Fourier Transform Infrared Spectroscopy Tracking of Fermentation of Oat and Pea Bases for Yoghurt-Type Products
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Setup
2.2. Preliminary Fermentation Study
2.3. Main Fermentation
2.3.1. Sample Preparation and Inoculation
2.3.2. Main Fermentation Trials
2.3.3. pH and Temperature Measurements
2.3.4. Spiking Experiments
2.4. FT-IR Measurements
2.5. Sampling and HPLC Analysis
2.6. Multivariate Data Analysis
3. Results and Discussion
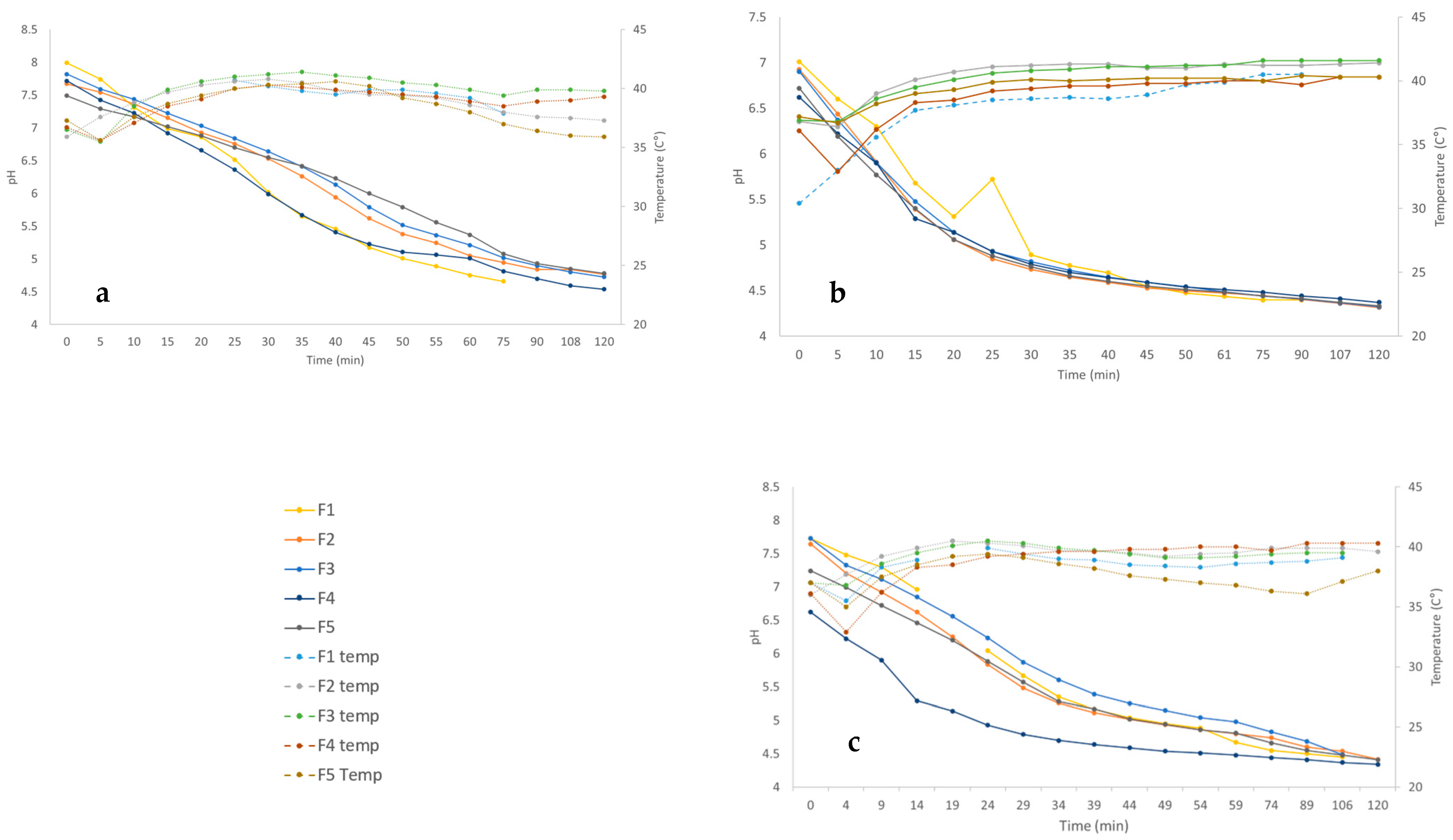
3.1. Acidification Dynamics
3.2. HPLC Quantification of Lactic Acid
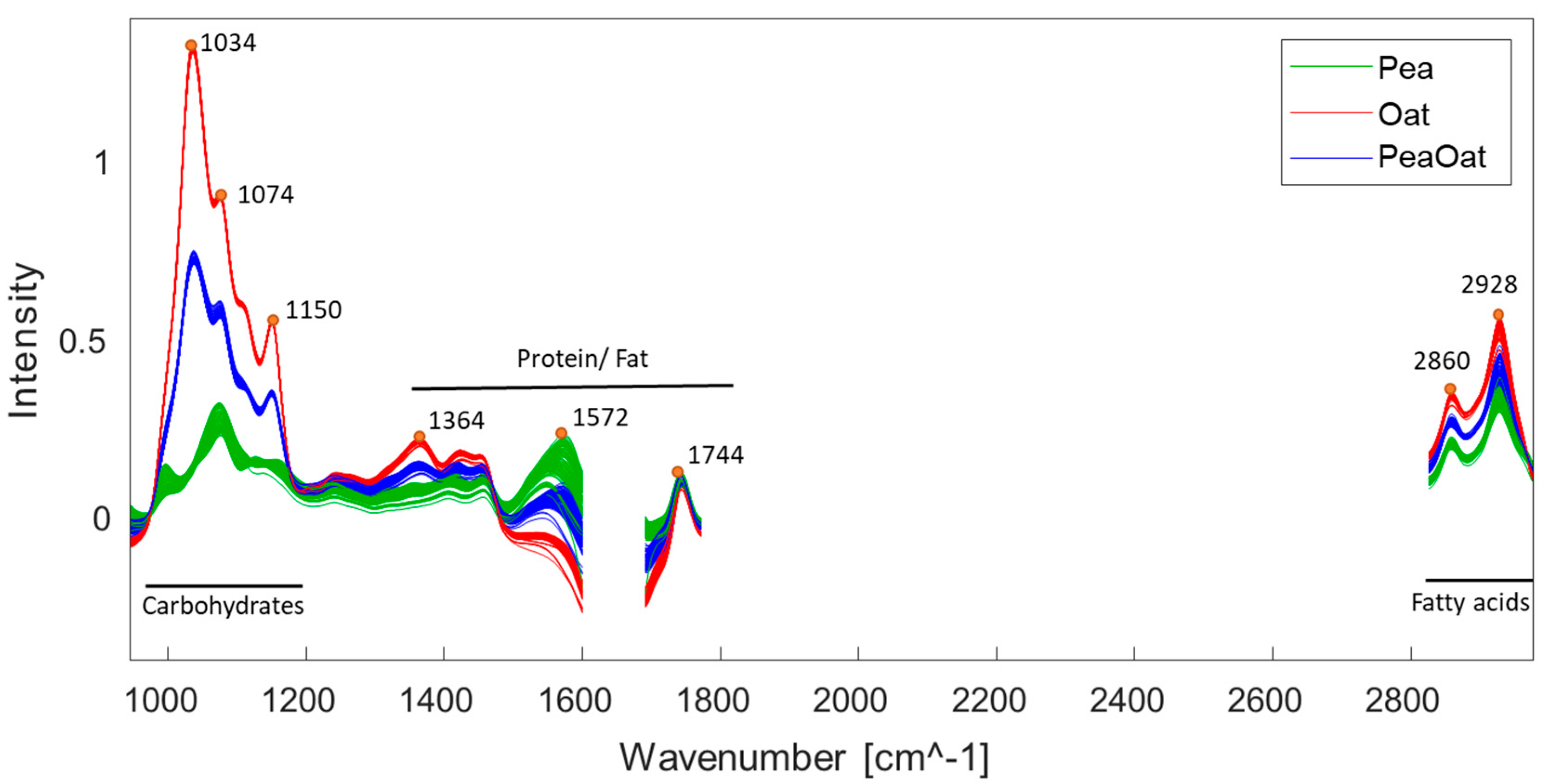
3.3. FT-IR Spectral Data Variation
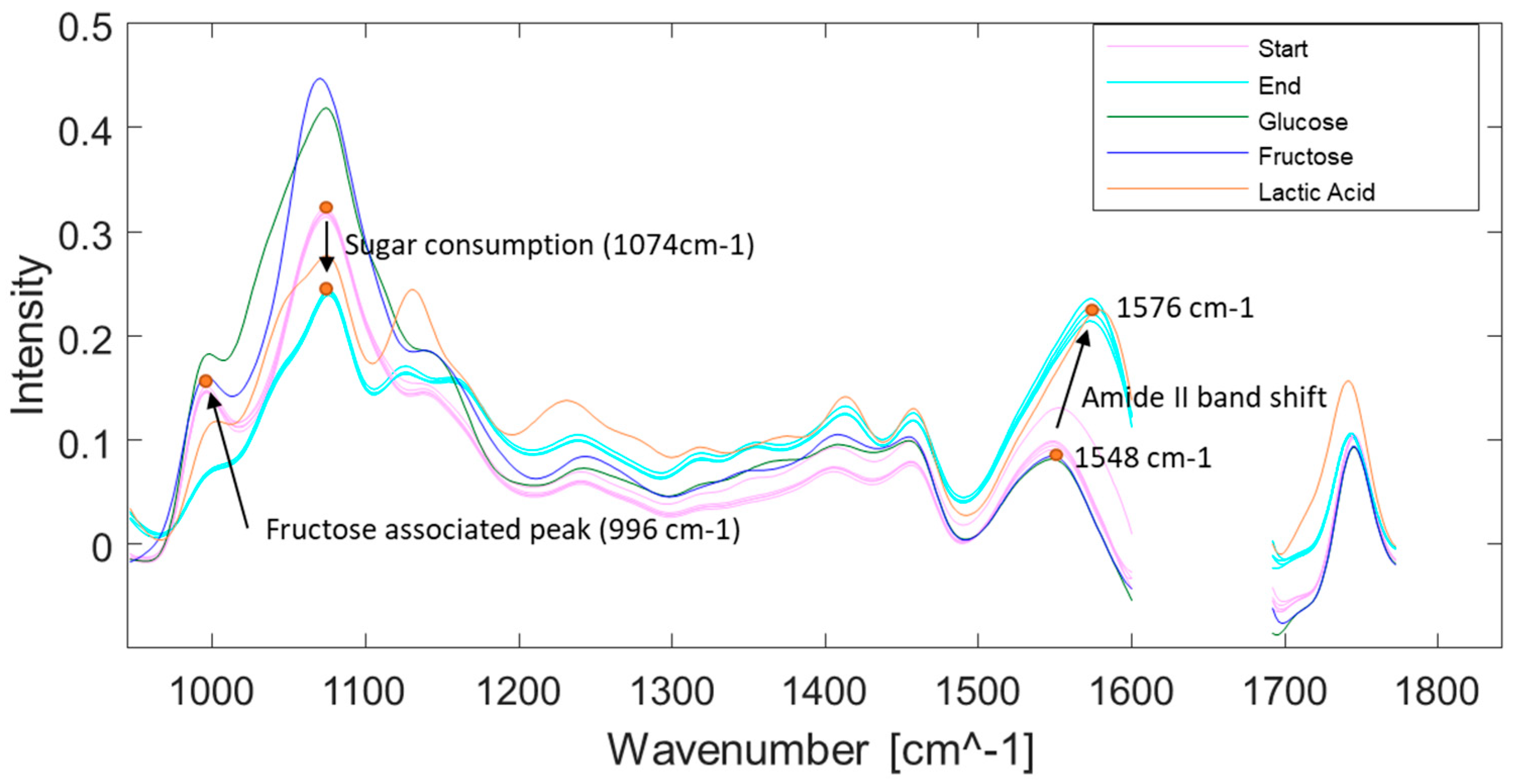
3.4. Spectral Development during Fermentation
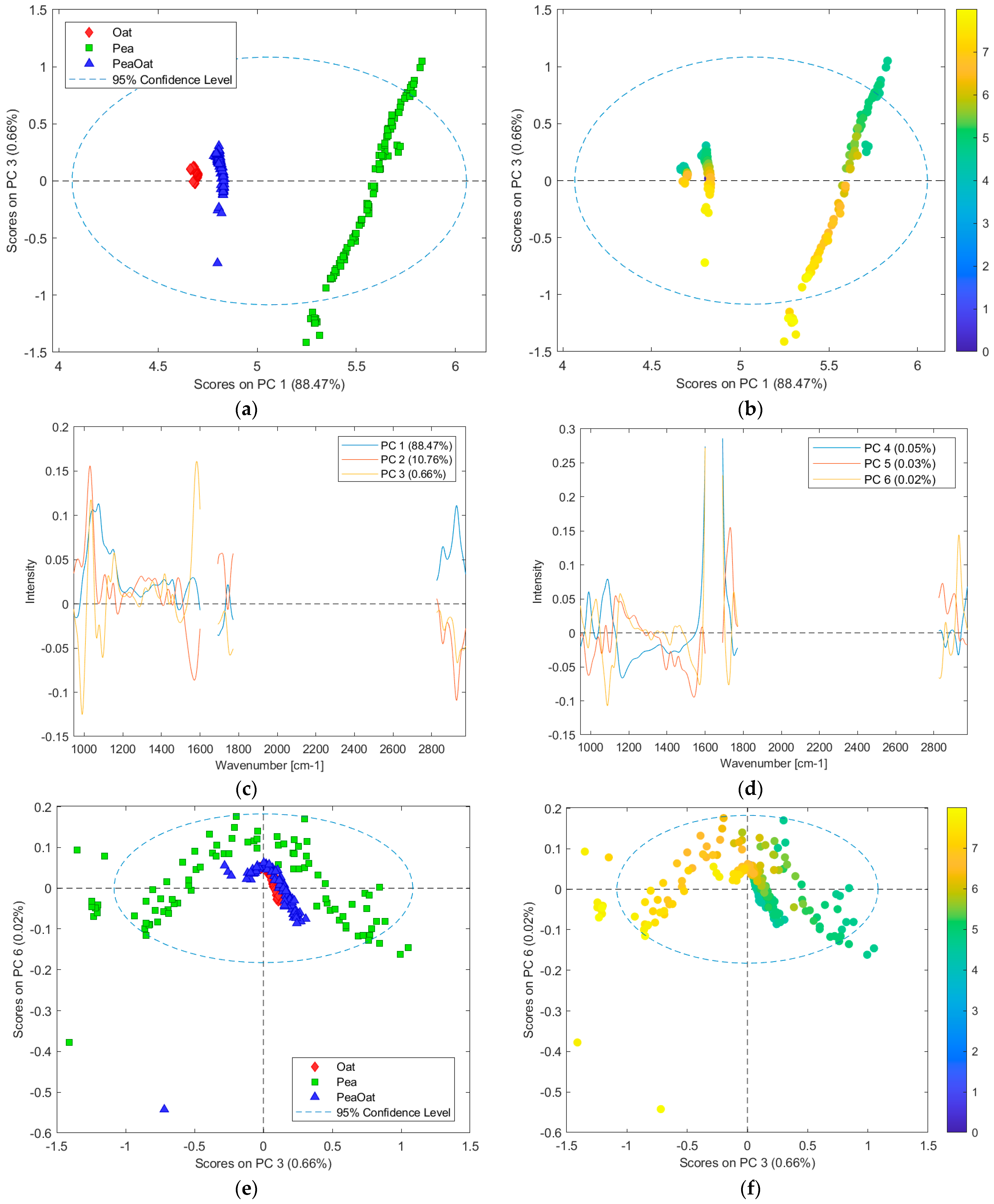
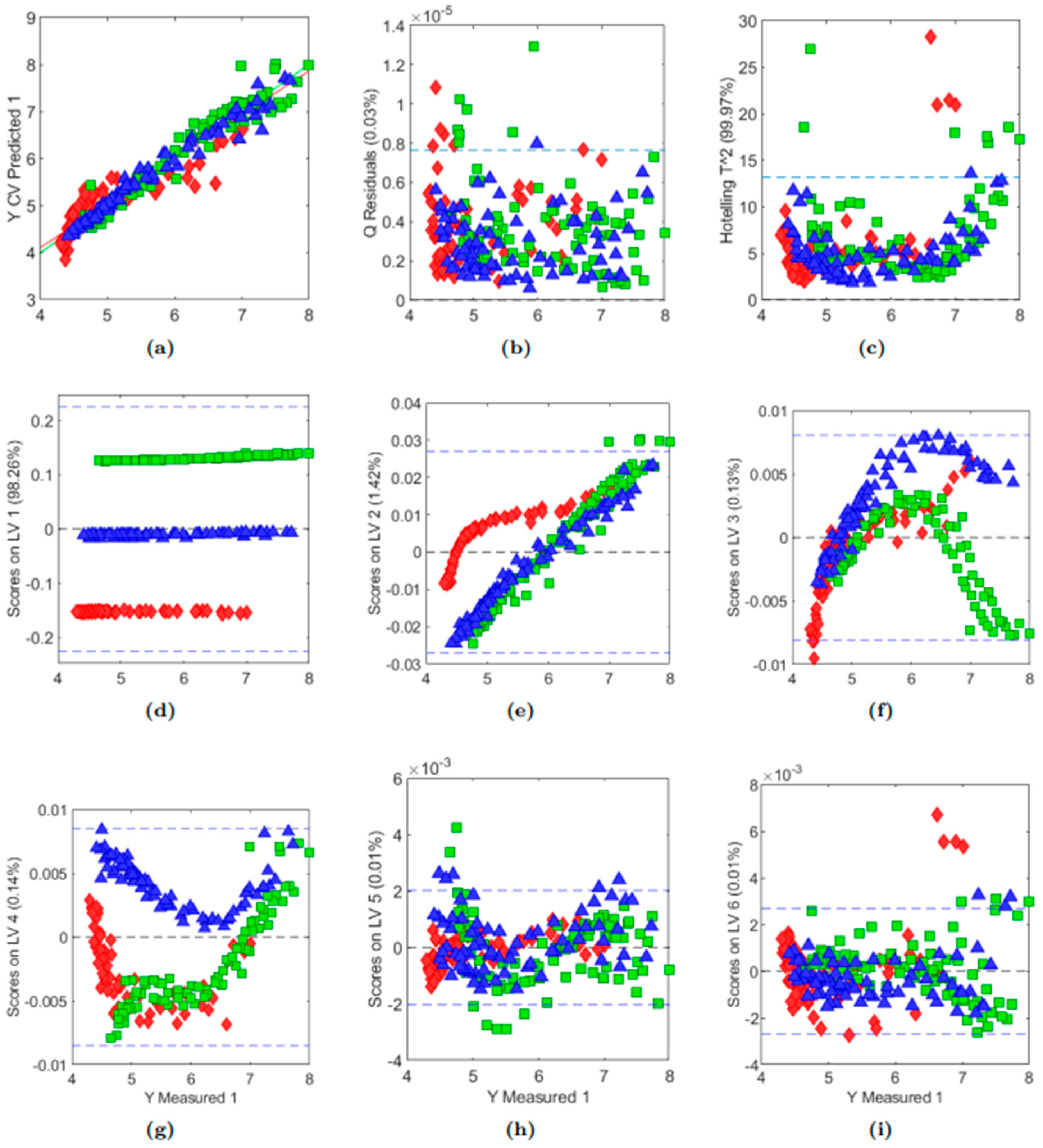
3.5. Investigating Captured Variance by PCA
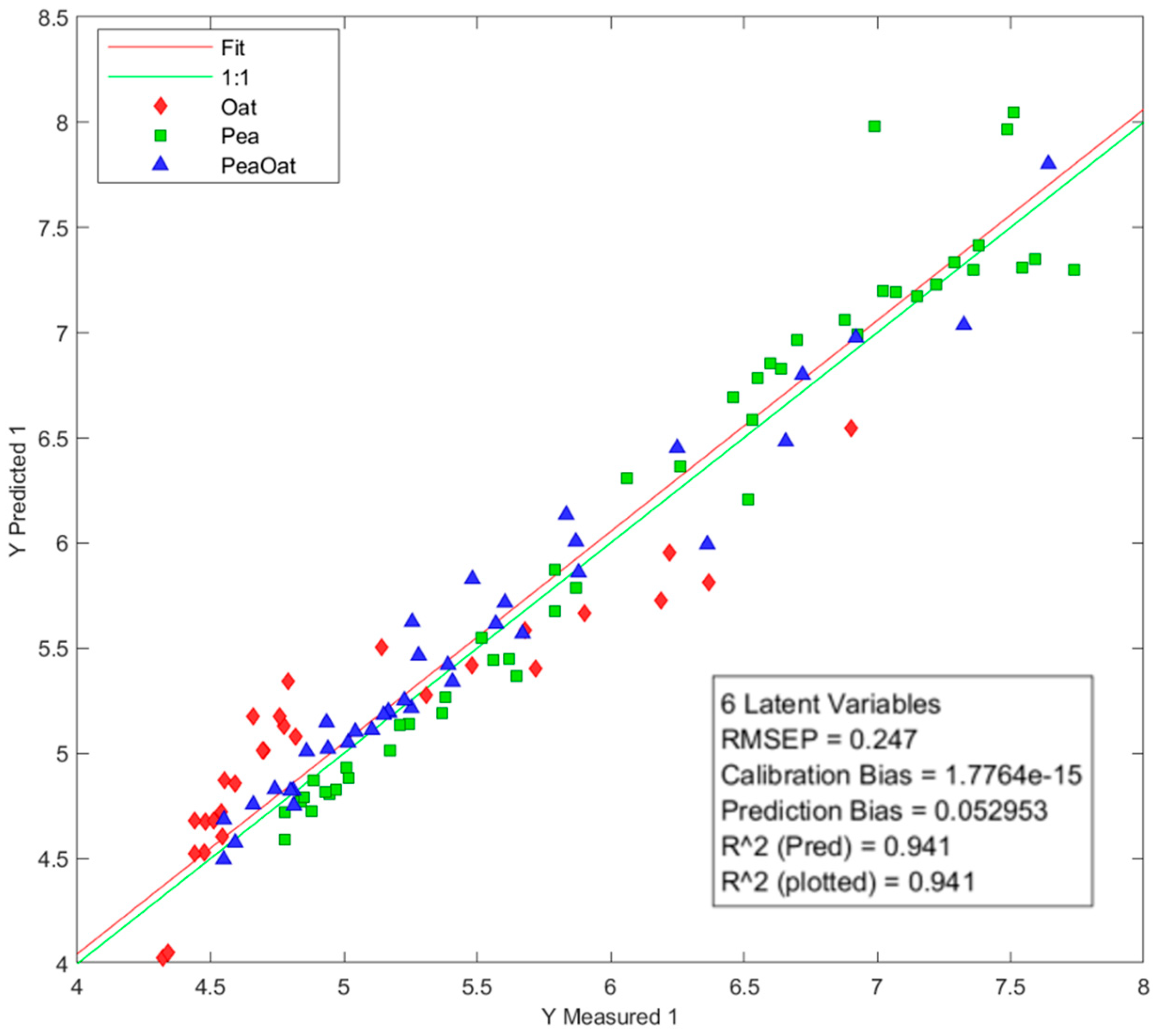
3.6. Prediction of pH by Partial Least Squares
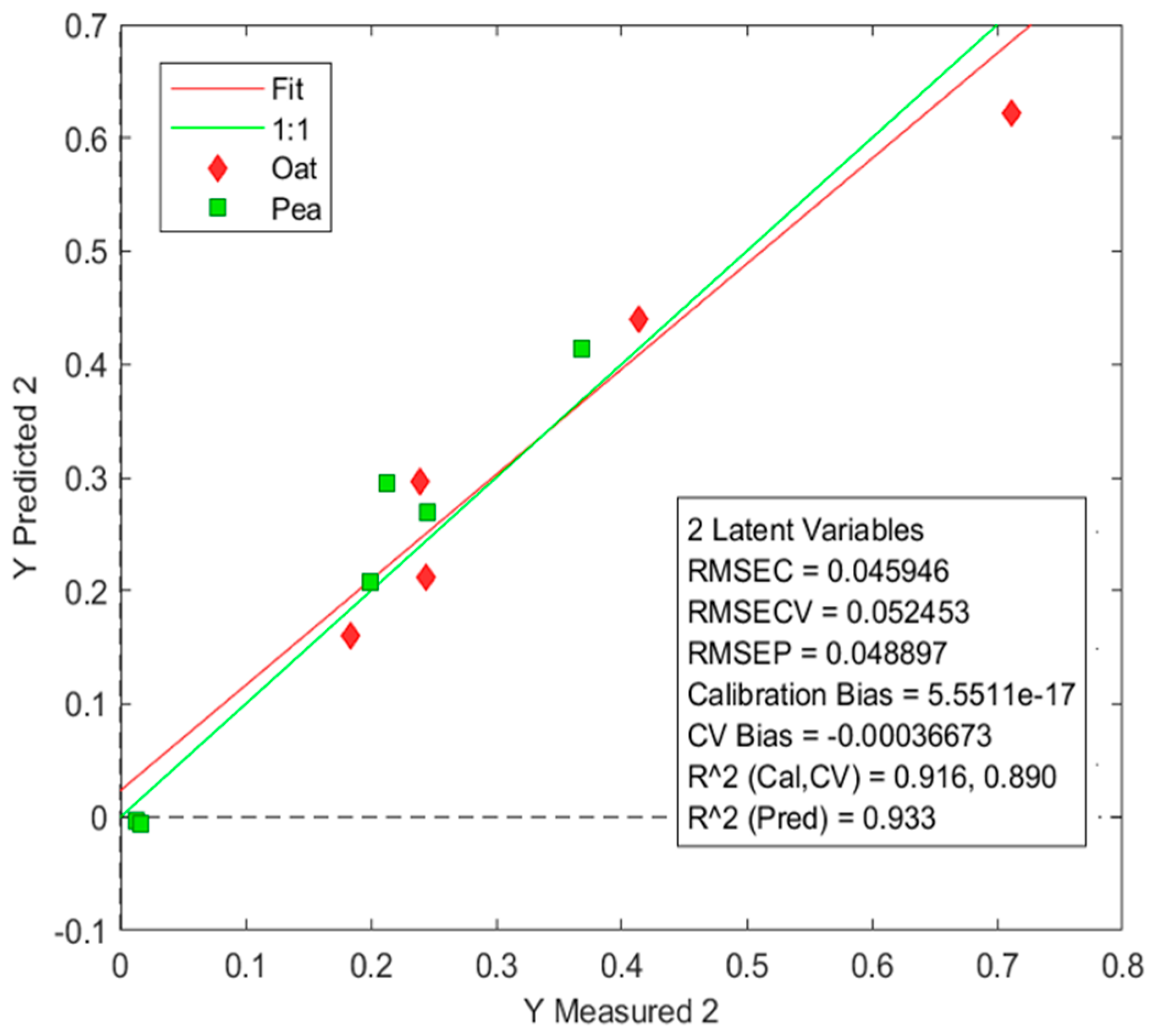
3.7. Prediction of Lactic Acid by Partial Least Squares
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aiking, H. Future protein supply. Trends Food Sci. Technol. 2011, 22, 112–120. [Google Scholar] [CrossRef]
- Pimentel, D.; Pimentel, M. Sustainability of meat-based and plant-based diets and the environment 1–3. Am. J. Clin. Nutr. 2003, 78, 660S–663S. Available online: https://academic.oup.com/ajcn/article/78/3/660S/4690010 (accessed on 31 December 2023). [CrossRef] [PubMed]
- Brückner-Gühmann, M.; Banovic, M.; Drusch, S. Towards an increased plant protein intake: Rheological properties, sensory perception and consumer acceptability of lactic acid fermented, oat-based gels. Food Hydrocoll. 2019, 96, 201–208. [Google Scholar] [CrossRef]
- Montemurro, M.; Pontonio, E.; Coda, R.; Rizzello, C.G. Plant-based alternatives to yogurt: State-of-the-art and perspectives of new biotechnological challenges. Foods 2021, 10, 316. [Google Scholar] [CrossRef] [PubMed]
- Zioga, E.; Tøstesen, M.; Kjaerulf Madsen, S.; Shetty, R.; Bang-Berthelsen, C.H. Bringing plant-based Cli-meat closer to original meat experience: Insights in flavor. Future Foods 2022, 5, 100138. [Google Scholar] [CrossRef]
- European Commission. Report form the Commission to the Council and the European Parliament on the Development of Plant Proteins in the European Union (tech.rep.). 2018. Available online: https://ec.europa.eu/info/sites/info/files/food-farming-fisheries/plants_and_plant_products/documents/report-plant-proteins-com2018-757-final_en.pdf (accessed on 31 December 2023).
- Wunsch, N.-G. Forecast Market Value of Non-Dairy Yogurt Worldwide from 2017 to 2027. 2020. Available online: https://www.statista.com/statistics/939300/global-non-dairy-yogurt-market-value-forecast/ (accessed on 24 November 2023).
- Fearnside, P.H. Soybean cultivation as a threat to the environment in Brazil. Environ. Conserv. 2001, 28, 23–38. [Google Scholar] [CrossRef]
- Mekonnen, H. The green, blue and grey water footprint of crops and derived crop products. Hydrol. Earth Syst. Sci. 2011, 15, 1577–1600. [Google Scholar] [CrossRef]
- Christensen, L.F.; García-Béjar, B.; Heiner Bang-Berthelsen, C.; Hansen, B. Extracellular microbial proteases with specificity for plant proteins in food fermentation. Int. J. Food Microbiol. 2022, 381, 109889. [Google Scholar] [CrossRef] [PubMed]
- Harper, A.R.; Dobson, R.C.J.; Morris, V.K.; Moggré, G.J. Fermentation of plant-based dairy alternatives by lactic acid bacteria. Microb. Biotechnol. 2022, 15, 1404–1421. [Google Scholar] [CrossRef]
- Mårtensson, O.; Öste, R.; Holst, O. Lactic Acid Bacteria in an Oat-based Non-dairy Milk Substitute: Fermentation Characteristics and Exopolysaccharide Formation. LWT 2000, 33, 525–530. [Google Scholar] [CrossRef]
- Tangyu, M.; Muller, J.; Bolten, C.J.; Wittmann, C. Fermentation of plant-based milk alternatives for improved flavour and nutritional value. Appl. Microbiol. Biotechnol. 2019, 103, 9263–9275. [Google Scholar] [CrossRef] [PubMed]
- Bocchi, S.; Rocchetti, G.; Elli, M.; Lucini, L.; Lim, C.-Y.; Morelli, L. The combined effect of fermentation of lactic acid bacteria and in vitro digestion on metabolomic and oligosaccharide profile of oat beverage. Food Res. Int. 2021, 142, 110216. [Google Scholar] [CrossRef] [PubMed]
- Molina, G.E.S.; Shetty, R.; Xiao, H.; Wätjen, A.P.; Tovar, M.; Bang-Berthelsen, C.H. Development of a novel lactic acid bacteria starter culture approach: From insect microbiome to plant-based fermentations. LWT 2022, 167, 113797. [Google Scholar] [CrossRef]
- Yakubu, H.G.; Kovacs, Z.; Toth, T.; Bazar, G. The recent advances of near-infrared spectroscopy in dairy production—A review. Crit. Rev. Food Sci. Nutr. 2020, 62, 810–831. [Google Scholar] [CrossRef] [PubMed]
- Bøge, C.; Anna, G.; Balling, S.; Harder, K.L. An NMR metabolomics approach to investigate factors affecting the yoghurt fermentation process and quality. Metabolites 2020, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, Y.; Tan, Z.; Huang, Z.X.; Zong, J.; Li, Q.F. On-line monitoring of key nutrients in yoghurt samples using digitally labelled Raman spectroscopy. Int. Dairy J. 2019, 96, 132–137. [Google Scholar] [CrossRef]
- Czaja, T.; Baranowska, M.; Mazurek, S.; Szostak, R. Determination of nutritional parameters of yoghurts by FT Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 196, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Arango, O.; Jos, A.; Castillo, M. Inline control of yoghurt fermentation process using a near infrared light backscatter sensor. J. Food Eng. 2020, 277, 109885. [Google Scholar] [CrossRef]
- Patra, T.; Olsen, K.; Rinnan, Å. A multivariate perspective on the stability of oat-based drinks assessed by spectroscopy. Food Hydrocoll. 2022, 131, 107831. [Google Scholar] [CrossRef]
- Ma, C.-Y.; Rout, M.K.; Mock, W.-Y. Study of Oat Globulin Conformation by Fourier Transform Infrared Spectroscopy. J. Agric. Food Chem. 2001, 49, 3328–3334. [Google Scholar] [CrossRef]
- Xu, L.; Yan, S.M.; Cai, C.B.; Wang, Z.J.; Yu, X.P. The feasibility of using near-infrared spectroscopy and chemometrics for untargeted detection of protein adulteration in yogurt: Removing unwanted variations in pure yogurt. J. Anal. Met. Chem. 2013, 2013, 201873. [Google Scholar] [CrossRef]
- Loudiyi, M.; Temiz, H.T.; Sahar, A.; Haseeb Ahmad, M.; Boukria, O.; Hassoun, A.; Aït-Kaddour, A. Spectroscopic techniques for monitoring changes in the quality of milk and other dairy products during processing and storage. Crit. Rev. Food Sci. Nutr. 2022, 62, 3063–3087. [Google Scholar] [CrossRef] [PubMed]
- Bonke, A.; Sieywerts, S.; Petersen, I.L. Amino acid composition of novel plant drinks from oat, lentil and pea. Foods 2020, 9, 429. [Google Scholar] [CrossRef] [PubMed]
- Iosca, G.; Fugaban, J.I.I.; Özmerih, S.; Wätjen, A.P.; Kaas, R.S.; Hà, Q.; Shetty, R.; Pulvirenti, A.; De Vero, L.; Bang-Berthelsen, C.H. Exploring the Inhibitory Activity of Selected Lactic Acid Bacteria against Bread Rope Spoilage Agents. Fermentation 2023, 9, 290. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627. [Google Scholar] [CrossRef]
- Engel, J.; Gerretzen, J.; Szymańska, E.; Jansen, J.J.; Downey, G.; Blanchet, L.; Buydens, L.M.C. Breaking with trends in pre-processing? TrAC Trends Analyt. Chem. 2013, 50, 96–106. [Google Scholar] [CrossRef]
- Dai, C.; Xu, X.; Huang, W.; Yan, P.; Hou, Y.; He, R.; Ma, H. Monitoring of critical parameters in thermophilic solid-state fermentation process of soybean meal using NIR spectroscopy and chemometrics. J. Food Meas. Charact. 2023, 17, 576–585. [Google Scholar] [CrossRef]
- Wang, X.; Kong, X.; Zhang, C.; Hua, Y.; Chen, Y.; Li, X. Comparison of physicochemical properties and volatile flavor compounds of plant-based yoghurt and dairy yoghurt. Food Res. Int. 2023, 164, 112375. [Google Scholar] [CrossRef] [PubMed]
- Salaün, F.; Mietton, B.; Gaucheron, F. Buffering capacity of dairy products. Int. Dairy J. 2005, 15, 95–109. [Google Scholar] [CrossRef]
- Laaksonen, O.; Kahala, M.; Marsol-Vall, A.; Blasco, L.; Järvenpää, E.; Rosenvald, S.; Virtanen, M.; Tarvainen, M.; Yang, B. Impact of lactic acid fermentation on sensory and chemical quality of dairy analogues prepared from lupine (Lupinus angustifolius L.) seeds. Food Chem. 2021, 346, 128852. [Google Scholar] [CrossRef]
- Part, N.; Kazantseva, J.; Rosenvald, S.; Kallastu, A.; Vaikma, H.; Kriščiunaite, T.; Pismennõi, D.; Viiard, E. Microbiological, chemical, and sensorial characterisation of commercially available plant-based yoghurt alternatives. Future Foods 2023, 7, 100212. [Google Scholar] [CrossRef]
- Tipson, S.R. Infrared Spectroscopy of Carbohydrates: A Review of the Literature; The National Bureau of Standards Monograph 110: Gaithersburg, MD, USA, 1968. [CrossRef]
- Zhi, F.; Yang, T.; Le, W.Q.; Jiang, B.; Wang, Z.P.; Zhang, J.; Chen, Y.Z. Isolation, structure and activity of a novel water-soluble polysaccharide from Dioscorea opposita Thunb. Int. J. Biol. Macromol. 2019, 133, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Jahanbin, K. Structural characterization of a new water-soluble polysaccharide isolated from Acanthophyllum acerosum roots and its antioxidant activity. Int. J. Biol. Macromol. 2018, 107, 1227–1234. [Google Scholar] [CrossRef]
- Lendl, B.; Schindler, R.; Frank, J.; Kellner, R.; Drott, J.; Laurell, T. Fourier Transform Infrared Detection in Miniaturized Total Analysis Systems for Sucrose Analysis. Anal. Chem. 1997, 69, 2877–2881. [Google Scholar] [CrossRef] [PubMed]
- Manolache, F.; Hanganu, A.; Duta, D.E.; Belc, N.; Marin, D.I. The Physico-chemical and Spectroscopic Composition Characterization of Oat Grains and Oat Oil Samples. Rev. Chim. 2013, 64, 45–48. Available online: http://www.revistadechimie.ro (accessed on 31 December 2023).
- Cast, J. 9 Infrared spectroscopy of lipids. In Developments in Oils and Fats, 1st ed.; Hamilton, J.R., Ed.; Springer Science+Business Media: Dordrecht, The Netherlands, 1995; pp. 224–269. [Google Scholar] [CrossRef]
- Luinge, H.J.; Hop, E.; Lutz, E.T.G.; Van Hemert, J.A.; De Jong, E.A.M. Determination of the fat, protein and lactose content of milk using Fourier transform infrared spectrometry. Anal. Chim. Acta 1993, 284, 419–433. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Harlé, O.; Falentin, H.; Niay, J.; Valence, F.; Courselaud, C.; Chuat, V.; Maillard, M.B.; Guédon, E.; Deutsch, S.M.; Thierry, A. Diversity of the metabolic profiles of a broad range of lactic acid bacteria in soy juice fermentation. Food Microbiol. 2020, 89, 103410. [Google Scholar] [CrossRef]
- Chumchuere, S.; Robinson, R.K. Selection of starter cultures for the fermentation of soya milk. Food Microbiol. 1999, 16, 129–137. [Google Scholar] [CrossRef]
- Kjaerulf Madsen, S.; Priess, C.; Peter, A.; Øzmerih, U.; Amin, M.M.; Bang-Bertelsen, C.H. Development of a yoghurt alternative, based on plant-adapted lactic acid bacteria, soy drink and the liquid fraction of brewers spent grain. FEMS Microbiol. Lett. 2021, 368, 93. [Google Scholar] [CrossRef]
- Pelton, J.T.; Mclean, L.R. Spectroscopic Methods for Analysis of Protein Secondary Structure. Anal. Biochem. 2000, 277, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Altan, A.; Yagci, S. Physicochemical characteristics and structural changes of fermented faba bean extrudates prepared by twin-screw extrusion. Food Chem. 2023, 411, 135502. [Google Scholar] [CrossRef]
- Wang, W.-Q.; Zhang, J.-L.Z.; Yu, Q.; Zhou, J.-Y.; Lu, M.-L.; Gu, R.-C.; Huang, Y. Structural and compositional changes of whey protein and blueberry juice fermented using Lactobacillus plantarum or Lactobacillus casei during fermentation. RSC Adv. 2021, 11, 26291–26302. [Google Scholar] [CrossRef]
- Tang, K.; Zhang, M.; Liu, D.; Li, Y.; Zhang, P. Changes in molecular structure of protein and carbohydrate in soybean products with different processing methods and their effects on nutrient degradation characteristics of the products. Czech J. Anim. Sci. 2020, 65, 233–246. [Google Scholar] [CrossRef]
- Arteaga, V.G.; Demand, V.; Kern, K.; Strube, A.; Szardenings, M.; Muranyi, I.; Eisner, P.; Schweiggert-Weisz, U. Enzymatic Hydrolysis and Fermentation of Pea Protein Isolate and Its Effects on Antigenic Proteins, Functional Properties, and Sensory Profile. Foods 2022, 11, 118. [Google Scholar] [CrossRef]
- Welman, A.D.; Maddox, I.S. Exopolysaccharides from lactic acid bacteria: Perspectives and challenges. Trends Biotechnol. 2003, 21, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Namli, S.; Sumnu, S.G.; Oztop, M.H. Microwave glycation of soy protein isolate with rare sugar (D-allulose), fructose and glucose. Food Biosci. 2021, 40, 100897. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, Y.; Ma, C.; McClements, D.J.; Liu, F.; Liu, X. Pea protein isolate-inulin conjugates prepared by pH-shift treatment and ultrasonic-enhanced glycosylation: Structural and functional properties. Food Chem. 2022, 384, 132511. [Google Scholar] [CrossRef]
- Soltanikazemi, M.; Mehdizadeh, S.A.; Heydari, M.; Faregh, S.M. Development of a smart spectral analysis method fothe determination of mulberry (Morus alba var. nigra L.) juice quality parameters using FT-IR spectroscopy. Food Sci. Nutr. 2023, 11, 1808–1817. [Google Scholar] [CrossRef]
- Parvarei, M.M.; Fazeli, M.R.; Mortazavian, A.M.; Nezhad, S.S.; Mortazavi, S.A.; Golabchifar, A.A.; Khorshidian, N. Comparative effects of probiotic and paraprobiotic addition on microbiological, biochemical and physical properties of yogurt. Food Res. Int. 2021, 140, 110030. [Google Scholar] [CrossRef]
- Khajehpour, M.; Dashnau, J.L.; Vanderkooi, J.M. Infrared spectroscopy used to evaluate glycosylation of proteins. Anal. Biochem. 2006, 348, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Du, S.; Deng, Q.; Tang, H.; Yang, C.; Wei, F.; Chen, H.; Quek, S.Y.; Zhou, A.; Liu, L. Study on the antioxidant activity and emulsifying properties of flaxseed gum-whey protein isolate conjugates prepared by Maillard reaction. Biol. Macromol. 2019, 153, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Jiang, W.; Chen, G.; Wang, Q.; Mcclements, D.J.; Liu, X.; Liu, F.; Ngai, T. Sonochemical effects on formation and emulsifying properties of zein-gum Arabic complexes. Food Hydrocoll. 2021, 114, 106557. [Google Scholar] [CrossRef]
- Li, B.; Lin, Y.; Yu, W.; Wilson, D.I.; Young, B.R. Application of Mechanistic Modelling and Machine Learning for Cream Cheese Fermentation pH Prediction; Wiley Online Library: Hoboken, NJ, USA, 2020. [Google Scholar] [CrossRef]


| Nutritional Value [100 mL−1] | Pea (Dryk) | Oat (Naturli) |
|---|---|---|
| Fat | 2.3 g | 2.9 g |
| -Saturated fatty acids | 0.2 g | 0.3 g |
| Carbohydrates | 1.5 g | 9.7 g |
| -Sugars | 1.5 g | 4.3 g |
| Protein | 2 g | 0.9 g |
| Ingredients | Water, pea protein (2.5%), rapeseed oil, sugar, acidity regulator: di calcium-phosphate, carriers: (calcium carbonate, calcium phosphate), gluten-free oat oil, salt, vitamins (D3, riboflavin, and B12) | Water, oat (13%), rapeseed oil, acidity regulator (tricalcium phosphate), sea salt, stabilizator (gellan gum), vitamins (D, B2 (riboflavin), and B12). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greulich, O.; Duedahl-Olesen, L.; Mikkelsen, M.S.; Smedsgaard, J.; Bang-Berthelsen, C.H. Fourier Transform Infrared Spectroscopy Tracking of Fermentation of Oat and Pea Bases for Yoghurt-Type Products. Fermentation 2024, 10, 189. https://doi.org/10.3390/fermentation10040189
Greulich O, Duedahl-Olesen L, Mikkelsen MS, Smedsgaard J, Bang-Berthelsen CH. Fourier Transform Infrared Spectroscopy Tracking of Fermentation of Oat and Pea Bases for Yoghurt-Type Products. Fermentation. 2024; 10(4):189. https://doi.org/10.3390/fermentation10040189
Chicago/Turabian StyleGreulich, Olivia, Lene Duedahl-Olesen, Mette Skau Mikkelsen, Jørn Smedsgaard, and Claus Heiner Bang-Berthelsen. 2024. "Fourier Transform Infrared Spectroscopy Tracking of Fermentation of Oat and Pea Bases for Yoghurt-Type Products" Fermentation 10, no. 4: 189. https://doi.org/10.3390/fermentation10040189
APA StyleGreulich, O., Duedahl-Olesen, L., Mikkelsen, M. S., Smedsgaard, J., & Bang-Berthelsen, C. H. (2024). Fourier Transform Infrared Spectroscopy Tracking of Fermentation of Oat and Pea Bases for Yoghurt-Type Products. Fermentation, 10(4), 189. https://doi.org/10.3390/fermentation10040189





