Unveiling Acetobacter syzygii from Tibetan Kefir Grain: Fermentation-Enhanced Anti-Tyrosinase, and Anti-Melanin
Abstract
1. Introduction
2. Materials and Methods
2.1. Acetobacter sp. Isolation and Identification
2.2. Enzymatic Activity of A. syzygii CCTCC M 2022983 by API
2.3. Genome Sequencing of A. syzygii CCTCC M 2022983
2.4. Cultivation and Fermentation Preparation of A. syzygii CCTCC M 2022983
2.5. Antioxidant Capacity of AFE and UFY
2.6. Anti-Tyrosinase Activity of AFE and UFY
2.7. Effect of AFE, UFY, Succinic Acid and Citric Acid on Cell Viability
2.8. Melanin Content of B16-F10 Cell
2.9. Western Blotting Analysis
2.10. Chemical Compound Identification of AFE and UFY
2.11. Enzyme Kinetic Analysis
2.12. Fluorescence Spectra Analysis
2.13. Inhibition of L-DOPA Auto-Oxidation
2.14. Nucleotide Sequence Accession Number
2.15. Statistical Analysis
3. Results
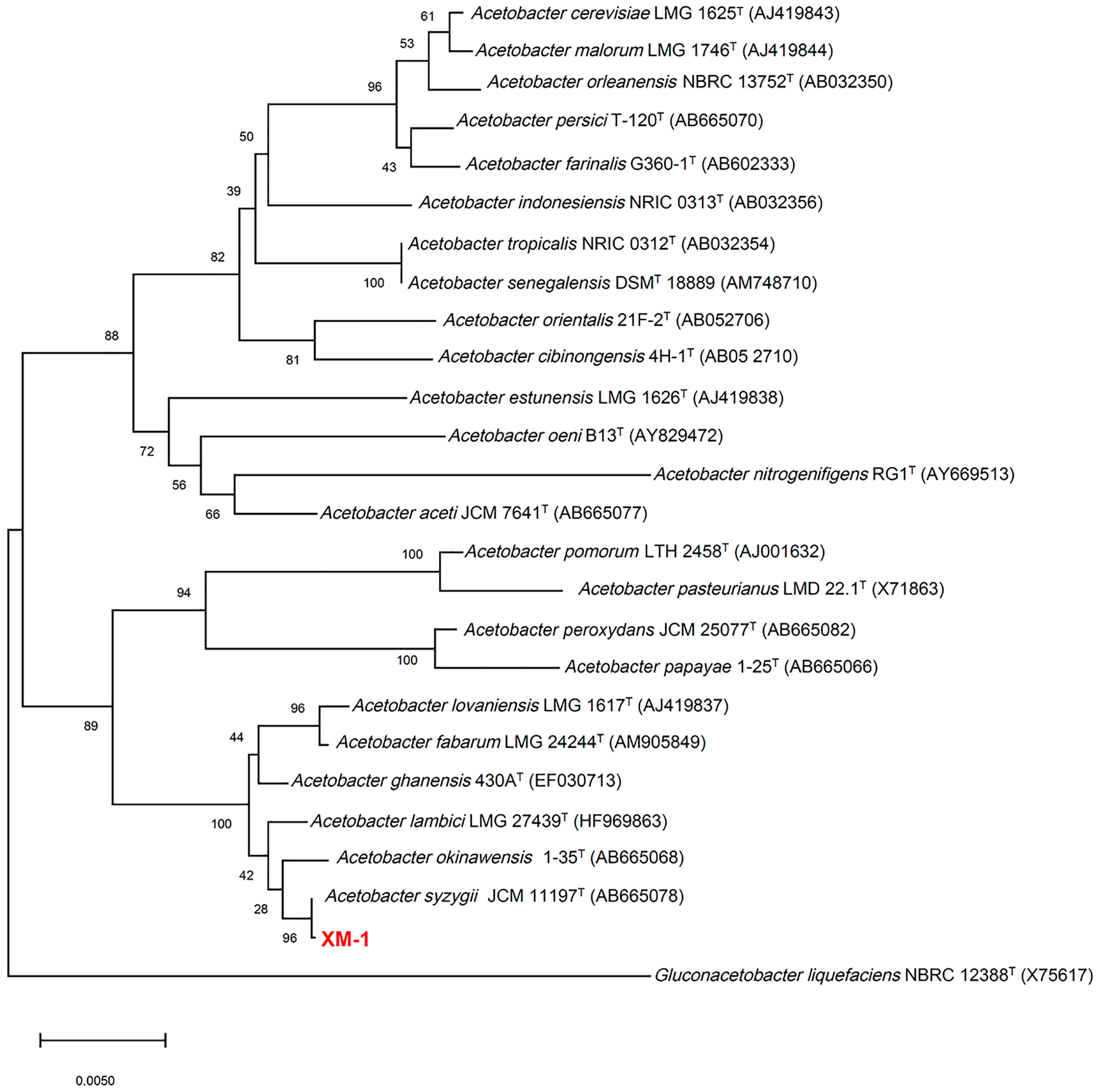
3.1. Identification and Characterization of A. syzygii CCTCC M 2022983
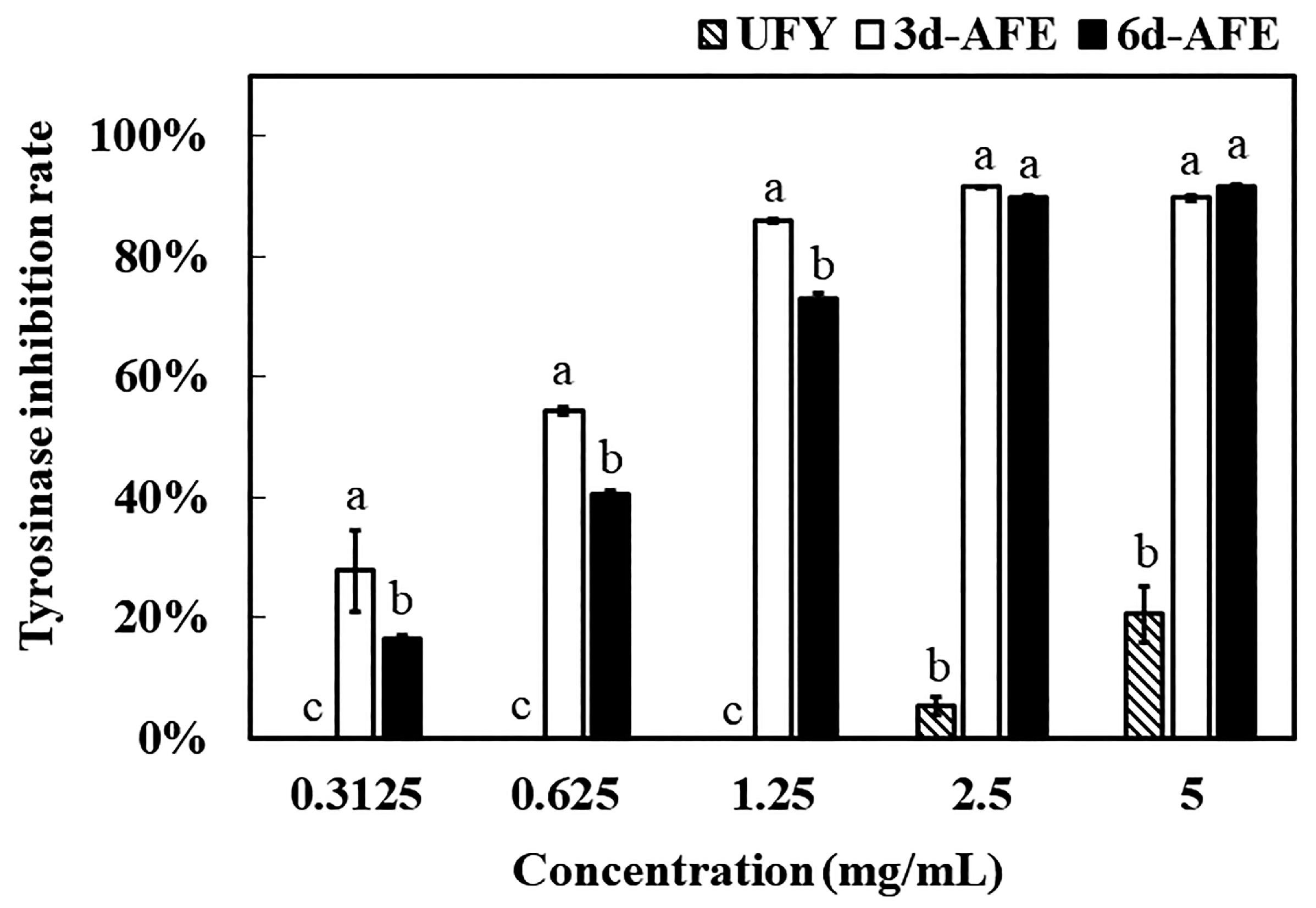
3.2. Antioxidant and Anti-Tyrosinase Capacity of AFE and UFY
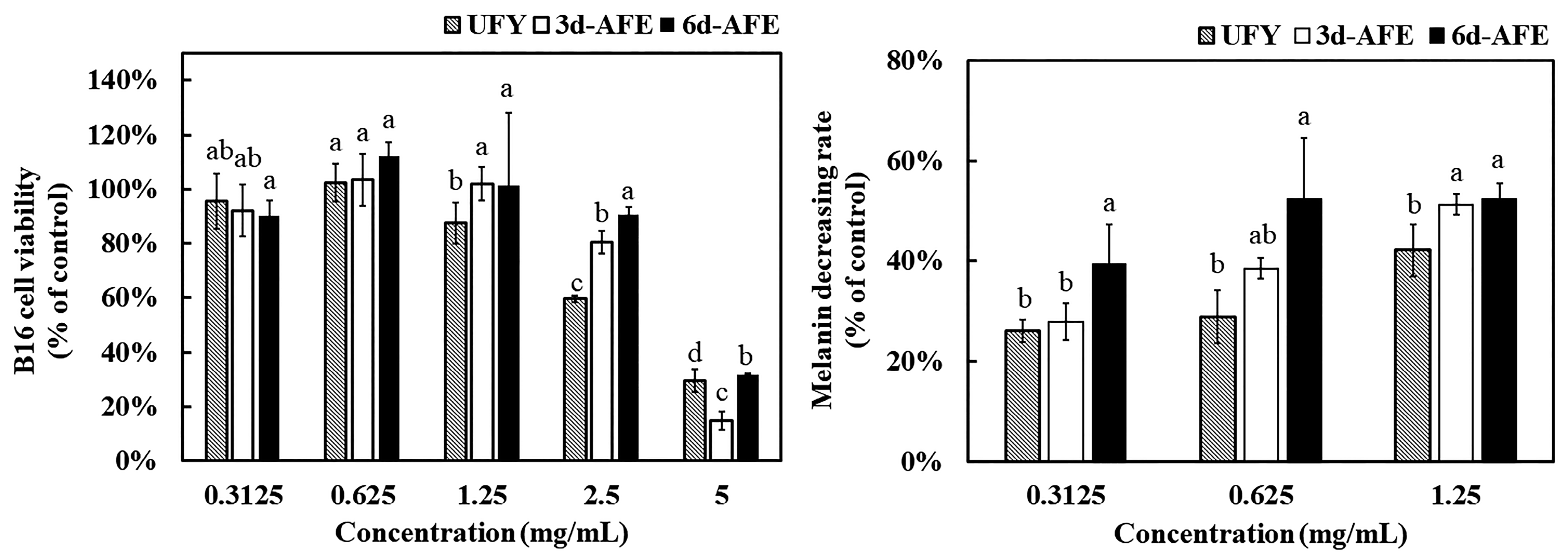
3.3. Melanin Inhibition of AFE and UFY
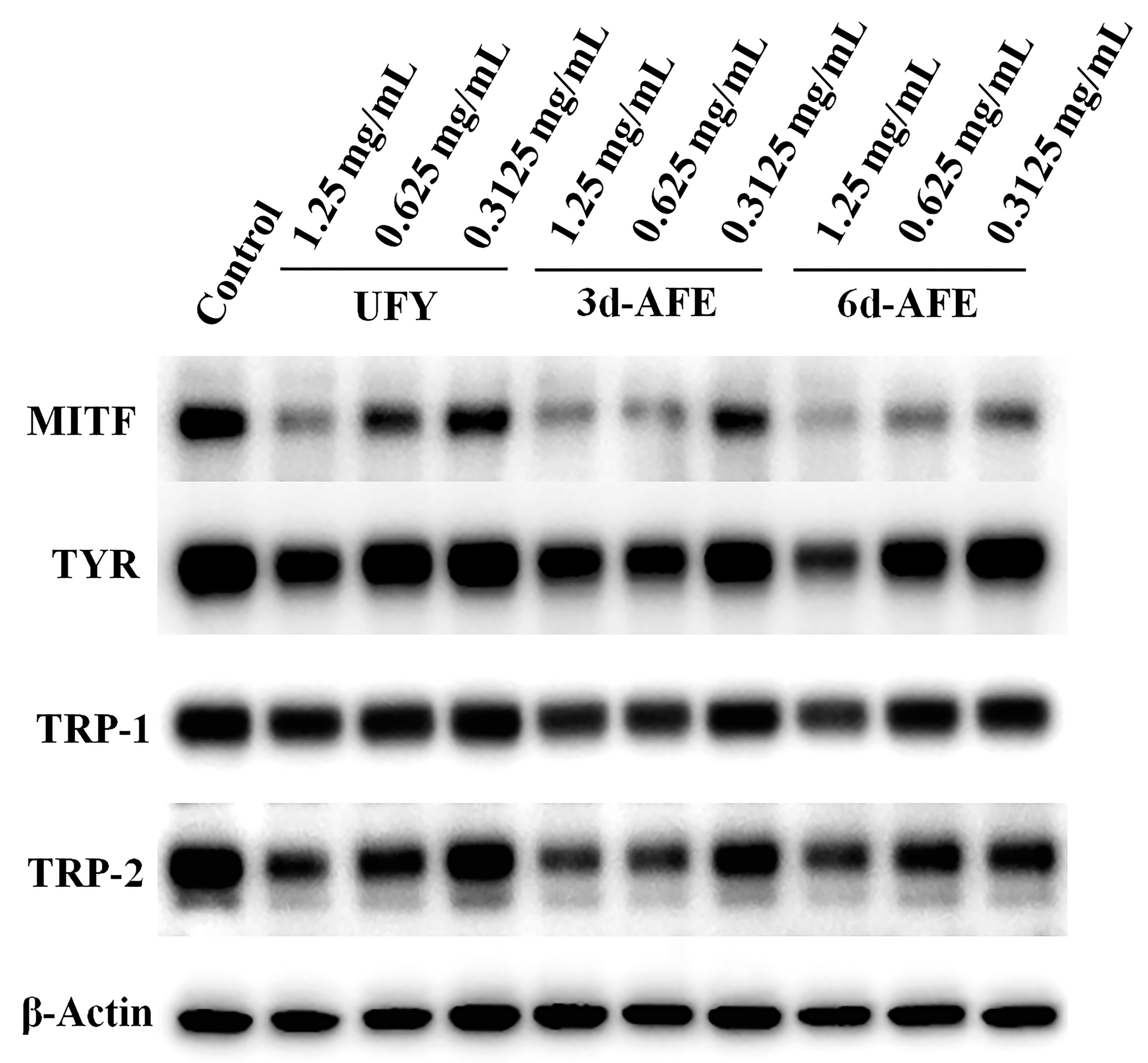
3.4. Western Blotting Analysis of B16-F10 Cells Treated with AFE and UFY
3.5. Chemical Compound Assay of AFE and UFY by LC-MS/MS
3.6. Anti-Tyrosinase Activity of Abundant Compounds in AFE
3.7. Effect of Succinic Acid and Citric Acid on Tyrosinase and L-DOPA
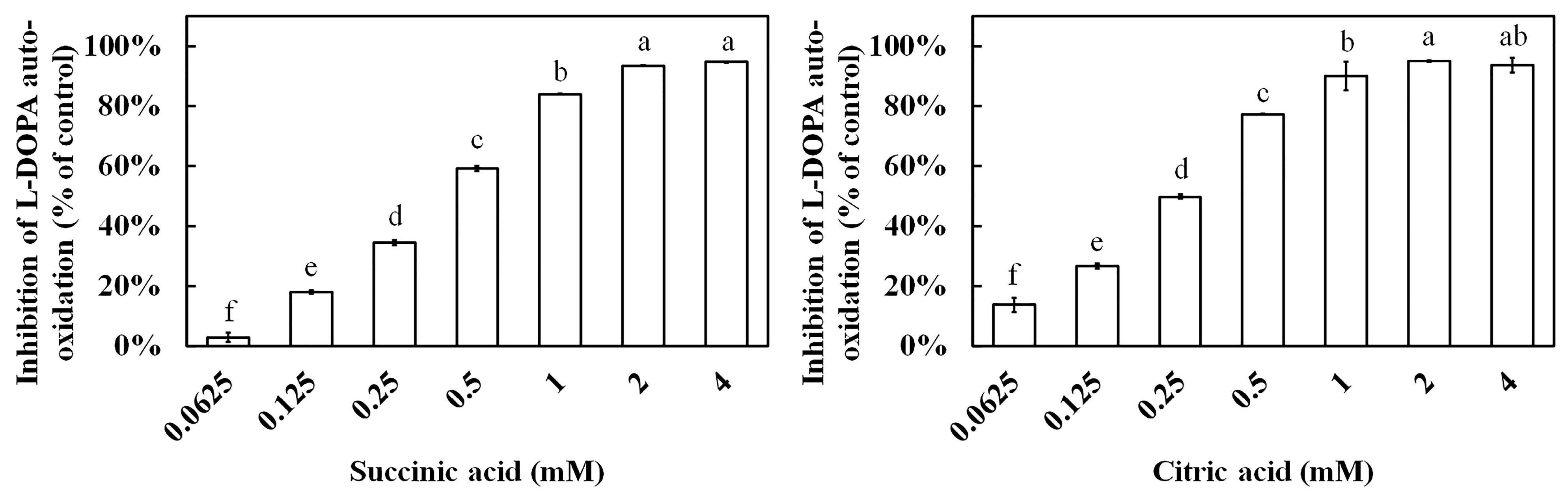
3.8. Anti-Melanin Assay of Succinic Acid and Citric Acid
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gunia-Krzyżak, A.; Popiol, J.; Marona, H. Melanogenesis inhibitors: Strategies for searching for and evaluation of active compounds. Curr. Med. Chem. 2016, 23, 3548–3574. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Fisher, D.E. Melanocyte biology and skin pigmentation. Nature 2007, 445, 843–850. [Google Scholar] [CrossRef]
- Naikoo, S.H.; Rashid, H.; Kumar, S.; Archoo, S.; Sheikh, U.A.; Lone, N.A.; Singh, P.P.; Tasduq, S.A. Inhibition of melanogenesis by 3-(1′-methyltetrahydropyridinyl)-2,4-6-trihydroxy acetophenone via suppressing the activity of cAMP response element-binding protein (CREB) and nuclear exclusion of CREB-regulated transcription coactivator 1 (CRTC1). Eur. J. Pharmacol. 2023, 952, 175734. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Shin, D.W.; Lim, B.O. Fermented Aronia melanocarpa inhibits melanogenesis through dual mechanisms of the PI3K/AKT/GSK-3β and PKA/CREB pathways. Molecules 2023, 28, 2981. [Google Scholar] [CrossRef]
- Wu, K.C.; Hseu, Y.C.; Shih, Y.C.; Sivakumar, G.; Syu, J.T.; Chen, G.L.; Lu, M.T.; Chu, P.C. Calycosin, a common dietary isoflavonoid, suppresses melanogenesis through the downregulation of PKA/CREB and p38 MAPK signaling pathways. Int. J. Mol. Sci. 2022, 23, 1358. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.S. An updated review of tyrosinase inhibitors. Int. J. Mol. Sci. 2009, 10, 2440–2475. [Google Scholar] [CrossRef]
- Allouche, J.; Rachmin, I.; Adhikari, K.; Pardo, L.M.; Lee, J.H.; McConnell, A.M.; Kato, S.; Fan, S.; Kawakami, A.; Suita, Y.; et al. NNT mediates redox-dependent pigmentation via a UVB- and MITF-independent mechanism. Cell 2021, 184, 4268–4283.e4220. [Google Scholar] [CrossRef]
- Zaidi, K.U.; Ali, A.S.; Ali, S.A. Purification and characterization of melanogenic enzyme tyrosinase from button mushroom. Enzym. Res. 2014, 2014, 120739. [Google Scholar] [CrossRef]
- Jin, Y.H.; Lee, S.J.; Chung, M.H.; Park, J.H.; Park, Y.I.; Cho, T.H.; Lee, S.K. Aloesin and arbutin inhibit tyrosinase activity in a synergistic manner via a different action mechanism. Arch. Pharm. Res. 1999, 22, 232–236. [Google Scholar] [CrossRef]
- Wang, W.; Yang, L.; Wang, W.; Zhang, J.; Engelhardt, U.H.; Jiang, H. Inhibitory activities of samples on tyrosinases were affected by enzyme species and sample addition methods. Int. J. Mol. Sci. 2023, 24, 6013. [Google Scholar] [CrossRef]
- Mann, T.; Gerwat, W.; Batzer, J.; Eggers, K.; Scherner, C.; Wenck, H.; Stäb, F.; Hearing, V.J.; Röhm, K.H.; Kolbe, L. Inhibition of human tyrosinase requires molecular motifs distinctively different from mushroom tyrosinase. J. Investig. Dermatol. 2018, 138, 1601–1608. [Google Scholar] [CrossRef] [PubMed]
- Lopes, T.I.B.; Coelho, R.G.; Honda, N.K. Inhibition of mushroom tyrosinase activity by orsellinates. Chem. Pharm. Bull. 2018, 66, 61–64. [Google Scholar] [CrossRef]
- Chai, W.M.; Lin, M.Z.; Song, F.J.; Wang, Y.X.; Xu, K.L.; Huang, J.X.; Fu, J.P.; Peng, Y.Y. Rifampicin as a novel tyrosinase inhibitor: Inhibitory activity and mechanism. Int. J. Biol. Macromol. 2017, 102, 425–430. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Guo, Z.; Tan, T.; Zhang, Y. Novel tyrosinase inhibitory peptide with free radical scavenging ability. J. Enzyme Inhib. Med. Chem. 2019, 34, 1633–1640. [Google Scholar] [CrossRef]
- Chen, M.Y.; Wu, H.T.; Chen, F.F.; Wang, Y.T.; Chou, D.L.; Wang, G.H.; Chen, Y.P. Characterization of Tibetan kefir grain-fermented milk whey and its suppression of melanin synthesis. J. Biosci. Bioeng. 2022, 133, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef]
- Hunt, M.; Newbold, C.; Berriman, M.; Otto, T.D. A comprehensive evaluation of assembly scaffolding tools. Genome Biol. 2014, 15, R42. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Zerbino, D.R.; McEwen, G.K.; Margulies, E.H.; Birney, E. Pebble and rock band: Heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PLoS ONE 2009, 4, e8407. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Blake, J.A.; Christie, K.R.; Dolan, M.E.; Drabkin, H.J.; Hill, D.P.; Ni, L.; Sitnikov, D.; Burgess, S.; Buza, T.; Gresham, C.; et al. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Reig, M.; Jaumot, J.; García-Reiriz, A.; Tauler, R. Evaluation of changes induced in rice metabolome by Cd and Cu exposure using LC-MS with XCMS and MCR-ALS data analysis strategies. Anal. Bioanal. Chem. 2015, 407, 8835–8847. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Shang, C.; Zhang, Y.; You, X.; Guo, N.; Wang, Y.; Fan, Y.; Liu, W. The effect of 7,8,4′-trihydroxyflavone on tyrosinase activity and conformation: Spectroscopy and docking studies. Lumin. J. Biol. Chem. Lumin. 2018, 33, 681–691. [Google Scholar] [CrossRef]
- Sriphochanart, W.; Krusong, W.; Mekkerdchoo, O.; Suwapanich, R.; Sriprom, P.; Tantratian, S. Impact of temperature on gluconic acid production during acetification by Acetobacter aceti. Biotechnol. Appl. Biochem. 2023, 70, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, R.; Siddartha, G.; Sundhar Reddy, C.H.; Harish, B.S.; Janaki Ramaiah, M.; Uppuluri, K.B. Antioxidant and anti-inflammatory levan produced from Acetobacter xylinum NCIM2526 and its statistical optimization. Carbohydr. Polym. 2015, 123, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Viana, R.O.; Magalhães-Guedes, K.T.; Braga, R.A., Jr.; Dias, D.R.; Schwan, R.F. Fermentation process for production of apple-based kefir vinegar: Microbiological, chemical and sensory analysis. Braz. J. Microbiol. 2017, 48, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yao, H.; Liu, Q.; Zheng, Z.; Cao, L.; Mu, D.; Wang, H.; Jiang, S.; Li, X. Producing acetic acid of Acetobacter pasteurianus by fermentation characteristics and metabolic flux analysis. Appl. Biochem. Biotechnol. 2018, 186, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Rhee, D.Y.; Bang, S.H.; Kim, S.Y.; Won, C.H.; Lee, M.W.; Choi, J.H.; Chang, S.E. The natural yeast extract isolated by ethanol precipitation inhibits melanin synthesis by modulating tyrosinase activity and downregulating melanosome transfer. Biosci. Biotechnol. Biochem. 2015, 79, 1504–1511. [Google Scholar] [CrossRef][Green Version]
- Sardar, T.; Maqbool, M.; Ishtiaq, M.; Mazhar, M.W.; El-Sheikh, M.A.; Casini, R.; Mahmoud, E.A.; Elansary, H.O. Synergistic influence of yeast extract and calcium oxide nanoparticles on the synthesis of bioactive antioxidants and metabolites in Swertia chirata in vitro callus cultures. Molecules 2023, 28, 4607. [Google Scholar] [CrossRef]
- Verhoef, S.; Ruijssenaars, H.J.; de Bont, J.A.; Wery, J. Bioproduction of p-hydroxybenzoate from renewable feedstock by solvent-tolerant Pseudomonas putida S12. J. Biotechnol. 2007, 132, 49–56. [Google Scholar] [CrossRef]
- Dong, L.M.; Jia, X.C.; Luo, Q.W.; Zhang, Q.; Luo, B.; Liu, W.B.; Zhang, X.; Xu, Q.L.; Tan, J.W. Phenolics from Mikania micrantha and their antioxidant activity. Molecules 2017, 22, 1140. [Google Scholar] [CrossRef]
- Gheibi, N.; Saboury, A.A.; Haghbeen, K.; Rajaei, F.; Pahlevan, A.A. Dual effects of aliphatic carboxylic acids on cresolase and catecholase reactions of mushroom tyrosinase. J. Enzym. Inhib. Med. Chem. 2009, 24, 1076–1081. [Google Scholar] [CrossRef]
- Yu, L. Inhibitory effects of (S)- and (R)-6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acids on tyrosinase activity. J. Agric. Food Chem. 2003, 51, 2344–2347. [Google Scholar] [CrossRef]
- Liu, W.; Zou, L.Q.; Liu, J.P.; Zhang, Z.Q.; Liu, C.M.; Liang, R.H. The effect of citric acid on the activity, thermodynamics and conformation of mushroom polyphenoloxidase. Food Chem. 2013, 140, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhou, L.; Liao, T.; Liu, J.; Yu, K.; Zou, L.; Zhou, W.; Liu, W. Comparing the effect of benzoic acid and cinnamic acid hydroxyl derivatives on polyphenol oxidase: Activity, action mechanism, and molecular docking. J. Sci. Food Agric. 2022, 102, 3771–3780. [Google Scholar] [CrossRef]
- Varela, M.T.; Ferrarini, M.; Mercaldi, V.G.; Sufi, B.D.S.; Padovani, G.; Nazato, L.I.S.; Fernandes, J.P.S. Coumaric acid derivatives as tyrosinase inhibitors: Efficacy studies through in silico, in vitro and ex vivo approaches. Bioorg. Chem. 2020, 103, 104108. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.; Zhou, L.; Liu, J.; Zou, L.; Dai, T.; Liu, W. Inhibitory mechanism of salicylic acid on polyphenol oxidase: A cooperation between acidification and binding effects. Food Chem. 2021, 348, 129100. [Google Scholar] [CrossRef]
- Mccord, J.D.; Kilara, A. Control of enzymatic browning in processed mushrooms (Agaricus bisporus). J. Food Sci. 1983, 48, 1479–1484. [Google Scholar] [CrossRef]
- Yin, S.J.; Si, Y.X.; Qian, G.Y. Inhibitory effect of phthalic acid on tyrosinase: The mixed-type inhibition and docking simulations. Enzym. Res. 2011, 2011, 294724. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.J.; Si, Y.X.; Chen, Y.F.; Qian, G.Y.; Lü, Z.R.; Oh, S.; Lee, J.; Lee, S.; Yang, J.M.; Lee, D.Y.; et al. Mixed-type inhibition of tyrosinase from Agaricus bisporus by terephthalic acid: Computational simulations and kinetics. Protein J. 2011, 30, 273–280. [Google Scholar] [CrossRef]
- Hwang, J.H.; Lee, B.M. Inhibitory effects of plant extracts on tyrosinase, L-DOPA oxidation, and melanin synthesis. J. Toxicol. Environ. Health Part A 2007, 70, 393–407. [Google Scholar] [CrossRef]
- Ismaya, W.T.; Rozeboom, H.J.; Weijn, A.; Mes, J.J.; Fusetti, F.; Wichers, H.J.; Dijkstra, B.W. Crystal structure of Agaricus bisporus mushroom tyrosinase: Identity of the tetramer subunits and interaction with tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Maekawa, A.; Todate, A.; Onodera, H.; Matsushima, Y.; Nagaoka, T.; Shibutani, M.; Ogasawara, H.; Kodama, Y.; Hayashi, Y. Lack of toxicity/carcinogenicity of monosodium succinate in F344 rats. Food Chem. Toxicol. 1990, 28, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Saleem, R.; Ahmad, M.; Naz, A.; Siddiqui, H.; Ahmad, S.I.; Faizi, S. Hypotensive and toxicological study of citric acid and other constituents from Tagetes patula roots. Arch. Pharmacal Res. 2004, 27, 1037–1042. [Google Scholar] [CrossRef] [PubMed]




| Compounds | mz | Retention Time (s) | Relative Abundance | Relative Ratio | |||
|---|---|---|---|---|---|---|---|
| UFY | 3d * | 6d * | 3d/UFY | 6d/UFY | |||
| Succinic acid | 118.03 | 142.2 | 9.46 × 108 ± 2.74 × 108 | 4.72 × 1010 ± 3.44 × 109 | 4.26 × 1010 ± 7.48 × 109 | 49.88 | 45.05 |
| Pipecolic acid | 129.08 | 151.3 | 5.83 × 109 ± 3.11 × 108 | 3.68 × 1010 ± 5.09 × 109 | 3.36 × 1010 ± 3.82 × 109 | 6.31 | 5.77 |
| 4-Acetylbutyrate | 130.06 | 317 | 4.10 × 108 ± 6.07 × 107 | 3.13 × 1010 ± 8.88 × 108 | 2.51 × 1010 ± 1.09 × 109 | 76.39 | 61.22 |
| Iminoarginine | 172.10 | 344.8 | 9.47 × 109 ± 1.15 × 109 | 2.78 × 1010 ± 1.09 × 109 | 2.42 × 1010 ± 1.08 × 109 | 2.94 | 2.55 |
| Citric acid | 192.03 | 68.4 | 5.92 × 109 ± 1.11 × 108 | 2.46 × 1010 ± 9.11 × 108 | 2.42 × 1010 ± 5.92 × 108 | 4.16 | 4.08 |
| N-Acetylleucine | 173.11 | 494.6 | 1.64 × 109 ± 3.17 × 107 | 1.31 × 1010 ± 2.46 × 108 | 1.25 × 1010 ± 5.93 × 108 | 7.99 | 7.62 |
| Isopentenyl adenosine | 335.16 | 512.3 | 1.33 × 1010 ± 1.37 × 109 | 1.04 × 1010 ± 6.76 × 108 | 1.03 × 1010 ± 3.01 × 108 | 0.78 | 0.77 |
| 3-Dehydrosphinganine | 299.28 | 779.9 | 1.23 × 106 ± 3.90 × 105 | 2.23 × 1010 ± 4.67 × 109 | 9.18 × 109 ± 3.49 × 109 | 18,100.09 | 7437.96 |
| Gluconic acid | 196.06 | 115.7 | 4.40 × 106 ± 7.49 × 105 | 9.09 × 109 ± 1.82 × 109 | 1.20 × 1010 ± 3.85 × 109 | 2064.51 | 2732.81 |
| Geranyl diphosphate | 314.07 | 197.3 | 2.75 × 1010 ± 7.82 × 109 | 1.00 × 1010 ± 6.24 × 108 | 9.20 × 109 ± 7.79 × 108 | 0.36 | 0.33 |
| L-Threo-2-pentulose | 150.05 | 115.4 | 4.85 × 107 ± 9.01 × 105 | 8.04 × 109 ± 3.61 × 109 | 8.65 × 109 ± 3.34 × 109 | 165.73 | 178.36 |
| N-Acetyl-L-phenylalanine | 207.09 | 522.8 | 9.67 × 108 ± 2.98 × 107 | 8.21 × 109 ± 4.91 × 107 | 7.72 × 109 ± 2.72 × 108 | 8.50 | 7.99 |
| 6-Hydroxyhexanoic acid | 132.08 | 278.8 | 4.63 × 109 ± 1.94 × 108 | 8.14 × 109 ± 3.07 × 109 | 9.17 × 109 ± 2.93 × 109 | 1.76 | 1.98 |
| 3-Hydroxymethylglutaric acid | 162.05 | 142.9 | 8.63 × 108 ± 3.58 × 107 | 7.60 × 109 ± 4.46 × 108 | 7.47 × 109 ± 5.07 × 108 | 8.81 | 8.65 |
| D-Mannose | 180.06 | 88.2 | 4.28 × 1010 ± 1.06 × 1010 | 6.42 × 109 ± 1.12 × 109 | 7.38 × 109 ± 1.26 × 109 | 0.15 | 0.17 |
| 4-Hydroxybenzoic acid | 138.03 | 279.6 | 1.54 × 108 ± 1.28 × 108 | 5.47 × 109 ± 4.32 × 109 | 7.12 × 109 ± 3.71 × 109 | 35.57 | 46.30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, L.; He, Q.; Xu, M.; Chen, F.-F.; Li, F.; Chen, Y.-P. Unveiling Acetobacter syzygii from Tibetan Kefir Grain: Fermentation-Enhanced Anti-Tyrosinase, and Anti-Melanin. Fermentation 2024, 10, 459. https://doi.org/10.3390/fermentation10090459
Zhong L, He Q, Xu M, Chen F-F, Li F, Chen Y-P. Unveiling Acetobacter syzygii from Tibetan Kefir Grain: Fermentation-Enhanced Anti-Tyrosinase, and Anti-Melanin. Fermentation. 2024; 10(9):459. https://doi.org/10.3390/fermentation10090459
Chicago/Turabian StyleZhong, Lin, Qi He, Meng Xu, Fang-Fang Chen, Fei Li, and Yu-Pei Chen. 2024. "Unveiling Acetobacter syzygii from Tibetan Kefir Grain: Fermentation-Enhanced Anti-Tyrosinase, and Anti-Melanin" Fermentation 10, no. 9: 459. https://doi.org/10.3390/fermentation10090459
APA StyleZhong, L., He, Q., Xu, M., Chen, F.-F., Li, F., & Chen, Y.-P. (2024). Unveiling Acetobacter syzygii from Tibetan Kefir Grain: Fermentation-Enhanced Anti-Tyrosinase, and Anti-Melanin. Fermentation, 10(9), 459. https://doi.org/10.3390/fermentation10090459






