Transcriptomic Analysis Reveals Opposing Roles of CEL1B in Sophorose- and Lactose-Induced Cellulase Expression in Trichoderma reesei Rut C30
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Construction of CEL1B Gene Knockout T. reesei
2.3. Cellulase Production
2.4. RNA Isolation and High-Throughput RNA-Seq
2.5. Determination of Copy Numbers by qPCR Analysis
2.6. Analytical Methods
3. Results
3.1. Gene Knockout and Phenotypic Analysis of CEL1B in Trichoderma reesei RUT C30
3.2. Impact of CEL1B Gene Knockout on Cellulase Production
3.3. Transcriptome Analysis
3.4. GH Family
3.5. Transporters
3.6. Transcription Factors
3.7. Endoplasmic Reticulum (ER) Protein Processing Pathway
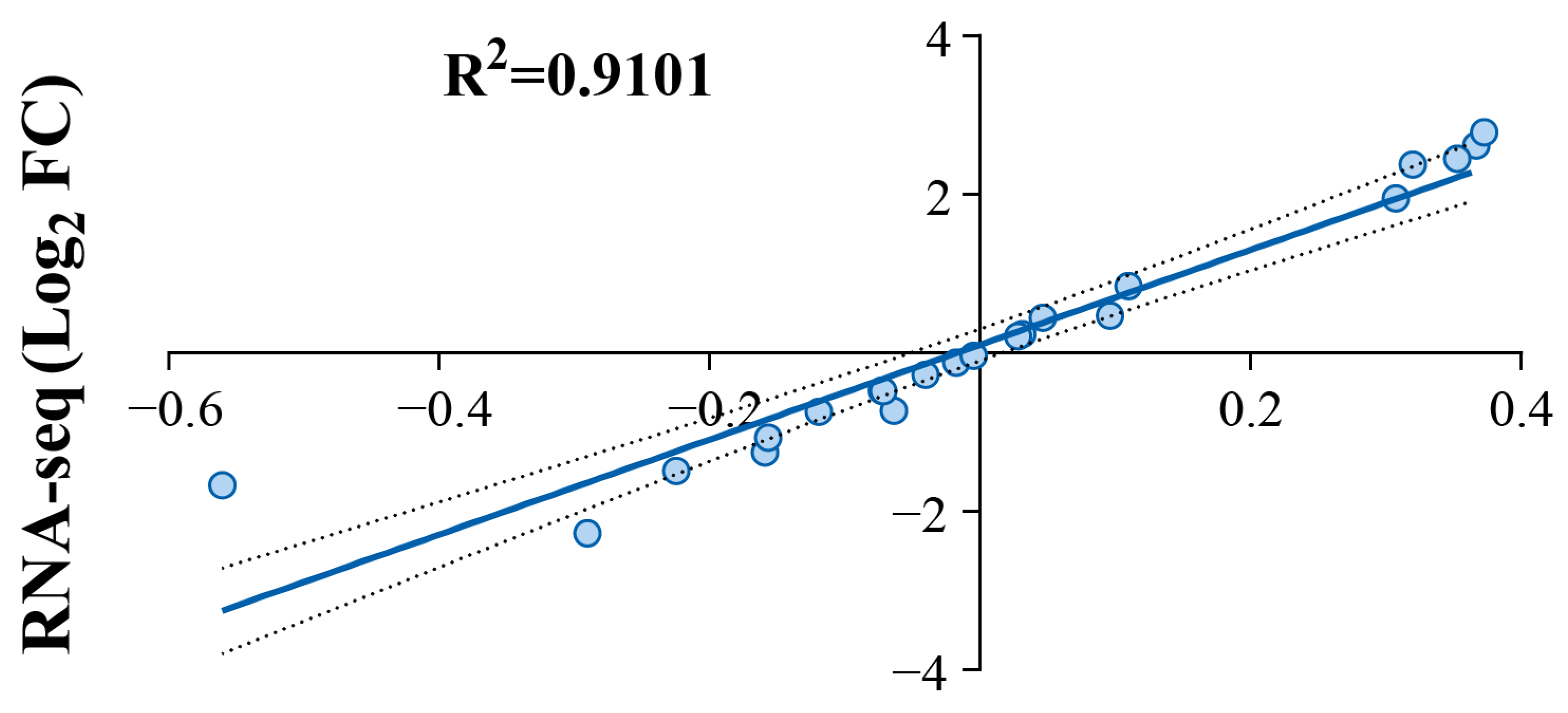
3.8. RT-qPCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MGD | Glucose–sophorose mixture |
| GH families | Glycoside Hydrolase families |
| T. reesei | Trichoderma reesei |
| SLF | Submerged liquid fermentation |
| Sophorose | β-1,2-linked disaccharide |
| CRISPR-Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats associated with Cas9 |
| sgRNA | single guide RNA |
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| KOG | Eukaryotic Orthologous Groups |
| SRA | Sequence Read Archive |
| qPCR | Quantitative Polymerase Chain Reaction |
| DNS | 3,5-dinitrosalicylic acid |
| pNPC | p-nitrophenyl-β-D-cellobioside |
| pNP | p-nitrophenol |
| CMC | Carboxymethyl cellulose |
| BCA | Bicinchoninic acid |
| SDS-PAGE | Sulfate polyacrylamide gel electrophoresis |
| 5-FOA | 5-fluoroorotic acid |
| PCA | Principal Component Analysis |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| TFs | Transcription factors |
| GNAT | Gcn5-related N-acetyltransferase |
| ER | Endoplasmic reticulum |
| ARS | Agricultural Research Service |
References
- Li, G.; Wang, R.; Pang, J.; Wang, A.; Li, N.; Zhang, T. Production of renewable hydrocarbon biofuels with lignocellulose and its derivatives over heterogeneous catalysts. Chem. Rev. 2024, 124, 2889–2954. [Google Scholar] [CrossRef] [PubMed]
- Machineni, L.; Deepanraj, B.; Chew, K.W.; Rao, A.G. Biohydrogen production from lignocellulosic feedstock: Abiotic and biotic methods. Renew. Sustain. Energy Rev. 2023, 182, 113344. [Google Scholar] [CrossRef]
- Shivhare, A.; Kumar, A.; Srivastava, R. Metal phosphate catalysts to upgrade lignocellulose biomass into value-added chemicals and biofuels. Green Chem. 2021, 23, 3818–3841. [Google Scholar] [CrossRef]
- Woiciechowski, A.L.; Neto, C.J.D.; de Souza Vandenberghe, L.P.; de Carvalho Neto, D.P.; Sydney, A.C.N.; Letti, L.A.J.; Karp, S.G.; Torres, L.A.Z.; Soccol, C.R. Lignocellulosic biomass: Acid and alkaline pretreatments and their effects on biomass recalcitrance—Conventional processing and recent advances. Bioresour. Technol. 2020, 304, 122848. [Google Scholar] [CrossRef]
- Das, N.; Jena, P.K.; Padhi, D.; Mohanty, M.K.; Sahoo, G. A comprehensive review of characterization, pretreatment and its applications on different lignocellulosic biomass for bioethanol production. Biomass Convers. Biorefinery 2021, 13, 1503–1527. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Li, H.; Zhao, Q.; Li, X.; Liu, G.; Lu, X.; Zhao, J.; Qu, Y. The combination of continuous-pulse feeding hydrolysates with soybean hulls induction during fed-batch fermentation of Trichoderma reesei b5 significantly elevated the cellulase production and its degradation ability on lignocellulosic biomass. Int. J. Biol. Macromol. 2025, 307, 142244. [Google Scholar] [CrossRef]
- Fonseca, L.M.; Parreiras, L.S.; Murakami, M.T. Rational engineering of the Trichoderma reesei RUT-C30 strain into an industrially relevant platform for cellulase production. Biotechnol. Biofuels 2020, 13, 93. [Google Scholar] [CrossRef]
- Al-Qassab, A.A.; Zakaria, M.R.; Yunus, R.; Salleh, M.A.M.; Mokhtar, M.N. Investigating process parameters to enhance (hemi)cellulolytic enzymes activity produced by Trichoderma reesei RUT-C30 using deoiled oil palm mesocarp fiber in solid-state fermentation. Int. J. Biol. Macromol. 2024, 276, 134030. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, Y.; Wang, Z.; Peng, N.; Ran, X.; Chen, T.; Liu, L.; Li, Y. The Influence of Trctf1 Gene Knockout by CRISPR–Cas9 on Cellulase Synthesis by Trichoderma reesei with Various Soluble Inducers. Fermentation 2023, 9, 746. [Google Scholar] [CrossRef]
- Aich, S.; Datta, S. Engineering of a highly thermostable endoglucanase from the GH7 family of bipolaris sorokiniana for higher catalytic efficiency. Appl. Microbiol. Biotechnol. 2020, 104, 3935–3945. [Google Scholar] [CrossRef] [PubMed]
- Contreras, F.; Pramanik, S.; Rozhkova, A.M.; Zorov, I.N.; Korotkova, O.; Sinitsyn, A.P.; Schwaneberg, U.; Davari, M.D. Engineering robust cellulases for tailored lignocellulosic degradation cocktails. Int. J. Mol. Sci. 2020, 21, 1589. [Google Scholar] [CrossRef] [PubMed]
- Marjamaa, K.; Rahikainen, J.; Karjalainen, M.; Maiorova, N.; Holopainen-Mantila, U.; Molinier, M.; Aro, N.; Nygren, H.; Mikkelson, A.; Koivula, A.; et al. Oxidative modification of cellulosic fibres by lytic polysaccharide monooxygenase AA9A from Trichoderma reesei. Cellulose 2022, 29, 6021–6038. [Google Scholar] [CrossRef]
- Adnan, M.; Ma, X.; Xie, Y.; Waheed, A.; Liu, G. Heterologously expressed cellobiose dehydrogenase acts as efficient electron-donor of lytic polysaccharide monooxygenase for cellulose degradation in Trichoderma reesei. Int. J. Mol. Sci. 2023, 24, 17202. [Google Scholar] [CrossRef]
- Jia, H.; Feng, X.; Huang, J.; Guo, Y.; Zhang, D.; Li, X.; Zhao, J. Recombinant family 1 carbohydrate-binding modules derived from fungal cellulase enhance enzymatic degradation of lignocellulose as novel effective accessory protein. Front. Microbiol. 2022, 13, 876466. [Google Scholar] [CrossRef]
- Li, Y.; Liu, C.; Bai, F.; Zhao, X. Overproduction of cellulase by Trichoderma reesei RUT C30 through batch-feeding of synthesized low-cost sugar mixture. Bioresour. Technol. 2016, 216, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Xu, Y.; Yu, X. From induction to secretion: A complicated route for cellulase production in Trichoderma reesei. Bioresour. Bioprocess. 2021, 8, 107. [Google Scholar] [CrossRef]
- Zhang, P.; Li, Q.; Chen, Y.; Peng, N.; Liu, W.; Wang, X.; Li, Y. Induction of cellulase production in Trichoderma reesei by a glucose–sophorose mixture as an inducer prepared using stevioside. RSC Adv. 2022, 12, 17392–17400. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.; Saito, H.; Nishiyama, R.; Yoshida, N.; Matsubayashi, T.; Teshima, Y.; Yamada, C.; Hiramatsu, S.; Yamada, K.; Kagawa, Y. Isolation of a cellulase hyperproducing mutant strain of Trichoderma reesei. Bioresour. Technol. Rep. 2021, 15, 100733. [Google Scholar] [CrossRef]
- Wang, H.; Pang, A.; Li, B.; Huo, L.; Wu, F.; Lin, F. Intracellular sugar transporters facilitate cellulase synthesis in Trichoderma reesei using lactose. Biomolecules 2023, 13, 295. [Google Scholar] [CrossRef]
- Li, C.; Lin, F.; Zhou, L.; Qin, L.; Li, B.; Zhou, Z.; Jin, M.; Chen, Z. Cellulase hyper-production by Trichoderma reesei mutant SEU-7 on lactose. Biotechnol. Biofuels 2017, 10, 228. [Google Scholar] [CrossRef]
- Ran, X.; Gao, Y.; He, X.; Wang, Z.; Mo, Y.; Li, Y. Enhanced glucose-1-phosphate production from corn stover using cellulases with reduced β-glucosidase activity via Trbgl1 gene knockout in Trichoderma reesei Rut C30. Enzyme Microb. Technol. 2024, 180, 110503. [Google Scholar] [CrossRef] [PubMed]
- Long, T.; Zhang, P.; Yu, J.; Gao, Y.; Ran, X.; Li, Y. Regulation of β-Disaccharide Accumulation by β-Glucosidase Inhibitors to Enhance Cellulase Production in Trichoderma reesei. Fermentation 2022, 8, 232. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, G.; Kou, Y.; Zhang, W.; Zhou, Q.; Chen, G.; Liu, W. Intracellular β-Glucosidases CEL1a and CEL1B Are Essential for Cellulase Induction on Lactose in Trichoderma reesei. Eukaryot. Cell 2014, 13, 1001–1013. [Google Scholar] [CrossRef]
- Pang, A.; Luo, Y.; Hu, X.; Zhang, F.; Wang, H.; Gao, Y.; Durrani, S.; Li, C.; Shi, X.; Wu, F.; et al. Transmembrane transport process and endoplasmic reticulum function facilitate the role of gene CEL1B in cellulase production of Trichoderma reesei. Microb. Cell Factories 2022, 21, 90. [Google Scholar] [CrossRef]
- Jung, K.W.; Jung, J.H.; Park, H.Y. Functional roles of homologous recombination and non-homologous end joining in DNA damage response and microevolution in cryptococcus neoformans. J. Fungi 2021, 7, 566. [Google Scholar] [CrossRef] [PubMed]
- Rassinger, A.; Gacek-Matthews, A.; Strauss, J.; Mach, R.L.; Mach-Aigner, A.R. Truncation of the transcriptional repressor protein Cre1 in Trichoderma reesei rut-C30 turns it into an activator. Fungal Biol. Biotechnol. 2018, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yu, B.; Zhang, K.; Ding, X.; Chen, D. Expression of a Trichoderma reesei beta-xylanase gene in Escherichia coli and activity of the enzyme on fiber-bound substrates. Protein Expr. Purif. 2009, 67, 1–6. [Google Scholar] [CrossRef]
- Ogawa, K.; Ohara, H.; Koide, T.; Toyama, N. Intraspecific hybridization of Trichoderma reesei by protoplast fusion. J. Ferment. Bioeng. 1989, 67, 3. [Google Scholar] [CrossRef]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Jalak, J.; Väljamäe, P. Mechanism of initial rapid rate retardation in cellobiohydrolase catalyzed cellulose hydrolysis. Biotechnol. Bioeng. 2010, 106, 871–883. [Google Scholar] [CrossRef]
- Pang, A.; Wang, H.; Luo, Y.; Yang, Z.; Liu, Z.; Wang, Z.; Li, B.; Yang, S.; Zhou, Z.; Lu, X.; et al. Dissecting cellular function and distribution of β-glucosidases in Trichoderma reesei. mBio 2021, 12, e03671-20. [Google Scholar] [CrossRef]
- Chai, S.; Zhu, Z.; Tian, E.; Xiao, M.; Wang, Y.; Zou, G.; Zhou, Z. Building a versatile protein production platform using engineered Trichoderma reesei. ACS Synth. Biol. 2021, 11, 486–496. [Google Scholar] [CrossRef]
- Li, Y.; Yu, J.; Zhang, P.; Long, T.; Mo, Y.; Li, J.; Li, Q. Comparative transcriptome analysis of Trichoderma reesei reveals different gene regulatory networks induced by synthetic mixtures of glucose and β-disaccharide. Bioresour. Bioprocess. 2021, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Fernandez, P.; Plaza-Vinuesa, L.; Lizasoain-Sánchez, S.; de Las, B.; Muñoz, R.; Jimeno, M.L.; García-Doyagüez, E.; Moreno, F.J.; Corzo, N. Hydrolysis of lactose and transglycosylation of selected sugar alcohols by LacA β-galactosidase from lactobacillus plantarum WCFS1. J. Agric. Food Chem. 2020, 68, 7040–7050. [Google Scholar] [CrossRef]
- Häkkinen, M.; Arvas, M.; Oja, M.; Aro, N.; Penttilä, M.; Saloheimo, M.; Pakula, T.M. Re-annotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb. Cell Factories 2012, 11, 134. [Google Scholar] [CrossRef]
- Florindo, R.N.; Souza, V.P.; Mutti, H.S.; Camilo, C.; Manzine, L.R.; Marana, S.R.; Polikarpov, I.; Nascimento, A.S. Structural insights into β-glucosidase transglycosylation based on biochemical, structural and computational analysis of two GH1 enzymes from trichoderma harzianum. New Biotechnol. 2018, 40, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, K.; Anwar, W.; Subhani, M.N.; Akhter, A.; Iftikhar, S.; Khan, H.A.A.; Shahid, A.A. Glucanase gene of trichoderma; new strategy for the management of root rot disease in chili. J. Soil Sci. Plant Nutr. 2023, 24, 354–370. [Google Scholar] [CrossRef]
- von Freiesleben, P.; Spodsberg, N.; Stenbæk, A.; Stålbrand, H.; Krogh, K.B.R.M.; Meyer, A.S. Boosting of enzymatic softwood saccharification by fungal GH5 and GH26 endomannanases. Biotechnol. Biofuels 2018, 11, 194. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Walton, J.D. Functional diversity for biomass deconstruction in family 5 subfamily 5 (GH5_5) of fungal endo-β1,4-glucanases. Appl. Microbiol. Biotechnol. 2017, 101, 4093–4101. [Google Scholar] [CrossRef]
- Christensen, S.J.; Badino, S.F.; Cavaleiro, A.M.; Borch, K.; Westh, P. Functional analysis of chimeric TrCel6A enzymes with different carbohydrate binding modules. Protein Eng. Des. Sel. 2019, 32, 401–409. [Google Scholar] [CrossRef]
- Haataja, T.; Gado, J.E.; Nutt, A.; Anderson, N.T.; Nilsson, M.; Momeni, M.H.; Isaksson, R.; Väljamäe, P.; Johansson, G.; Payne, C.M.; et al. Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: Insights from crystal structures and MD simulation. FEBS J. 2022, 290, 379–399. [Google Scholar] [CrossRef]
- Haviland, Z.K.; Nong, D.; Zexer, N.; Tien, M.; Anderson, C.T.; Hancock, W.O. Lignin impairs Cel7A degradation of in vitro lignified cellulose by impeding enzyme movement and not by acting as a sink. Biotechnol. Biofuels 2024, 17, 7. [Google Scholar] [CrossRef]
- Zhang, J.; Hong, Y.; Li, K.; Sun, Y.; Yao, C.; Ling, J.; Zhong, Y. Enhancing the production of a heterologous trametes laccase (LacA) by replacement of the major cellulase CBH1 in Trichoderma reesei. J. Ind. Microbiol. Biotechnol. 2023, 50, kuad002. [Google Scholar] [CrossRef]
- Zhang, J.; Li, K.; Sun, Y.; Yao, C.; Liu, W.; Liu, H.; Zhong, Y. An efficient CRISPR/Cas9 genome editing system based on a multiple sgRNA processing platform in Trichoderma reesei for strain improvement and enzyme production. Biotechnol. Biofuels Bioprod. 2024, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Wada, M.; Nishiguchi, H.; Takimura, Y.; Ishii, J. Inducer-free recombinant protein production in Trichoderma reesei: Secretory production of endogenous enzymes and heterologous nanobodies using glucose as the sole carbon source. Microb. Cell Factories 2023, 22, 103. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Wang, Y.; Liu, G.; Zuo, B.; Zhang, Y.; Wang, Y.; Liu, W.; Liu, X.; Zhong, Y. The effects of deletion of cellobiohydrolase genes on carbon source-dependent growth and enzymatic lignocellulose hydrolysis in Trichoderma reesei. J. Microbiol. 2020, 58, 687–695. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, J.; Han, J.; Mei, Y.; Fang, H. Improved consolidated bioprocessing for itaconic acid production by simultaneous optimization of cellulase and metabolic pathway of neurospora crassa. Biotechnol. Biofuels 2024, 17, 57. [Google Scholar] [CrossRef]
- Lee, D.S.; Song, Y.; Lee, Y.G.; Bae, H.J. Comparative evaluation of adsorption of major enzymes in a cellulase cocktail obtained from Trichoderma reesei onto different types of lignin. Polymers 2022, 14, 167. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, R.; Liu, H.; Li, X.; Shen, L.; Zhang, W.; Song, X.; Liu, W.; Liu, X.; Zhong, Y. Development of a powerful synthetic hybrid promoter to improve the cellulase system of Trichoderma reesei for efficient saccharification of corncob residues. Microb. Cell Factories 2022, 21, 5. [Google Scholar] [CrossRef] [PubMed]
- Fitz, E.; Gamauf, C.; Seiboth, B.; Wanka, F. Deletion of the small GTPase rac1 in Trichoderma reesei provokes hyperbranching and impacts growth and cellulase production. Fungal Biol. Biotechnol. 2019, 6, 16. [Google Scholar] [CrossRef]
- Karlsson, J.; Siika-aho, M.; Tenkanen, M.; Tjerneld, F. Enzymatic properties of the low molecular mass endoglucanases Cel12A (EG III) and Cel45A (EG V) of Trichoderma reesei. J. Biotechnol. 2002, 99, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.C.; Agger, J.W.; Wichmann, J.; Meyer, A.S. Oxidative cleavage and hydrolytic boosting of cellulose in soybean spent flakes by Trichoderma reesei Cel61A lytic polysaccharide monooxygenase. Enzym. Microb. Technol. 2017, 98, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Pang, A.; Wang, H.; Luo, Y.; Zhang, F.; Wu, F.; Zhou, Z.; Lu, Z.; Lin, F. Investigating the cellular functions of β-glucosidases for synthesis of lignocellulose-degrading enzymes in Trichoderma reesei. Eng. Microbiol. 2023, 3, 100105. [Google Scholar] [CrossRef]
- Jia, H.; Sun, W.; Li, X.; Zhao, J. Cellulose induced protein 1 (Cip1) from Trichoderma reesei enhances the enzymatic hydrolysis of pretreated lignocellulose. Microb. Cell Factories 2021, 20, 136. [Google Scholar] [CrossRef]
- Shen, L.; Yan, A.; Wang, Y.; Wang, Y.; Liu, H.; Zhong, Y. Tailoring the expression of Xyr1 leads to efficient production of lignocellulolytic enzymes in Trichoderma reesei for improved saccharification of corncob residues. Biotechnol. Biofuels 2022, 15, 142. [Google Scholar] [CrossRef]
- Keller, M.B.; Badino, S.F.; Blossom, B.M.; McBrayer, B.; Borch, K.; Westh, P. Promoting and impeding effects of lytic polysaccharide monooxygenases on glycoside hydrolase activity. ACS Sustain. Chem. Eng. 2020, 8, 14117–14126. [Google Scholar] [CrossRef]
- Wang, H.; Shi, X.; Huo, L.; Tu, J.; Li, C.; Wu, F.; Lin, F. The discovery of novel ER-localized cellobiose transporters involved in cellulase biosynthesis in Trichoderma reesei. J. Basic Microbiol. 2024, 65, e2400573. [Google Scholar] [CrossRef]
- Antoniêto, A.C.C.; de Paula, R.G.; Castro, L.d.S.; Silva-Rocha, R.; Persinoti, G.F.; Silva, R.N. Trichoderma reesei CRE1-mediated Carbon Catabolite Repression in Response to Sophorose Through RNA Sequencing Analysis. Curr. Genomics 2016, 17, 119–131. [Google Scholar] [CrossRef]
- Havukainen, S.; Pujol-Giménez, J.; Valkonen, M.; Hediger, M.A.; Landowski, C.P. Functional characterization of a highly specific l-arabinose transporter from Trichoderma reesei. Microb. Cell Factories 2021, 20, 177. [Google Scholar] [CrossRef]
- Sloothaak, J.; Tamayo-Ramos, J.A.; Odoni, D.I.; Laothanachareon, T.; Derntl, C.; Mach-Aigner, A.R.; Santos, V.A.P.M.D.; Schaap, P.J. Identification and functional characterization of novel xylose transporters from the cell factories Aspergillus niger and Trichoderma reesei. Biotechnol. Biofuels 2016, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, X.; Qin, L.; Wu, H.; Su, X.; Dong, Z. A novel major facilitator transporter TrSTR1 is essential for pentose utilization and involved in xylanase induction in Trichoderma reesei. Biochem. Biophys. Res. Commun. 2015, 460, 663–669. [Google Scholar] [CrossRef]
- Saloheimo, A.; Rauta, J.; Stasyk, O.V.; Sibirny, A.A.; Penttilä, M.; Ruohonen, L. Xylose transport studies with xylose-utilizing saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl. Microbiol. Biotechnol. 2007, 74, 1041–1052. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, Y.; Chen, G.; Liu, W. Identification of the structural determinants for efficient glucose transport via segment swapping between two fungal glucose transporters. RSC Adv. 2017, 7, 25109–25117. [Google Scholar] [CrossRef]
- Yan, S.; Xu, Y.; Yu, X. Role of cellulose response transporter-like protein CRT2 in cellulase induction in Trichoderma reesei. Biotechnol. Biofuels 2023, 16, 118. [Google Scholar] [CrossRef]
- Wang, H.; Pang, A.; Wang, W.; Li, B.; Li, C.; Wu, F.; Lin, F. Discovery of ER-localized sugar transporters for cellulase production with lac1 being essential. Biotechnol. Biofuels 2022, 15, 132. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, R.; Lv, W.; Zhang, W.; Meng, X.; Liu, W. Functional characterization of sugar transporter CRT1 reveals differential roles of its C-terminal region in sugar transport and cellulase induction in Trichoderma reesei. Microbiol. Spectr. 2022, 10, e0087222. [Google Scholar] [CrossRef]
- Havukainen, S.; Valkonen, M.; Koivuranta, K.; Landowski, C.P. Studies on sugar transporter CRT1 reveal new characteristics that are critical for cellulase induction in Trichoderma reesei. Biotechnol. Biofuels 2020, 13, 158. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Meng, X.; Liu, L.; Li, Z.; Jia, K.; Liu, W. Simultaneous in vivo assembly and targeted genome integration of gene clusters in Trichoderma reesei. ACS Synth. Biol. 2025, 14, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Sveholm, E.; Mattila, H.; Aro, N.; Valkonen, M.; Paasela, T.; Pakula, T.M. Transcriptomic and metabolic changes in Trichoderma reesei caused by mutation in xylanase regulator 1 (xyr1). Biotechnol. Biofuels 2024, 17, 106. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Z.; Fang, Y.; Yang, R.; Pu, Y.; Meng, X.; Liu, W. Small GTPase Rab7 is involved in stress adaptation to carbon starvation to ensure the induced cellulase biosynthesis in Trichoderma reesei. Biotechnol. Biofuels 2024, 17, 55. [Google Scholar] [CrossRef]
- Maués, D.B.; Maraschin, J.C.; Duarte, D.Â.; Antoniêto, A.C.C.; Silva, R.N. Overexpression of the Transcription Factor Azf1 Reveals Novel Regulatory Functions and Impacts β-Glucosidase Production in Trichoderma reesei. J. Fungi 2023, 9, 1173. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Zhang, W.; Meng, X.; Liu, W. Single mutation in transcriptional activator Xyr1 enhances cellulase and xylanase production in Trichoderma reesei on glucose. J. Agric. Food Chem. 2023, 71, 11993–12003. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Zhang, W.; Meng, X.; Liu, W. A novel fusion transcription factor drives high cellulase and xylanase production on glucose in Trichoderma reesei. Bioresour. Technol. 2023, 370, 128520. [Google Scholar] [CrossRef]
- Li, N.; Qiu, Z.; Cai, W.; Shen, Y.; Wei, D.; Chen, Y.; Wang, W. The ras small GTPase RSR1 regulates cellulase production in Trichoderma reesei. Biotechnol. Biofuels 2023, 16, 87. [Google Scholar] [CrossRef]
- Li, N.; Li, J.; Chen, Y.; Shen, Y.; Wei, D.; Wang, W. Mechanism of Zn2+ regulation of cellulase production in Trichoderma reesei Rut-C30. Biotechnol. Biofuels Bioprod. 2023, 16, 73. [Google Scholar] [CrossRef]
- Arai, T.; Ichinose, S.; Shibata, N.; Kakeshita, H.; Kodama, H.; Igarashi, K.; Takimura, Y. Inducer-free cellulase production system based on the constitutive expression of mutated XYR1 and ACE3 in the industrial fungus Trichoderma reesei. Sci. Rep. 2022, 12, 19445. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, W.; Liu, P.; Lin, A.; Fan, X.; Wu, C.; Li, N.; Wei, L.; Wei, D. The novel repressor Rce2 competes with Ace3 to regulate cellulase gene expression in the filamentous fungus Trichoderma reesei. Mol. Microbiol. 2021, 116, 1298–1314. [Google Scholar] [CrossRef]
- Sun, Y.; Qian, Y.; Zhang, J.; Yao, C.; Wang, Y.; Liu, H.; Zhong, Y. Development of a novel expression platform for heterologous protein production via deleting the p53-like regulator Vib1 in Trichoderma reesei. Enzym. Microb. Technol. 2022, 155, 109993. [Google Scholar] [CrossRef]
- Chen, X.; Song, B.; Liu, M.; Qin, L.; Dong, Z. Understanding the role of Trichoderma reesei Vib1 in gene expression during cellulose degradation. J. Fungi 2021, 7, 613. [Google Scholar] [CrossRef]
- Ullah, S.F.; Oreb, M.; Boles, E.; Srivastava, V.; Seidl-Seiboth, V.; Seiboth, B.; Kappel, L. N-acetylglucosamine sensing in the filamentous soil fungus Trichoderma reesei. FEBS J. 2025, 292, 3072–3090. [Google Scholar] [CrossRef]
- Fekete, E.; Karaffa, L.; Aghcheh, R.K.; Németh, Z.; Fekete, É.; Orosz, A.; Paholcsek, M.; Stágel, A.; Kubicek, C.P. The transcriptome of lae1 mutants of Trichoderma reesei cultivated at constant growth rates reveals new targets of LAE1 function. BMC Genom. 2014, 15, 447. [Google Scholar] [CrossRef] [PubMed]
- Xin, Q.; Gong, Y.; Lv, X.; Chen, G.; Liu, W. Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, conidiation, and cellulase gene expression. Curr. Microbiol. 2013, 67, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Cao, Y.; Lv, X.; Wang, L.; Li, C.; Zhang, W.; Chen, G.; Liu, W. A copper-responsive promoter replacement system to investigate gene functions in Trichoderma reesei: A case study in characterizing SAGA genes. Appl. Microbiol. Biotechnol. 2016, 101, 2067–2078. [Google Scholar] [CrossRef]
- Shen, L.; Gao, J.; Wang, Y.; Li, X.; Liu, H.; Zhong, Y. Engineering the endoplasmic reticulum secretory pathway in Trichoderma reesei for improved cellulase production. Enzym. Microb. Technol. 2021, 152, 109923. [Google Scholar] [CrossRef]
- Heydenreich, R.M.J. The Impact of the Unfolded Protein Response on Enzyme Secretion in Trichoderma reesei. Ph.D. Thesis, Technische Universität Wien, Vienna, Austria, 2020. Available online: https://repositum.tuwien.at/handle/20.500.12708/15474 (accessed on 19 April 2025).
- Neves-da-Rocha, J.; Santos-Saboya, M.J.; Lopes, M.E.; Rossi, A.; Martinez-Rossi, N. Insights and perspectives on the role of proteostasis and heat shock proteins in fungal infections. Microorganisms 2023, 11, 1878. [Google Scholar] [CrossRef]
- Zhang, F.; Li, J.; Champreda, V.; Liu, C.; Bai, F.; Zhao, X. Global reprogramming of gene transcription in Trichoderma reesei by overexpressing an artificial transcription factor for improved cellulase production and identification of Ypr1 as an associated regulator. Front. Bioeng. Biotechnol. 2020, 8, 649. [Google Scholar] [CrossRef] [PubMed]


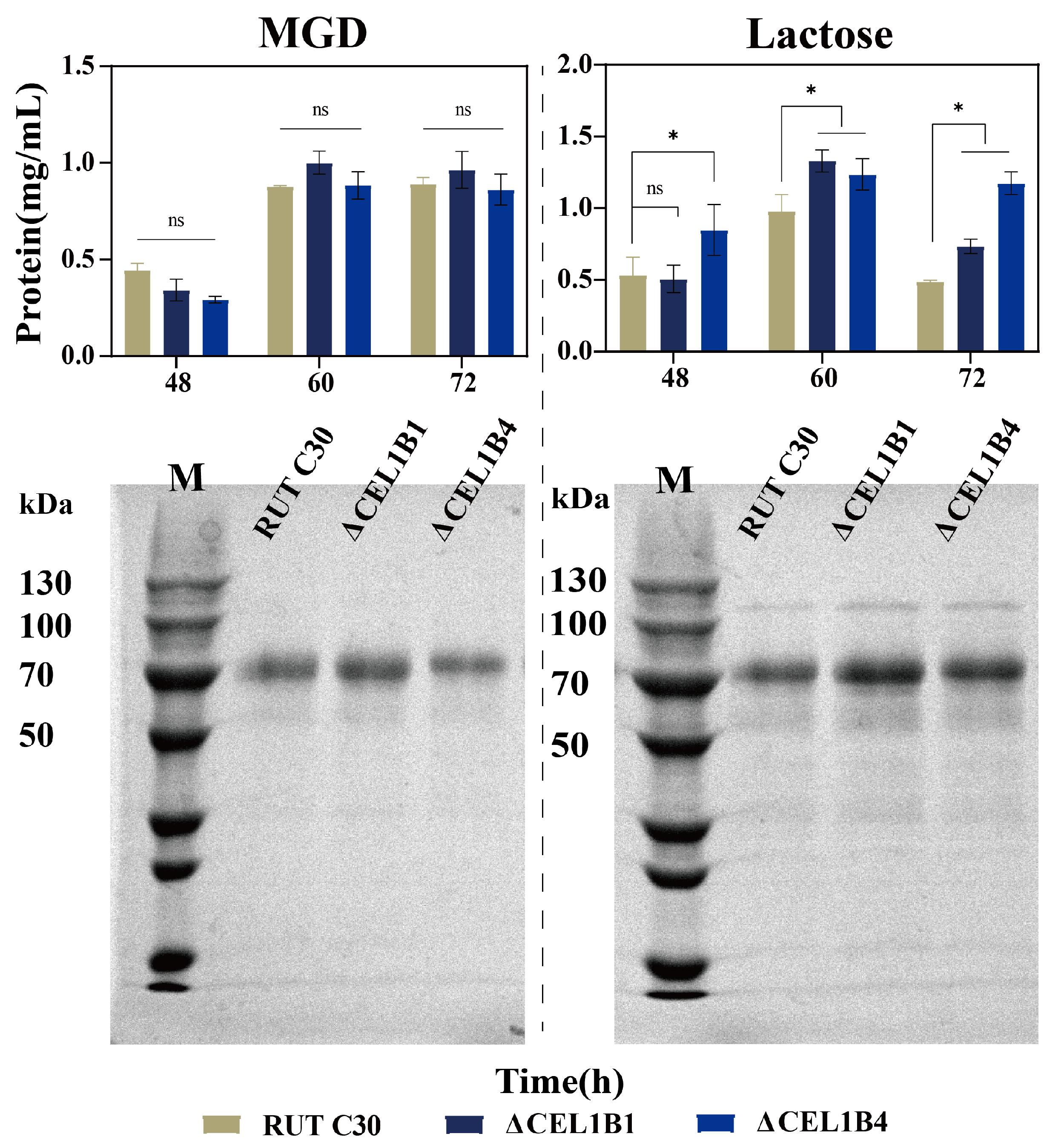
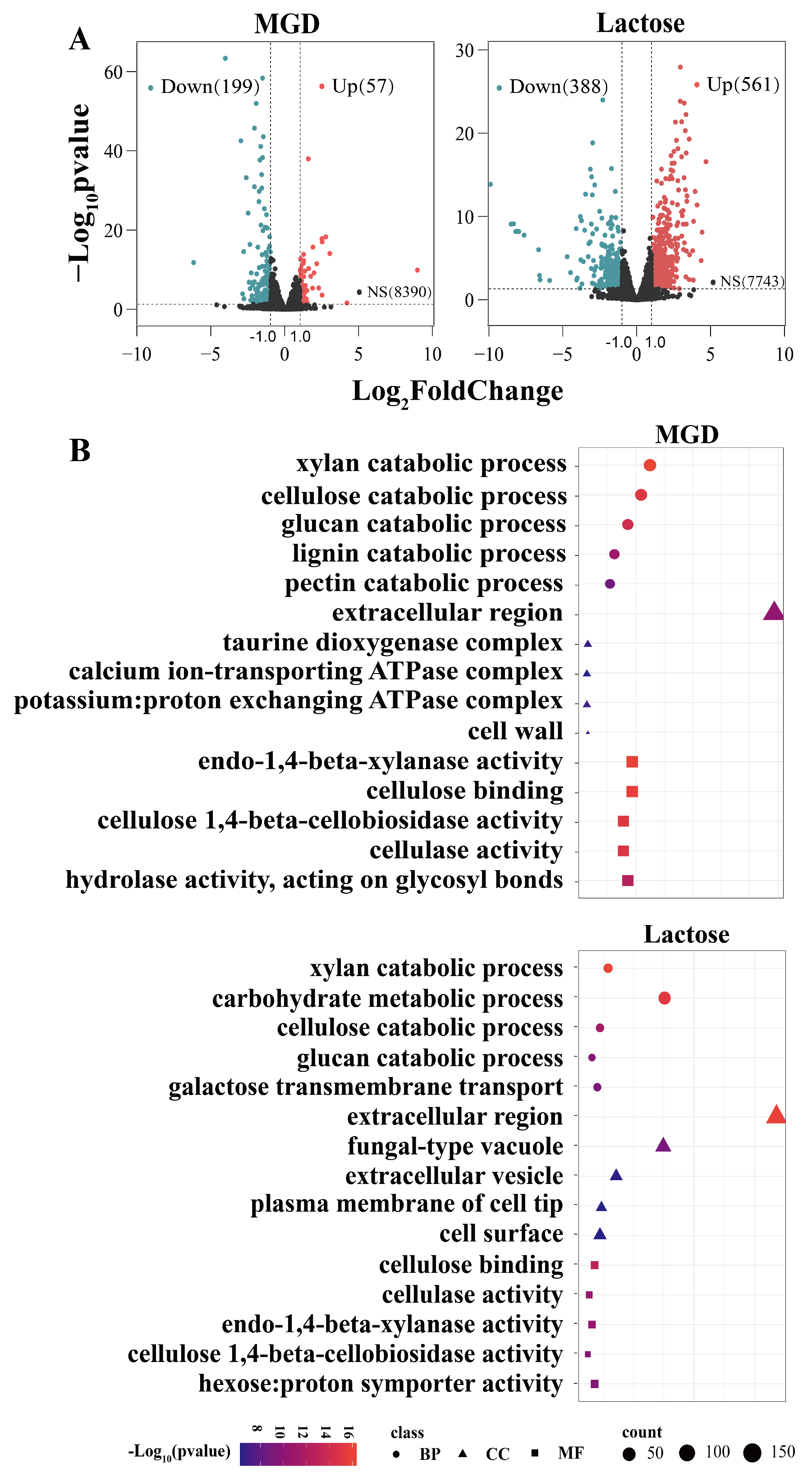
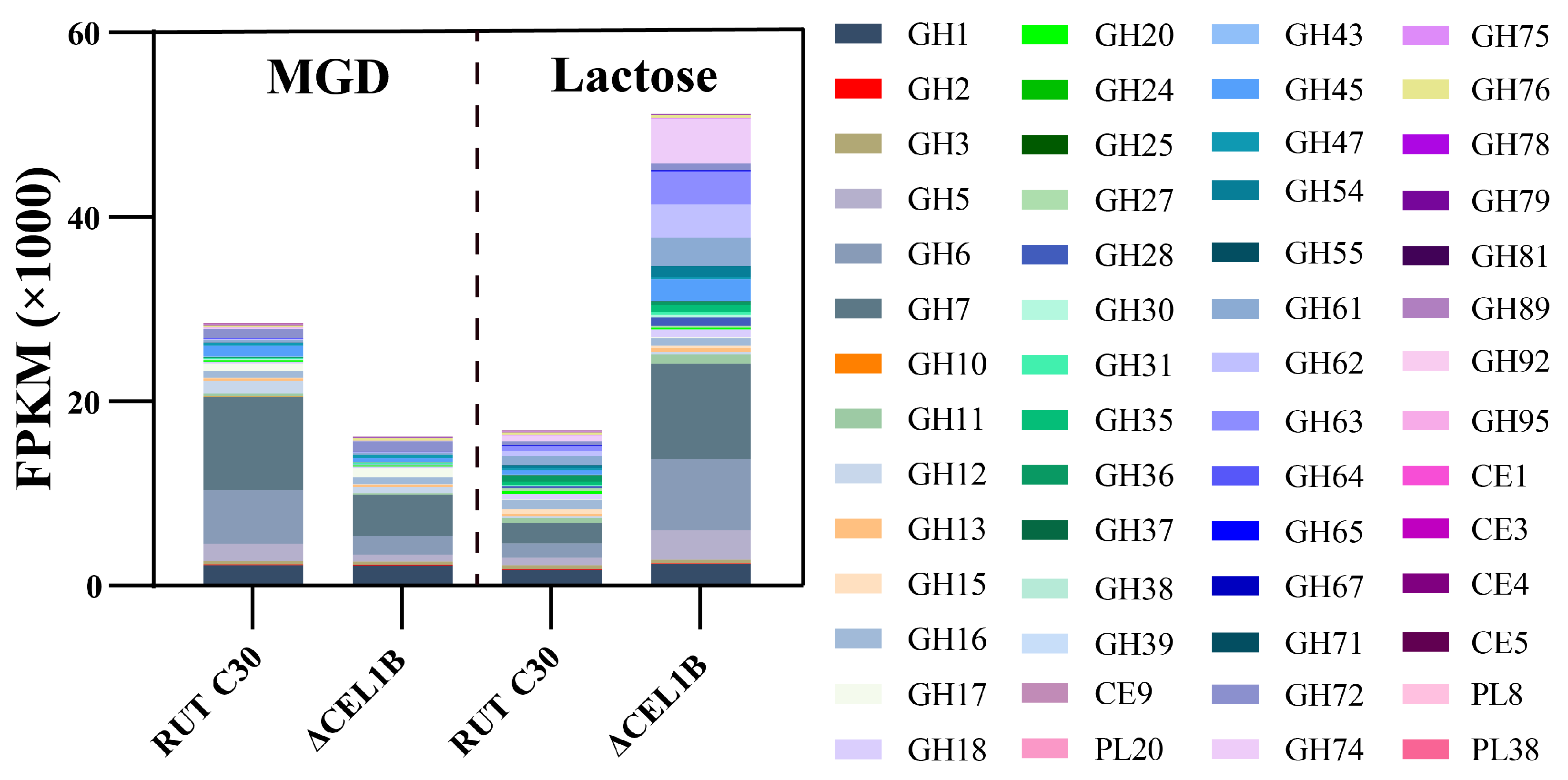
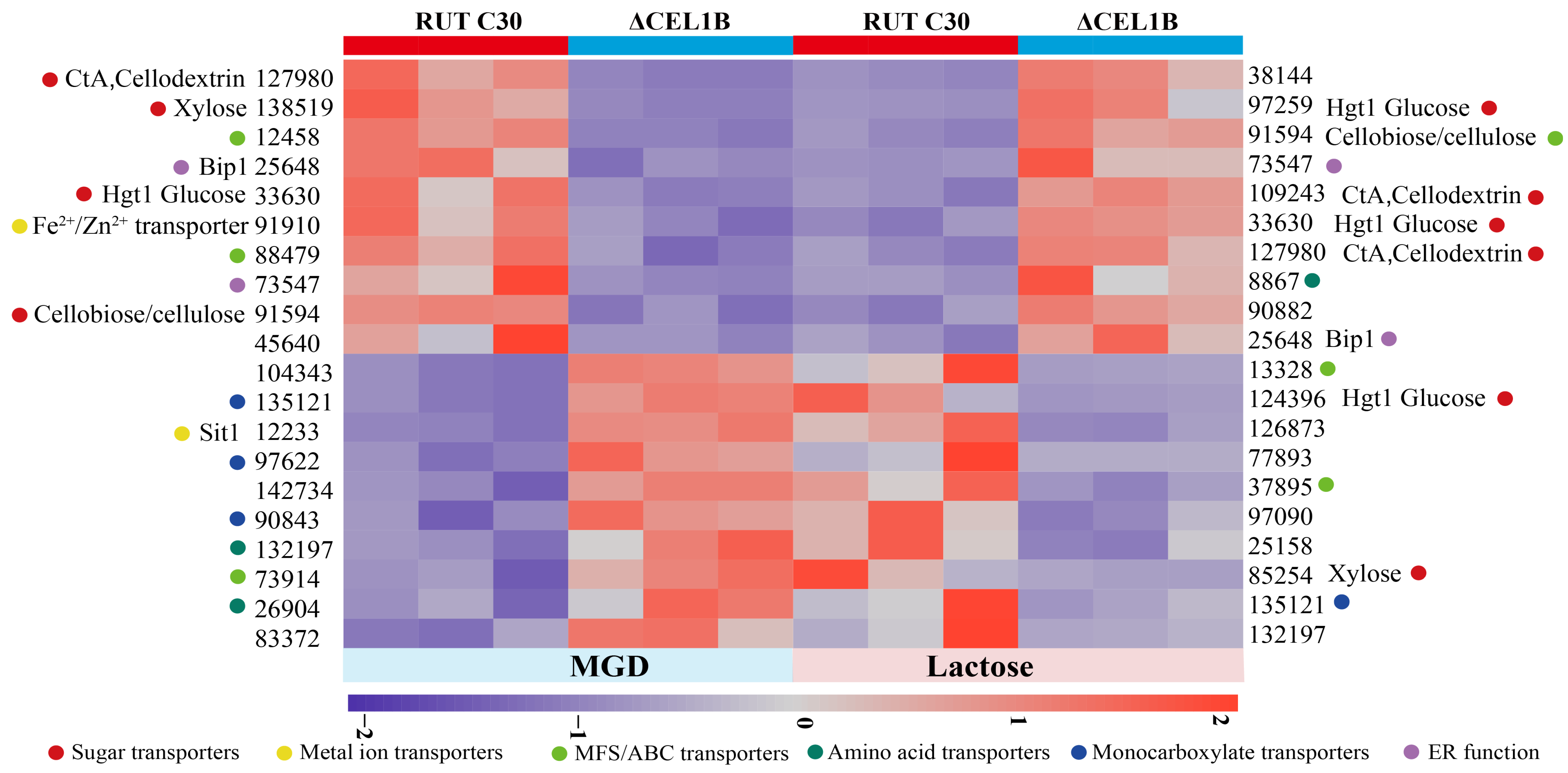
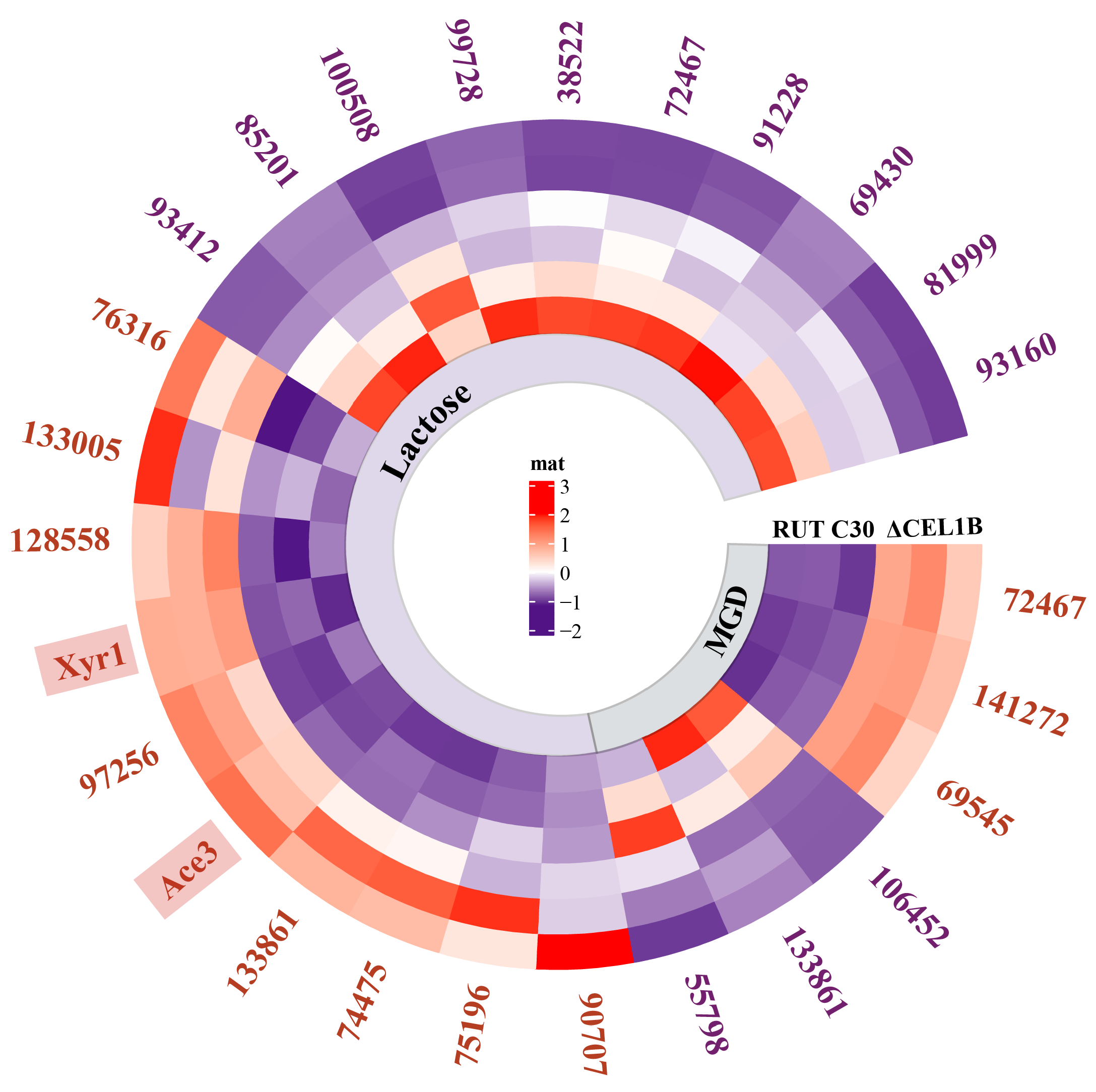
| Category | Gene ID | Description/Name | MGD Log2FC | Lac Log2FC |
|---|---|---|---|---|
| Cellulase | 125125 | Exoglucanase I/CEL7A | −1.3 | 2.4 |
| 122470 | Exoglucanase II/CEL6A | −1.7 | 2.6 | |
| 5304 | Endoglucanase I/CEL7B | −1.6 | 3.4 | |
| 72489 | Endoglucanase III/CEL5A | −1.7 | 2.5 | |
| 124438 | Endoglucanase II/CEL12A | −1.4 | 3.3 | |
| 74496 | Endoglucanase V/CEL45A | NS | NS | |
| 136547 | β-glucosidase/CEL3A | −0.8 | 3.0 | |
| 82126 | β-glucosidase/CEL3H | −1.6 | NS | |
| Auxiliary protein | 104220 | Swollenin | −1.5 | 2.3 |
| 139633 | Endoglucanase IV (AA9) | NS | 2.0 | |
| 122518 | Endoglucanase VII (AA9) | −1.2 | 1.8 | |
| 121449 | Cellulose binding domain/CIP1 | −1.5 | 2.2 | |
| 125575 | Cellulose binding domain/CIP2 | −1.6 | 2.3 |
| Gene ID | Name | MGD Up/Dwon | Lac Up/Dwon | Description |
|---|---|---|---|---|
| 97259 | Hgt1 | NS | Up | Glucose transporter |
| 109243 | Crt1 | NS | Up | Cellulose sensor |
| 136988 | Stp1 | NS | Up | Cellobiose transporter |
| 138519 | Str1 | Down | Up | Xylose transporter |
| 33630 | Xlt1 | Down | Up | Xylose transporter |
| 38765 | NS | Up | Xylose transporter | |
| 91594 | Down | Up | Cellobiose/Xylose transporter | |
| 127980 | Down | Up | Cellodextrin transporter | |
| 55710 | NS | Up | Permease of the major facilitator superfamily | |
| 137001 | Crt2 | NS | Up | Cellulose sensor |
| 137795 | NS | Up | Cellobiose transporter | |
| 79984 | NS | Up | Cellobiose transporter | |
| 124396 | Hxt1 | NS | Down | Glucose transporter |
| Gene ID | Name | Positive/Negative-Acting | MGD Up/Down | MGD Log2FC | Lac Up/Down | Lac Log2FC |
|---|---|---|---|---|---|---|
| 98788 | Xyr1 | Positive | NS | −0.1 | Up | 1.7 |
| 98455 | Ace3 | Positive | NS | −0.7 | Up | 1.8 |
| 125610 | Vib1 | Positive | NS | −0.2 | Up | 1.3 |
| 122363 | Ace1 | Negative | NS | 0.2 | NS | −0.6 |
| 32395 | Ace2 | Positive | NS | −0.1 | NS | 0.5 |
| 93466 | Hap2 | Positive | NS | 0.1 | NS | −0.1 |
| 24298 | Hap3 | Positive | NS | −0.1 | NS | 0.2 |
| 95791 | Pacc | Negative | NS | 0.6 | NS | −0.2 |
| 140814 | Area | Positive | NS | 0.3 | NS | 0.1 |
| 91236 | Bglr | Positive | NS | 0.3 | NS | 0.1 |
| 68701 | Clr-1 | Positive | NS | 0.3 | NS | 0.3 |
| 76250 | Clr-2 | Positive | NS | −0.2 | NS | 0.7 |
| 93861 | Ypr1 | Positive | NS | −0.5 | NS | 0.8 |
| 112571 | Tmac1 | Positive | NS | 0.2 | NS | −0.3 |
| 23706 | Cre1 | Negative | NS | −0.6 | NS | 0.7 |
| 6520 | Rce1 | Negative | NS | −0.2 | NS | 0.7 |
| Gene ID | Name | MGD Up/Dwon | Lac Up/Dwon | Description |
|---|---|---|---|---|
| 124246 | Fes1 | NS | Up | Hsp70 nucleotide exchange factor |
| 121926 | NS | Up | Heat shock protein 70 kDa | |
| 102206 | NS | Up | Heat shock protein 90 | |
| 122594 | NS | Up | DnaJ domain-containing protein | |
| 25648 | Bip1 | Down | Up | Heat shock protein 70 kDa |
| 83445 | ERManI | NS | Up | 1, 2-α-mannosidase |
| 137683 | NEF | NS | Up | Armadillo/beta-catenin-like repeat-containing protein |
| 101105 | ERManI | NS | Up | 1, 2-α-mannosidase |
| 116265 | SPCS1 | Down | NS | Microsomal signal peptidase 12 kDa subunit |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Fan, J.; He, X.; Cheng, J.; Zhang, X.; Tian, T.; Li, Y. Transcriptomic Analysis Reveals Opposing Roles of CEL1B in Sophorose- and Lactose-Induced Cellulase Expression in Trichoderma reesei Rut C30. Fermentation 2025, 11, 439. https://doi.org/10.3390/fermentation11080439
Wang L, Fan J, He X, Cheng J, Zhang X, Tian T, Li Y. Transcriptomic Analysis Reveals Opposing Roles of CEL1B in Sophorose- and Lactose-Induced Cellulase Expression in Trichoderma reesei Rut C30. Fermentation. 2025; 11(8):439. https://doi.org/10.3390/fermentation11080439
Chicago/Turabian StyleWang, Lu, Junping Fan, Xiao He, Jian Cheng, Xinyan Zhang, Tian Tian, and Yonghao Li. 2025. "Transcriptomic Analysis Reveals Opposing Roles of CEL1B in Sophorose- and Lactose-Induced Cellulase Expression in Trichoderma reesei Rut C30" Fermentation 11, no. 8: 439. https://doi.org/10.3390/fermentation11080439
APA StyleWang, L., Fan, J., He, X., Cheng, J., Zhang, X., Tian, T., & Li, Y. (2025). Transcriptomic Analysis Reveals Opposing Roles of CEL1B in Sophorose- and Lactose-Induced Cellulase Expression in Trichoderma reesei Rut C30. Fermentation, 11(8), 439. https://doi.org/10.3390/fermentation11080439







