned goat kids under shrub-grassland grazing and indoor feeding. Animals 2020, 10, 176. [Google Scholar] [CrossRef] [PubMed]
| Ingredient | % DM |
|---|---|
| Distillers dried grains | 15 |
| Agave duranguensis bagasse | 6 |
| Ground corn | 9 |
| Soybean paste | 18 |
| Cottonseed flour | 15 |
| Wheat bran | 17 |
| Huizache (A. farnesiana leaves) | 15 |
| Huizache (A. schaffneri leaves) | 5 |
| Pelletized Concentrate | Grazed Forage | |
|---|---|---|
| DM | 96.33 ± 0.033 | 92.23 ± 0.033 |
| OM | 93.26 ± 0.028 | 85.00 ± 0.058 |
| CP | 25.47 ± 0.581 | 23.17 ± 0.589 |
| EE | 3.5 ± 0.001 | 5.37 ± 0.145 |
| NDF | 31.1 ± 0.173 | 53.30 ± 1.124 |
| ADF | 12.03 ± 0.260 | 34.33 ± 0.549 |
| HEM | 19.07 ± 0.433 | 18.83 ± 0.433 |
| CEL | 9.97 ± 0.0333 | 26.03 ± 0.617 |
| LIG | 1.87 ± 0.203 | 3.57 ± 0.318 |
| TC | 64.3 ± 0.608 | 57.97 ± 1.198 |
| NSC | 33.17 ± 0.751 | 3.182 ± 1.610 |
| A | 6.14 ± 1.112 | 2.29 ± 0.736 |
| B | 55.22 ± 0.282 | 49.95 ± 1.093 |
| Kd | 0.045 ± 0.0064 | 0.070 ± 0.002 |
| Before Supplementation | After Supplementation | Difference (Mean) | t-Value | p-Value | SEM | |
|---|---|---|---|---|---|---|
| pH | 6.85 | 6.13 | 0.7267 | 4.27 | 0.0508 | 0.1703 |
| N-NH3 (mM) | 3.89 | 3.74 | 0.1575 | 1.96 | 0.1885 | 0.0802 |
| Total gas (mL/g) | 60.96 | 88.34 | −27.38 | −3.63 | 0.0682 | 7.542 |
| CH4 (mL/g) | 9.06 | 4.96 | 4.099 | 7.17 | 0.0189 | 0.572 |
| CO2 (mL/g) | 51.89 | 83.38 | −31.48 | −3.89 | 0.0602 | 8.096 |
| CO2/CH4 | 5.85 | 16.8 | −10.95 | −6.95 | 0.0201 | 1.5767 |
| %CH4 | 15.14 | 5.63 | 9.5066 | 4.49 | 0.0463 | 2.1192 |
| Supplementation | A (%) | B (%) | Kd (%/h) | Non-Degradable (%) | Degradability Potential (%) | |
|---|---|---|---|---|---|---|
| DM | Before | 2.3 | 50 | 0.07 | 47.76 | 52.24 |
| After | 4.1 | 38.2 | 0.03 | 57.69 | 42.31 | |
| Difference (mean) | −1.805 | 11.735 | 0.0369 | |||
| t-value | −2.12 | 3.63 | 8.89 | |||
| p-value | 0.1244 | 0.036 | 0.003 | |||
| SEM | 1.0553 | 3.2312 | 0.0042 | |||
| CP | Before | 36.3 | 52.8 | 0.08 | 10.85 | 89.15 |
| After | 39 | 29.7 | 0.06 | 31.31 | 68.69 | |
| Difference (mean) | −2.645 | 23.111 | 0.0212 | |||
| t-value | −2.17 | 4.65 | 0.98 | |||
| p-value | 0.119 | 0.0188 | 0.3995 | |||
| SEM | 1.2216 | 4.9754 | 0.0216 | |||
| NDF | Before | 3.5 | 58.9 | 0.06 | 37.61 | 62.39 |
| After | 2.8 | 45.4 | 0.03 | 51.8 | 48.2 | |
| Difference (mean) | 0.6925 | 13.494 | 0.03 | |||
| t-value | 0.71 | 3.45 | 5.57 | |||
| p-value | 0.5288 | 0.0409 | 0.0114 | |||
| SEM | 0.975 | 3.9092 | 0.0416 | |||
| ADF | Before | 1.9 | 57.5 | 0.06 | 40.59 | 59.41 |
| After | 3.4 | 47.5 | 0.02 | 49.16 | 50.84 | |
| Difference (mean) | −1.468 | 10.032 | 0.0376 | |||
| t-value | −1.32 | 1.63 | 4.22 | |||
| p-value | 0.2787 | 0.2008 | 0.0243 | |||
| SEM | 1.1129 | 6.1403 | 0.0089 | |||
| HEM | Before | 15.7 | 54 | 0.57 | 30.24 | 69.76 |
| After | 6.5 | 49.5 | 0.02 | 44.05 | 55.95 | |
| Difference (mean) | 9.2548 | 4.5553 | 0.0358 | |||
| t-value | 6.76 | 1.72 | 19.32 | |||
| p-value | 0.0066 | 0.1845 | 0.0003 | |||
| SEM | 1.37 | 2.6534 | 0.0019 |
| Genus | Before Supplementation (%) | After Supplementation (%) | Difference (Mean) | t-Value | p-Value | SEM |
|---|---|---|---|---|---|---|
| Xylanibacter | 46.31 | 35.82 | 13.02 | 17.94 | 0.0004 | 0.7261 |
| Prevotella | 18.24 | 21.08 | −4.08 | −2.70 | 0.0738 | 1.5118 |
| Aristaella | 10.59 | 5.82 | 3.49 | 4.72 | 0.018 | 0.7397 |
| Enterococcus | 7.13 | 5.75 | 3.04 | 2.63 | 0.078 | 1.1548 |
| Butyrivibrio | 1.96 | 4.30 | −2.39 | −5.75 | 0.0104 | 0.4154 |
| Treponema | 1.31 | 3.85 | −2.69 | −7.17 | 0.0056 | 0.3752 |
| Segatella | 1.21 | 5.32 | −4.42 | −3.33 | 0.0446 | 1.3246 |
| Ruminococcus | 0.89 | 1.33 | −0.32 | −0.51 | 0.6457 | 0.6285 |
| Selenomonas | 0.36 | 2.30 | −2.03 | −3.85 | 0.0309 | 0.5255 |
| Anaerostipes | 0.33 | 0.37 | −0.13 | −0.25 | 0.8213 | 0.5073 |
| Olsenella | 0.21 | 0.00 | 0.28 | 1.70 | 0.1884 | 0.1621 |
| Thermococcus | 0.52 | 0.00 | 0.60 | 2.66 | 0.0766 | 0.2241 |
| Fibrobacter | 0.14 | 0.34 | −0.23 | −1.00 | 0.391 | 0.2325 |
| Hoylesella | 0.33 | 0.39 | −0.14 | −0.27 | 0.8058 | 0.531 |
| Simiaoa | 0.30 | 0.31 | −0.31 | −1.00 | 0.391 | 0.3125 |
| Paraprevotella | 0.33 | 0.43 | −0.43 | −1.00 | 0.391 | 0.4325 |
| Genus | Before Supplementation (%) | After Supplementation (%) | Difference | t-Value | p-Value | SEM |
|---|---|---|---|---|---|---|
| Methanobrevibacter | 72.67 | 51.44 | 20.41 | 4.37 | 0.0221 | 4.6656 |
| Methanomethylophilus | 24.63 | 43.26 | −17.82 | −3.40 | 0.0424 | 5.2384 |
| Methanosphaera | 1.27 | 3.65 | −2.38 | −3.91 | 0.0297 | 0.6088 |
| Candidatus Methanoplasma | 0.86 | 1.50 | −0.73 | −1.06 | 0.3663 | 0.683 |
| Acinetobacter | 0.07 | 0.02 | 0.05 | 1.25 | 0.2999 | 0.04 |
| Methanobacterium | 0.08 | 0.04 | 0.06 | 1.57 | 0.2142 | 0.0366 |
| Paenibacillus | 0.03 | 0.01 | 0.02 | 1.63 | 0.201 | 0.0122 |
| Methanogenium | 0.01 | 0.02 | 0.00 | 0.00 | 1.000 | 0.0071 |
| Methanoculleus | 0.01 | 0.02 | −0.01 | −1.41 | 0.2522 | 0.0071 |
| Xylanibacter | 0.01 | 0.00 | 0.01 | 1.57 | 0.2152 | 0.0048 |
| Haloferax | 0.28 | 0.00 | 0.28 | 1.00 | 0.391 | 0.28 |
| Amplicon | Before Supplementation | After Supplementation | Difference | t-Value | p-Value | SEM |
|---|---|---|---|---|---|---|
| 16S | 4.46 | 4.47 | −0.0120 | −0.2000 | 0.8550 | 0.0596 |
| mcrA | 3.5 | 3.13 | 0.3651 | 1.2000 | 0.3169 | 0.3048 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Velázquez, D.S.; Ramos-Rosales, D.F.; Murillo-Ortiz, M.; Páez-Lerma, J.B.; Rojas-Contreras, J.A.; Araiza-Ponce, K.A.; Reyes-Jáquez, D. Changes in Ruminal Dynamics and Microbial Populations Derived from Supplementation with a Protein Concentrate for Cattle with the Inclusion of Non-Conventional Feeding Sources. Fermentation 2025, 11, 438. https://doi.org/10.3390/fermentation11080438
Torres-Velázquez DS, Ramos-Rosales DF, Murillo-Ortiz M, Páez-Lerma JB, Rojas-Contreras JA, Araiza-Ponce KA, Reyes-Jáquez D. Changes in Ruminal Dynamics and Microbial Populations Derived from Supplementation with a Protein Concentrate for Cattle with the Inclusion of Non-Conventional Feeding Sources. Fermentation. 2025; 11(8):438. https://doi.org/10.3390/fermentation11080438
Chicago/Turabian StyleTorres-Velázquez, Diana Sofía, Daniel Francisco Ramos-Rosales, Manuel Murillo-Ortiz, Jesús Bernardo Páez-Lerma, Juan Antonio Rojas-Contreras, Karina Aide Araiza-Ponce, and Damián Reyes-Jáquez. 2025. "Changes in Ruminal Dynamics and Microbial Populations Derived from Supplementation with a Protein Concentrate for Cattle with the Inclusion of Non-Conventional Feeding Sources" Fermentation 11, no. 8: 438. https://doi.org/10.3390/fermentation11080438
APA StyleTorres-Velázquez, D. S., Ramos-Rosales, D. F., Murillo-Ortiz, M., Páez-Lerma, J. B., Rojas-Contreras, J. A., Araiza-Ponce, K. A., & Reyes-Jáquez, D. (2025). Changes in Ruminal Dynamics and Microbial Populations Derived from Supplementation with a Protein Concentrate for Cattle with the Inclusion of Non-Conventional Feeding Sources. Fermentation, 11(8), 438. https://doi.org/10.3390/fermentation11080438







