Abstract
Despite advances in treatments and therapies, cardiovascular diseases (CVDs) remain one of the leading causes of death worldwide. The discovery that most of the human genome, although transcribed, does not encode proteins was crucial for focusing on the potential of long non-coding RNAs (lncRNAs) as essential regulators of cell function at the epigenetic, transcriptional, and post-transcriptional levels. This class of non-coding RNAs is related to the pathophysiology of the cardiovascular system. The different expression profiles of lncRNAs, in different contexts of CVDs, change a great potential in their use as a biomarker and targets of therapeutic intervention. Furthermore, regular physical exercise plays a protective role against CVDs; on the other hand, little is known about its underlying molecular mechanisms. In this review, we look at the accumulated knowledge on lncRNAs and their functions in the cardiovascular system, focusing on the cardiovascular pathology of arterial hypertension, coronary heart disease, acute myocardial infarction, and heart failure. We discuss the potential of these molecules as biomarkers for clinical use, their limitations, and how the manipulation of the expression profile of these transcripts through physical exercise can begin to be suggested as a strategy for the treatment of CVDs.
1. Introduction
Cardiovascular diseases (CVDs) are the leading cause of death worldwide. It is estimated that approximately 18.5 million people die annually on account of these diseases, with a third of these people dying under the age of 70 years [1]. Identifying those most affected by CVDs and ensuring they receive the appropriate treatment can prevent premature deaths. Furthermore, the development of new therapeutic strategies and biomarkers with the potential to predict the progression of CVDs is fundamental to reducing mortality worldwide. CVDs can be defined as disorders that affect the heart or blood vessels such as heart failure, coronary heart disease, cerebrovascular disease, peripheral arterial disease, and congenital heart disease [1].
The main risk factors associated with these diseases are smoking, excessive alcohol consumption, sedentary lifestyle, obesity, high blood cholesterol, among others. Individuals at risk of developing CVDs may, therefore, have increased blood pressure, glucose, and triglycerides as well as overweight and obesity. Among the many risk factors that predispose to the development and progression of CVDs, a sedentary lifestyle, supported by consistently low levels of physical activity, represents a major contributor to CVDs. On the other hand, regular exercise is associated with health benefits and a lower risk of disease [2,3]. Several studies have demonstrated that increased physical activity promotes a reduction in all-cause mortality and can increase life expectancy, affecting a strongly link to a decline in the risk of developing CVDs, in part by promoting weight loss, blood pressure control as well as improving blood lipid profile and insulin sensitivity [2,4]. For these reasons, physical activity has been recommended worldwide for CVD prevention and treatment. Despite the benefits of regular physical exercise, the molecular mechanisms by which they occur are still poorly understood.
In recent decades, a research effort has been aimed at identifying the major physiological, biochemical, and molecular contributors to the cardiovascular benefits of exercise. This research resulted in advances obtained from observational studies and interventions in both human and animal models. The Encyclopedia of DNA Elements (ENCODE) [5], a project realized in 2012, challenged the central dogma of biology and the interpretation of what is considered a functional region of the human genome [6]. From the use of high-throughput genomic platforms, it was discovered that the coding transcripts (i.e., mRNAs) represent less than 3% of the genome, while everything else represents transcripts that have little or no ability to synthesize protein. These transcripts are called non-coding RNAs (ncRNAs) [7]. For a long time, these non-coding transcripts were neglected and treated as “junk of the DNA” [8,9], “transcription noise” [10,11,12], or even as “dark matter of the genome” [13,14]; however, evidence shows that ncRNAs are not only functionally active as RNA molecules but are also one of the major regulatory networks of gene expression at the epigenetic, transcriptional, and even post-transcriptional levels (for more information, see References [9,15,16,17,18]).
According to the number of nucleotides (nt), ncRNAs can be divided into two large classes: those with fewer than 200 nt, called small non-coding RNAs such as microRNAs (miRNAs), and those with more than 200 nt, called long non-coding RNAs (lncRNAs). Among them, miRNAs are better understood and act mainly in post-transcriptional control as protein synthetic silencers binding to their target mRNA on which they induce translation degradation or repression [19]. The identification of stable miRNAs in body fluids, a strong indicator of cell–cell communication via circulating RNAs, suggested for the first time the possibility of non-coding transcripts serving as diagnostic and prognostic biomarkers for several diseases [20,21] as well as the possibility of their being used as therapeutic targets and monitoring of physical performance induced by exercise training [22,23,24,25]. On the other hand, lncRNAs can modulate gene expression at multiple levels and in an even more complex way than a regulation made by miRNAs [26,27,28]. However, lncRNAs have only recently attracted the attention of researchers, and knowledge about them, including their potential as a biomarker and therapeutic target, is still limited [29,30,31,32].
According to the LncRNA Disease v2.0 database (www.rnanut.net/lncrnadisease, accessed on 16 September 2021), there are currently more than 205,959 associations between lncRNAs and diseases including CVDs. As knowledge about these associations grows, so does the interest in investigating the influence of exercise training on the modulation of the expression of these transcripts and the possibility of using the health benefits as potential therapeutic targets [33,34,35]. In this review, after a simple presentation of lncRNAs, their biogenesis, classifications, and mechanisms of action, we discuss the current understanding of the actions performed by these ncRNAs in cardiac pathophysiology. Finally, we review the most recent publications on the role of lncRNAs in cardiac adaptations induced by exercise training, addressing the therapeutic potential of lncRNAs in clinical applications for CVDs and propose considerations regarding the present and future of research in this area.
2. Long Non-Coding RNAs
Differently from what was previously thought, the complexity of organisms may originate not in the number of coding genes that the species has but in the number and diversity of non-coding genes [36,37]. It is estimated that the more complex the organism, the greater the ratio between the number of ncRNAs and the number of mRNAs present in its genome [37]. Pioneers in ncRNA studies have already stated that being “junk” is not synonymous with being useless [38,39,40].
The arrival of state-of-the-art sequencing technology expanded the frontiers of knowledge by bringing with it, in the middle of the 2000s, the RNA-seq [41]. Through a large-scale sequencing methodology and bioinformatics tools, this technique allows not only to quantify RNA levels more precisely than techniques such as microarrays but also allows to trace the profile of a transcriptome, identifying which molecules of RNA are being expressed [41]. With the RNA-seq technique, it is possible to visualize the real dimension and intricacies involved behind the human transcriptome in a quick and less expensive way.
Each year, more lncRNAs are discovered, and according to version 6.0 of NONCODE, a database dedicated to ncRNAs, especially lncRNAs, the human species contains approximately 173,112 lncRNA transcripts [42], while the current number of transcripts with protein-coding capacity corresponds to approximately half, i.e., 86,757 (based on GENCODE, version 38 [43]). However, the naming, categorization, validation as well as the annotations of lncRNAs are still not complete.
3. Definition
The definition of lncRNAs is still ongoing. At present, there is no universal description method for these transcripts; thus, there are multiple synonyms describing similar or slightly different molecules to lncRNAs which creates some confusion as to what is or is not an lncRNA [29]. Based on the basic characteristics of lncRNAs, they are generally defined as those transcribed with more than 200 nt that do not have the capacity to encode proteins. However, such a definition that uses a criterion based on the length of the transcript has been shown to be increasingly arbitrary, as high-throughput sequencing and computational analyses contribute to the characterization of ncRNAs, since it was defined taking into account only the small ncRNA isolation protocol already well characterized [44]. Contrary to what was initially thought, some lncRNAs contain small open reading frames; that is, they have the coding potential for peptides, albeit reduced [45,46].
4. Biogenesis
There is a great similarity regarding the biogenesis of mRNAs and lncRNAs [28,47,48]. lncRNAs are encoded within the genome, suggesting that their transcription might be tightly coordinated with the transcription of other genes including protein-coding genes. Similar to protein-coding genes, most lncRNAs are transcribed by RNA polymerase II (RNA PolII). This enzyme, unlike RNA Pols I and III, has a high processing capacity capable of ensuring the stability of the non-coding transcript and, therefore, its functionality [29]. They also undergo post-transcriptional modifications of capping with methylguanosine at the 5’ end of their still immature transcript and polyadenylation at the 3’ end and, in various situations, they undergo alternative splicing and chemical modifications in their bases. However, since the lncRNA class houses a large diversity of transcripts, there are exceptions to those of non-canonical formations, where they do not resemble the processing of mRNAs such as circular RNAs (circRNAs) [49,50,51] and sno-lncRNAs [52].
5. Classification
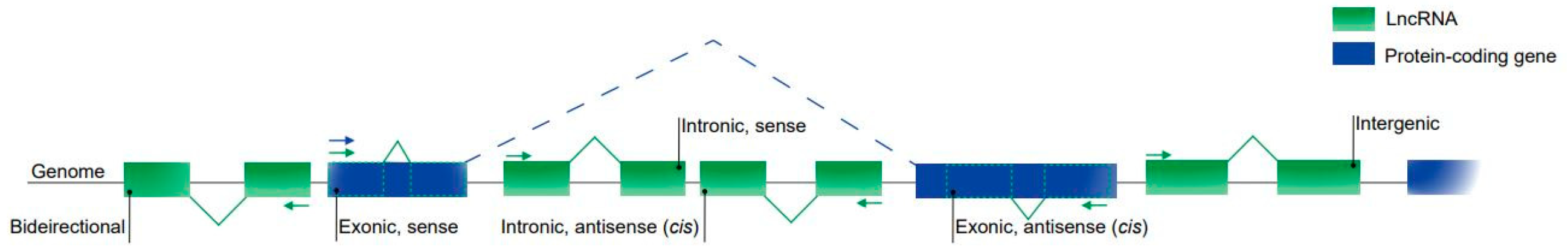
There is still no consensus on the classification of lncRNAs [32,53], although the most common method considers their genomic location in relation to their proximity to the coding genes (Figure 1).
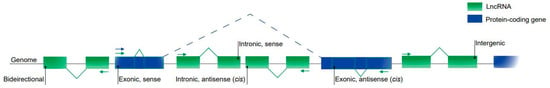
Figure 1.
Classification of lncRNAs based on their genomic location. Intergenic lncRNAs are located between two protein-coding genes. Exonic and intronic lncRNAs are located entirely in the exonic and intronic regions of a protein-coding gene, respectively. Sense lncRNAs are transcribed from the same strand and in the same direction as the protein-coding genes; they can be both exonic and intronic. Antisense lncRNAs are transcribed from the opposite strand of the protein-coding genes and can also be both exonic and intronic. Bidirectional lncRNAs are located within the promoter region of a protein-coding gene.
Then, lncRNAs can be classified as intergenic, those located between two protein-coding genes, representing the vast majority of lncRNA transcripts; intronic or exonic, they are located in the introns or exons of a protein-coding gene, respectively. These classes can also be divided into opposite forms (on the opposite strand) to a coding gene and as bidirectional promoters, those located within the promoters in the opposite direction to the encoding gene. Other studies have identified new classifications for lncRNAs such as the enhancer classification (for a review, see References [54,55]).
6. Mechanisms of Action
Although the primary structure is an important property for the function of a transcript, whether coding or not, the position in the genome as well as the secondary and tertiary structure of lncRNAs are better conserved interspecies than their amino acid sequence. The fact that lncRNAs exhibit poor conservation in their primary structures does not imply a loss of RNA function, rather it suggests a role in the changes in complexity a species [56]. Based on their molecular mechanisms of action, lncRNAs can be divided into signal, decoy, guide, and scaffold [28,29,57,58,59].
lncRNAs have lower levels of expression compared to mRNAs. However, lncRNAs have a more specific expression pattern than mRNAs in response to various stimuli [60,61]. Such specificity, which varies according to cell type, tissue, the subcellular compartment in which they are found, and also according to the stage of development or disease [62,63], suggests that the control of the expression of these molecules is under significant transcriptional control. Thus, lncRNAs end up playing a role as a molecular signal, signal lncRNAs [64,65,66,67,68], given that each transcript is expressed in a very specific time–space context. Once they are expressed, they can play roles as biomarkers of biological or pathological events such as the development of cardiac tissue [55,69] and CVDs [69,70], respectively. lncRNAs also could act as a molecular sponge for regulatory factors to repress transcription. To do this, the transcript simply binds to its targets, which can be RNA-binding proteins, such as transcription factors or chromatin-modifying enzymes as well as RNA sequences such as miRNAs. As a consequence, the so-called decoy lncRNAs [71,72,73,74,75,76] end up limiting the availability of regulatory factors and, therefore, preventing their interaction with their targets. Another mechanism of molecular action is based on the ability of lncRNAs to bind to regulatory proteins or to those enzymatically active, such as transcription factors and chromatin modifiers, and guide them to specific sites in the genome to control the expression of a target gene. They can also bind directly to other transcripts with regulatory functions or even to DNA itself. Thus, they guide changes in chromatin and, therefore, in gene expression in both cis and trans. The lncRNAs with this function are called guide lncRNAs [77,78,79,80,81,82,83]. They can also support the assembly of structures with several molecular components through the simultaneous binding of their domains to different transcriptional activator or repressor effector molecules. The lncRNAs that act through this mechanism are scaffold lncRNAs [84,85,86].
It is noteworthy that these mechanisms are not mutually exclusive. On the contrary, a single lncRNA is capable of acting under more than one molecular mechanism of action at the same time [57], leading to the activation or repression of gene expression at its different levels of regulation.
7. Functions of LncRNAs in the Heart
Studies to identify the expression of heart-specific lncRNAs revealed that some of these transcripts already present a differential expression profile in cardiovascular development as well as in pathological conditions that affect this organ [34,55,70,87,88,89,90]. The cardiogenic lncRNA cardiac mesoderm enhancer-associated non-coding RNA (CARMEN), for example, is a crucial regulator of human cardiac precursor cell fate, differentiation, and homeostasis [91]. Another important example is fetal-lethal non-coding developmental regulatory RNA (FENDRR [92]), an intergenic lncRNA with high expression in the lateral mesoderm, the germinal leaflet that gives rise to the heart. By binding to the polymeric histone remodeling complex PRC2 and TrxG/MLL, FENDRR, as an lncRNA guide, acts by modulating the chromatin state [93]. Another study involving knockout mice for a set of lncRNA, including this transcript, which has its ortholog in humans, demonstrated the importance of FENDRR not only for perfect heart development and, consequently, perfect functionality but also for embryo survival [94]. Since then, hundreds of lncRNAs have had their expression profile traced in the heart involved in a range of cellular processes, including cardiomyocytes, endothelial, and smooth muscle cells, and in fibroblasts [55,95,96,97]. Despite these findings, the regulation of cardiac pathways by lncRNAs, that is, the characterization of the functional roles of these transcripts, is still poorly understood.
8. LncRNAs in Cardiovascular Diseases
As mentioned before, lncRNAs can be correlated with many human diseases [98,99,100,101] including CVDs. The first association between lncRNA and heart disease came from genetic studies in which it was discovered that the locus enriched in single nucleotide polymorphisms involved with myocardial infarction susceptibility was not actually a protein-coding locus but coding for an ncRNA, which the discoverers named MIAT (myocardial infarction-associated transcript) [102]. Since then, several studies have reported associations between lncRNAs and CVDs (Table 1).

Table 1.
List of lncRNAs involved in cardiovascular diseases.
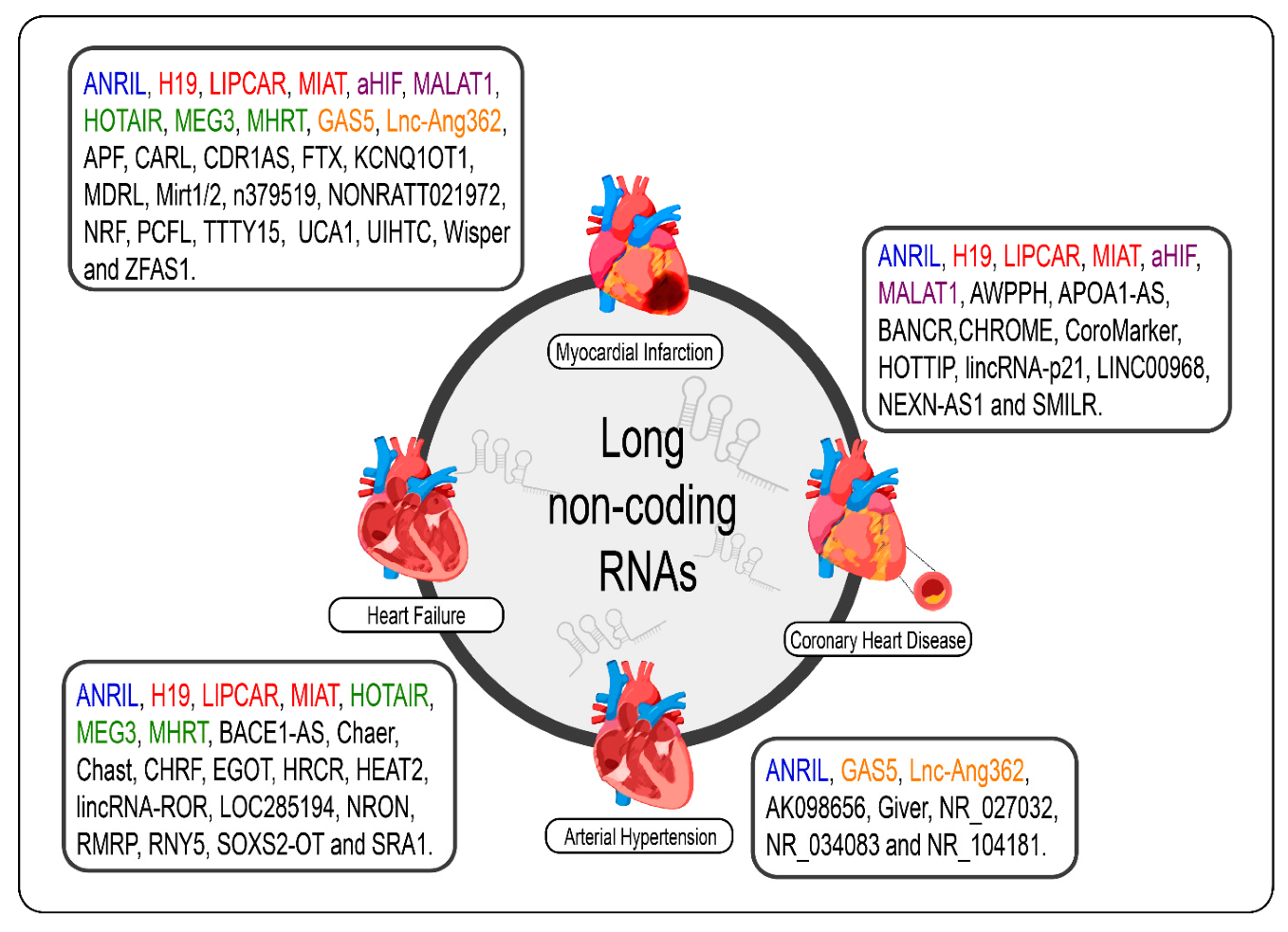
Given the specificity of expression of lncRNAs, it would be careless to think that the dysregulation of the expression of these molecules in cardiac pathological processes, even if the molecular mechanism behind them is not exactly understood, was a mere coincidence [69]. The poor conservation of these interspecies transcripts, however, makes it difficult to translate findings in rodent models for human applications; however, several studies have shown promising results regarding the prognosis of CVDs and new therapies from the modulation of cardiac lncRNAs [33,34,70,89,90,172,173]. We summarize the lncRNAs and the CVDs (Figure 2).
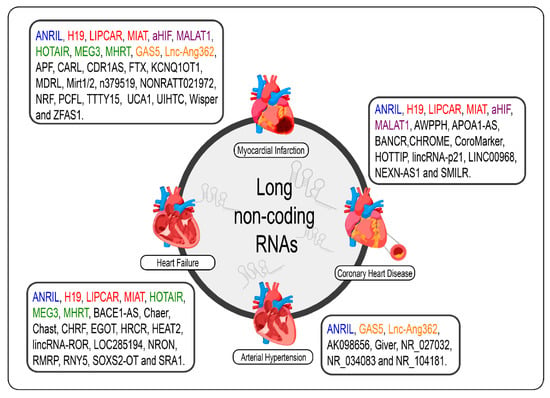
Figure 2.
lncRNAs are differentially expressed in cardiovascular diseases such as heart failure, myocardial infarction, coronary artery disease, and arterial hypertension. The lncRNAs marked in blue are the same present in myocardial infarction, coronary artery disease, heart failure, and arterial hypertension; those marked in red are the same present in myocardial infarction, coronary artery disease, and heart failure; those marked in green are the same present in myocardial infarction and heart failure, those marked in orange are present in myocardial infarction and arterial hypertension; and the purple present in myocardial infarction and coronary artery disease.
9. Arterial Hypertension
Arterial hypertension (AH) is among the most common CVDs and is a major risk factor, if not the main one, for the development of other diseases that affect this system. Vascular smooth muscle cells (VSMCs), as a contractile element of the vessel wall, are responsible for regulating vascular tone and resistance. In this sense, abnormalities in the processes of proliferation, migration, and differentiation of these cells consequently contribute to the pathogenesis of hypertension [174]. Despite its severity, the molecular mechanism of the pathogenesis of this disease has not been precisely defined, given its complex etiology. However, although in much smaller proportions than what is currently available on miRNAs [175], few studies have identified lncRNAs specifically expressed in these cells and, therefore, involved in the pathophysiology of AH through the regulation of vascular tone [176,177].
One of these studies performed in VSMCs of rats treated with angiotensin II (Ang II)—a peptide hormone inducing vasoconstriction, inflammation, fibrosis, and hypertrophy/hyperplasia—identified a variety of lncRNAs with differential expression, among them, lnc-Ang362, one of the few lncRNAs conserved in humans. miRNAs-221 and -222, previously related to both the regulation of VSMC proliferation and the actions of Ang II on endothelial cells (ECs), were also overexpressed in response to treatment with Ang II and appeared to be co-transcribed with the lncRNA [145]. In fact, silencing lnc-Ang362 decreased the expression levels of these miRNAs as well as decreased VSMC proliferation [145]. Another experiment, this time with growth arrest-specific 5 (GAS5), an lncRNA expressed mainly in ECs and VSMCs, showed that this lncRNA, by regulating the functions of ECs and VSMCs via signaling by β-catenin, plays a fundamental role in vascular remodeling in hypertension, which is a determining process in the prognosis of the disease [124]. The GAS5 knockdown mainly aggravated the hypertensive condition in rat models spontaneously hypertensive from the increase in blood arterial pressure and, together with that, it exacerbated the pathological arterial vascular remodeling, a common sequel in hypertensive humans [124]. GAS5 has also been described as an inhibitor of PDGF-bb-induced proliferation and migration of VSMCs by competitively binding to miRNA-21 and removing the inhibitory effect of this miRNA on its target PDCD4, a crucial regulator of apoptosis and proliferation of VSMCs [125]. a growth factor and proinflammatory cytokine-induced vascular cell-expressed RNA (Giver) is another example of an lncRNA that by induction of Ang II via the NR4A3 nuclear receptor mediates the expression of inflammatory genes and promotes increased oxidative stress and proliferation of VSMCs [129]. In the arteries of hypertensive patients related to this study, Giver had a significantly elevated expression profile, whereas when these patients were treated with angiotensin-converting enzyme inhibitors and angiotensin receptor blockers, a marked reduction in this lncRNA was observed [129].
Another study identified AK098656 as an lncRNA responsible for promoting increased proliferation and migration of VSMCs, that is, the synthetic phenotype of VSMCs, through increased synthesis of matrix proteins and degradation of contractile proteins in hypertensive patients [105]. According to the study, this lncRNA mediates the stability of cytoskeletal proteins through its scaffold mechanism to induce VSMCs differentiation and, consequently, causes resistance artery remodeling thus leading to an increase in blood pressure. In vivo, mice overexpressing this lncRNA spontaneously developed hypertension, marked by the synthetic VSMCs phenotype and the narrowing of resistance arteries [105]. These same rats also appeared to be suffering from mild cardiac hypertrophy [105]. The results of a genetic study in Turkish hypertensive patients suggested that polymorphisms rs10757274, rs2383207, rs10757278, and rs1333049 within the lncRNA antisense non-coding RNA in the INK4 locus (ANRIL), also known as CDKN2B-AS1, could confer increased susceptibility to the development of AH [107].
10. Coronary Heart Disease
Atherosclerosis is a chronic, progressive inflammatory process common in several CVDs and is characterized by the formation and deposition of fibrofatty plaques in the wall of arteries [178]. This accumulation of atheromatous plaques results in the narrowing and stiffening of these vessels, such that, over time, blood flow is partially or totally blocked, leading to tissue ischemia [178]. When atherosclerosis occurs in the arteries that supply the heart (i.e., the coronary arteries), it is then called coronary heart disease (CHD). Considering that several lncRNAs are responsible for conducting the functions of ECs, VSMCs, vascular inflammation, and cell metabolism, the dysregulation of the expression of specific non-coding long transcripts is involved with the development and progression of atherosclerosis and, consequently, CHD [179].
Among the findings, it was reported that high levels of H19 expression were associated with the diagnosis of CHD [133] and linked to hyperhomocysteinemia, an important risk factor for the development of CHD [180], and to polymorphisms also associated with the risk of developing CHD in the Chinese population [134]. The ANRIL lncRNA, discovered from genomic association studies, is yet another example of a transcript identified as directly related to the risk and severity of this disease. It is suggested that SNPs in this lncRNA contribute to susceptibility to CHD [106]. In VSMCs, lincRNA-p21 undoes the link between p53, which has a transitional target, and its mouse double minute 2 inhibitor (MDM2), leading to regulatory effects on cell proliferation and apoptosis [83]. In patients with CHD, this lncRNA has been shown to have low expression in coronary artery tissues [181]. Furthermore, polymorphisms in lincRNA-p21 were also associated with an increased risk of CHD [142]. A Brazilian study suggested the involvement of MIAT and MALAT1 in the pathogenesis of CHD in the Brazilian population [149]. According to the data obtained, both lncRNAs demonstrated high plasma expression levels in patients with CHD [149]. However, while MIAT, which has previously been associated with vascular endothelial dysfunction, does appear to be related to CHD, the data on MALAT1 is still inconclusive.
More examples of lncRNAs that have been shown to be associated with CHD are BRAF-activated non-protein coding RNA (BANCR) [115], cholesterol homeostasis regulator of miRNA expression (CHROME) [121], HOXA transcript at the distal tip (HOTTIP) [138], LINC00968 [143], nexilin F-actin binding protein antisense RNA 1 (NEXN-AS1) [160], and smooth muscle-induced lncRNA enhances replication (SMILR)m [165].
11. Acute Myocardial Infarction
Acute myocardial infarction (MI) is an interruption in blood flow, in any coronary artery, that induces a reduction in blood supply. These interruptions induce some portion of the cardiac muscle tissue to suffer an ischemic process, receiving less oxygen to meet their needs leading to metabolic diseases. It has been demonstrated that there are lncRNAs directly or indirectly involved in the occurrence of MI and that their expression levels have potential signaling for the disease [108,158].
MIAT has been characterized as an lncRNA with a pro-fibrotic function after MI [156]. Acting like a sponge on important miRNAs with anti-fibrotic function, such as miRNA-24, the action of MIAT not only led to myocardial degeneration by fibrosis but also, consequently, promoted adverse remodeling in the heart of infarcted rats [156]. On the other hand, the MIAT knockdown prior to MI contributed to the preservation of cardiac function by decreasing the extent of matrix deposition and interstitial fibrosis [156]. Other publications have now raised the pro-hypertrophic potential of MIAT in cardiomyocytes from its action as a sponge on miRNAs-150 [157] and -93 [182]. In a mouse model, shortly after MI induction, it was identified that two lncRNAs had a significantly high level of expression in the heart: myocardial infarction-associated transcript 1 (Mirt1) and 2 (Mirt2) [158]. Unlike MIAT, these transcripts have no direct relationship with infarct size and seem to play a protective role in cardiac ventricular function through intracellular communication via paracrine signaling. Its expression levels are correlated with the expression of genes known to be involved with the preservation of the ejection fraction and, therefore, with processes that affect left ventricular remodeling after MI such as extracellular matrix turnover, fibrosis, apoptosis, and mainly inflammation [158,183].
Some others lncRNAs have been proposed as MI biomarkers. The dysregulation of plasma LIPCAR levels in samples from patients with ventricular remodeling after infarction, for example, suggests an association, although it may not be causal of this lncRNA with the disease [140]. The finding that lncRNA urothelial cancer-associated 1 (UCA1) stimulates cardiomyocyte apoptosis, contributing to the progression of cardiac injury in rats with cardiac injury induced by ischemia-reperfusion injury, raised interest in its potential as a biomarker and also as a potential therapeutic target [167,168]. In the cardiac response to the ischemic process, the lncRNA aHIF appears as an mRNA destabilizer, and its translation product is the hypoxia-inducing factor 1α (HIF-1α), the master regulator of the cellular response such as angiogenesis and hypoxia [103,108]. In the blood of patients affected by MI, aHIF was highly expressed in relation to its expression levels in healthy patients [108]. The finding, consistent with the fact that HIF-1α is induced by hypoxia, suggests that there is likely an interaction between the mRNA HIF-1α and the lncRNA aHIF in regulating the post-ischemic angiogenesis process [103,108]. In MI, the inhibition of the autophagic process in an ischemic context can play a cardioprotective role itself, given that, from the moment that autophagosome accumulation is observed, especially when in reperfusion, autophagia ceases to be cardioprotective and starts to promote the death of cardiomyocytes. With low expression in the plasma of these patients, HOX transcript antisense RNA (HOTAIR) is another example of an lncRNA with a cardioprotective function in MI and a potential predictor for the diagnosis of MI [137]. HOTAIR regulates apoptosis through the sponge mechanism over miRNA-1, an miRNA already known to be at low levels of expression in infarcted myocardial tissue [137].
During ischemia-reperfusion injury, autophagy promoting factor (APF), highly expressed in this scenario, was responsible for the sequestration of miRNA-188-3p [112]. This miRNA targets the ATG7 gene, an autophagy promoter. miRNA sequestration, therefore, promoted the upregulation of ATG7 expression [112]. This study proposed APF and miRNA-188-3p as therapeutic targets for autophagy inhibition aiming at cardioprotection in MI [112]. Moreover, in this context of cell death, several studies have shown the participation of lncRNAs in regulating the death of viable cardiomyocytes by apoptosis in MI. Cardiac apoptosis-related lncRNA (CARL) [116] and mitochondrial dynamic-related lncRNA (MDRL) [150] are examples of these lncRNAs and were transcriptionally repressed after MI. The overexpression of these in vivo resulted in smaller-sized infarcts. The same consequence was observed from the silencing of Wisp2 super-enhancer-associated RNA (Wisper) after MI [170]. CARL and MDRL, by inhibiting miRNA-539 or miRNA-361, which are pro-apoptotic miRNAs, respectively, inhibited mitochondrial fission and apoptosis of cardiomyocytes. Like these lncRNAs, 5 prime to Xist (FTX), after reperfusion injury and ischemia, was found to have low expression levels and, in vitro, was described with anti-apoptotic action through the regulation of the apoptosis repressor BCL2L2 via sequestration of miRNA-29b-1-5p [123].
Many other lncRNAs have been implicated in the pathophysiology of MI, including ANRIL [108,109,110], CDR1 Antisense RNA (CDR1AS) [117], GAS5 [126,127,128], H19 [130,131,132], KCNQ1OT1 [108], lnc-Ang362 [146], metastasis associated lung adenocarcinoma transcript 1 (MALAT1) [108,132,147], maternally expressed gene 3 (MEG3) [151], myosin heavy chain-associated RNA transcripts (MHRT) [153], n379519 [159], necrosis-related factor (NRF) [163], NONRATT021972 [161], pro-cardiac fibrotic lncRNA (PCFL) [164], testis-specific transcript Y-linked 15 (TTTY15) [166], upregulated in hypothermia-treated cardiomyocytes (UIHTCs) [169], and zinc finger antisense 1 (ZFAS1) [117,171,184]. Therefore, the intrinsic relationship between lncRNAs and MI is evident.
12. Heart Failure
Heart failure (HF) refers to the heart’s inability to adequately perform its function as a blood pump due to the fact of several pathological processes of cardiac remodeling that affect its ability to contract and/or relax [185,186]. Similar to what has been reported for the other CVDs mentioned above, several lncRNAs have been identified behind the mechanisms involved with the progression of maladaptive cardiac remodeling in HF [111,172,187].
Myosin heavy-chain-associated RNA transcripts (MHRT) is a cluster of specific lncRNAs that partially overlap the Myh7 locus, which by harboring the human gene encoding β-myosin heavy chain is critical for cardiac function as an ejector pump [154,188]. MHRT is highly expressed in the heart under physiological conditions, given its protective role in reducing or even inhibiting the expression of genes involved with changes in pathological aspects of heart muscle contractility [154,155]. However, under stress conditions, such as what occurs in HF induced by transverse aortic constriction, a significant decrease in MHRT expression was reported and, concomitantly with a change in the Myh6 isoform (α) for Myh7(β), a well-known feature of HF development [154,189]. Under physiological conditions, MHRT inhibits its silencing by directly interacting with Brg1, the chromatin remodeling factor, in the region of its bidirectional promoter shared with Myh6 [154]. However, under cardiac stress situations, the expression of Brg1 exceeds that of MHRT, so that Brg1 is free to promote its remodeling function on chromatin and, thus, promote the exchange of the heavy chain from α-myosin to β-myosin [154]. The role of MHRT as a protector is validated when, from the restoration of its expression level, the protection of the heart against pathological hypertrophy and, therefore, against HF is noticed [154].
With a specific function in cardiomyocytes, cardiac hypertrophy-related factor (CHRF) was the first lncRNA to be related to HF. In in vitro experiments in cardiomyocytes, overexpression of CHRF induced a pathological process of hypertrophy and, in vivo, it induced apoptosis [120,139]. By sequestering the miRNA-489, this lncRNA prevents the interaction of the microRNA with its target Myd88 (myeloid differentiation primary response gene Myd88), an indispensable gene in the regulation of cardiac hypertrophy [120]. The hypertrophy-associated epigenetic regulator (CHAER) cardiac lncRNA, abundantly expressed in the heart of mice with HF, is another transcript involved in the control of cardiac remodeling [118]. This lncRNA interacts with PCR2 with the purpose of inhibiting the methylation of lysine residues in histones that are found in the promoter regions of pro-hypertrophic genes, making them amenable to expression. When silenced, it was observed that the pathological cardiac hypertrophic process was alleviated. Similarly, cardiac hypertrophy-associated transcript (CHAST) is characterized as a pro-hypertrophic lncRNA [119]. In both mouse hypertrophic cardiac models and in the cardiac tissue of patients with aortic stenosis, characterized by the compensatory development of cardiac hypertrophy, it was observed that CHAST was highly expressed [119]. In these mice, CHAST overexpression induced cardiomyocyte hypertrophy both in vivo and in vitro, while its suppression proved cardio-protective by preventing hypertrophy and preserving cardiac function in these animals [119].
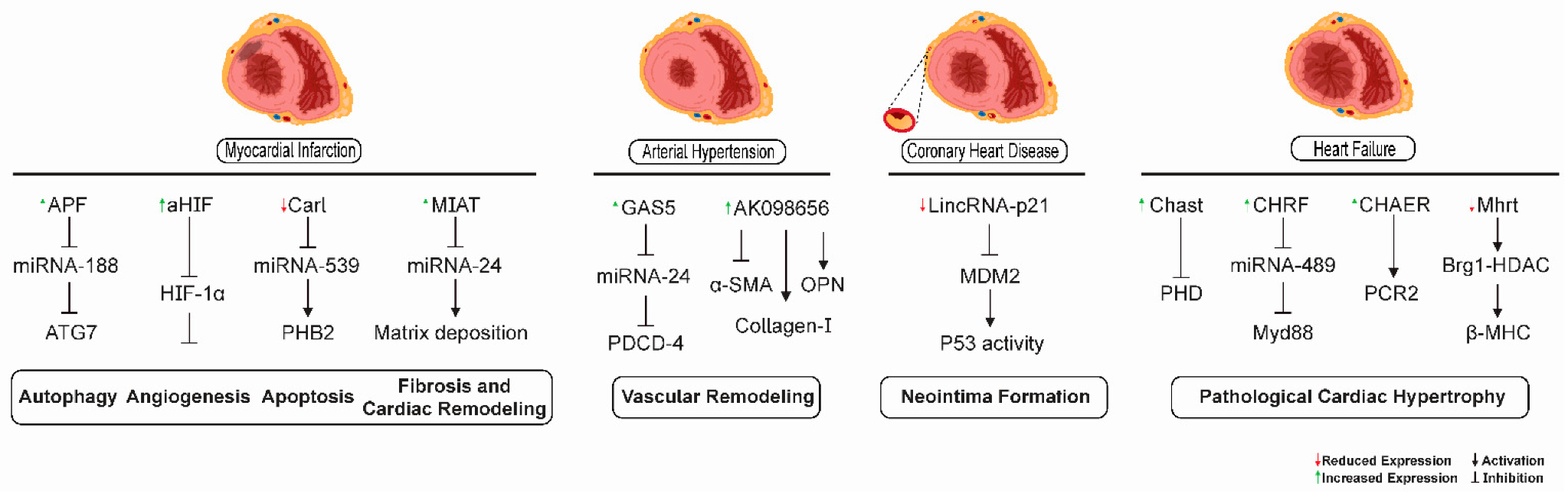
Some of the signaling mechanisms mediated by lncRNAs participating in different pathological processes in the heart and vascular tissues in heart failure, myocardial infarction, coronary heart disease, and arterial hypertension are depicted in Figure 3.
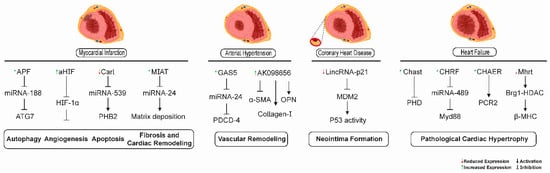
Figure 3.
lncRNA-mediated signaling mechanisms as possible therapeutic targets for cardiovascular diseases. lncRNAs have been shown to participate in different pathological processes in the heart and vascular tissues in heart failure, myocardial infarction, coronary heart disease, and arterial hypertension. lncRNAs APF, CARL, aHIF, and MIAT were involved in autophagy, angiogenesis, apoptosis, and cardiac fibrosis processes related to myocardial infarction, respectively. The lncRNAs GAS5 and AK0986656 promoted vascular remodeling in arterial hypertension. The lncRNA lincRNA-p21 was related to neointima formation in coronary heart disease. The lncRNAs MHRT, CHAER, CHRF, and CHAST could serve as promising targets for the retardation of pathological cardiac hypertrophy in heart failure.
13. LncRNAs as Biomarkers and Potential Therapeutic Targets for Cardiovascular Diseases
Offering a treatment that specifically meets the needs of a patient is the goal of personalized cardiovascular medicine, which considers aspects, such as clinical, genetic, genomic, and even environmental data, related to the patient in question [190,191,192]. However, despite technological advances, the current tools and methods available still lack precision. Circulating biomarkers have been shown to be considerable indicators regarding disease progression, as they simplify and guide clinical decisions so that medical intervention is no longer a generalized pattern applied to any patient and begins to adapt to the individual’s needs. Traditionally, proteins and peptides derived from the heart are the main circulating molecules to be explored in terms of their usefulness as biomarkers for the diagnosis of CVDs, so much so that, routinely, the expression of troponin T and B-type natriuretic peptide are investigated for diagnosis or prognosis [193]. Currently, a range of studies have proposed the use of circulating mRNAs and miRNAs as cardiac biomarkers in personalized medicine [20,21,194,195,196]. Recently, since lncRNAs are differentially expressed in a specific time–space context, participate in intracellular communication, and are associated with CVDs, it has been suggested that these lncRNA may be the newest class of non-coding biomarkers for non-invasive clinical use for the diagnosis and prognosis of CVDs. In support of this possibility, it has already been shown that lncRNAs can be detected in extracellular body fluids such as serum, plasma, and urine [102,140,197,198,199,200]. In addition to being encapsulated in exosomes, extracellular vesicles [122,201], and apoptotic bodies [202], circulating lncRNAs can associate with proteins and maintain the stability of their conserved transcript. Therefore, they have decisive characteristics of a biomarker: easy access, stability, and specific expression patterns in the context of CVDs.
The first study to test the use of an lncRNA as a biomarker for CVDs proposed LIPCAR as an example, since its plasma levels were shown to be related to left ventricular remodeling soon after MI as well as to the increased chances of progression to a condition of HF [140]. Those patients who developed HF as a result of an MI had a high level of LIPCAR expression in plasma compared to those patients who after the MI did not suffer from ventricular remodeling [140]. Furthermore, the magnitude of LIPCAR expression was also shown to be associated with the prediction of cardiovascular mortality in those patients with chronic HF [140]. More recently, the non-coding repressor of NFAT (NRON) and MHRT were proposed as independent predictive factors for HF, as they were found in high plasma levels in patients affected by this pathology [155]. Furthermore, others lncRNAs, such as ANRIL [111], antisense transcript of β-secretase-1 (BACE1-AS) [114], eosinophil granule ontogeny transcript (EGOT) [111], H19 [135], heart disease-associated transcript 2 (HEAT2) [136], HOTAIR [111], long intergenic non-protein coding RNA, regulator of reprogramming (lincRNA-ROR) [144], LOC285 [111], ribonuclease mitochondrial RNA processing (RMRP) [111], Ro60-associated Y5 (RNY5) [111], SOX2 overlapping transcript (SOX2-OT) [111], and steroid receptor RNA activator 1 (SRA1) [111] showed variations in their expression levels in the context of HF, which eventually suggests their potential as biomarkers for this disease.
It is not only in HF that lncRNAs have been found with the potential to serve as a biomarker. Even though the biomarker potential of lncRNAs in arterial hypertension is still a little explored subject [176], some circulating lncRNAs, such as GAS5, AK098656, NR_027032, NR_034083, and NR_104181, were differentially expressed in hypertensive patients compared to healthy individuals, suggesting a potential role of these lncRNAs as biomarkers in this disease. GAS5 is an lncRNA expressed mainly in ECs and VSMCs and was found with low expression in the plasma of hypertensive patients and in arteries and retinas of a spontaneously hypertensive rat model [124]. On the other hand, the lncRNA AK098656 stood out as positively regulated in the plasma of hypertensive patients and, mainly, the predominance of its expression was located in VSMCs [105]. Compared with normotensive patients, the lncRNAs NR_027032 and NR_034083 presented with significantly low levels in peripheral blood leukocytes from patients with essential hypertension, whereas the opposite was observed for the levels of NR_104181; that is, this lncRNA presented itself with significantly reduced levels in these patients [162]. In CHD, CoroMarker was also proposed as a promising biomarker, since it was found to be dysregulated, but stabilized in vesicles, in plasma samples from these patients [122]. aHIF [104], APOA1 antisense RNA (APOA1-AS) [104], lncRNA associated with poor prognosis of hepatocellular carcinoma (AWPPH) [113], KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) [104], and LIPCAR [141] are others prominent lncRNAs to be used in clinical settings as biomarkers for diagnosis and monitoring of CHD. The molecular mechanism of these lncRNAs as biomarkers in the pathophysiology of CHD, however, is still something to be better explored. In another study, this time involving peripheral blood mononuclear cells from patients with MI, MIAT presented with reduced levels of expression and was associated with ST-segment elevation in these patients, suggesting the potential biomarker of this lncRNA [197]. In addition, lncRNAs can be detected in whole blood samples from patients with MI [108]. The study responsible for this finding identified that ANRIL and KCNQ1OT1 could be used in favor of disease prognosis as predictors of ventricular dysfunction after MI [108]. Despite the evident incremental advantage to be conferred by lncRNAs in addition to the already available biomarkers, there is still much more work to be conducted until they reach clinical application.
Considering all of the above regarding the regulatory role of lncRNAs in the cardiovascular system and the observation that the dysregulation of the expression of these transcripts has been observed in CVDs, it seems appropriate to consider them as molecular targets in therapies [173,203,204,205]. Through counter-regulation, the restoration of physiological expression levels of lncRNAs can occur both through gain-of-function strategies (for those lncRNAs that are significantly impaired) and through loss-of-function strategies (for those overexpressed). Among the strategies involved in decreasing RNA expression levels is the RNA interference technique, which, using small interfering RNAs (siRNAs), is able to decrease the expression levels of lncRNAs located mainly in the cytoplasm [206,207] and, alternatively, with the use of siRNAs, there are GapmeRs that can be used in a technique based on the complementarity of single-stranded antisense oligonucleotides in the gene to be silenced [33,152,170,208,209,210]. In the opposite direction, plasmids and viral vectors have been used for the overexpression of lncRNAs [116,150,211]. Recently, gene-editing tools, such as the CRISPR system, have been proposed as attractive approaches in terms of their high therapeutic potential [212,213]. In fact, the modulation of expression levels of cardiac-specific lncRNAs in vivo has shown promising results in terms of improving cardiac dysfunction and even delaying the progression of other pathological processes in the heart already affected by some diseases. Despite this, there are obstacles to be overcome in the process of translating these lncRNA modulation strategies for the treatment of CVDs addressed here: (i) drug delivery mechanisms, which vary according to the technique used, to the heart, vessel, specific cell type, or even cell compartment; (ii) affinity of the therapeutic molecule to the target lncRNA as well as the integrity of its stability in circulation; (iii) pharmacokinetic and pharmacodynamic characterization of the drug in question; (iv) assessment of the systemic consequences of target modulation; (v) evaluation of the effectiveness and duration of responses. Resolving these issues is critical to the success of lncRNAs as new therapeutic targets for the treatment of CVDs [214].
14. Cardiac LncRNAs and Exercise Training
Regular physical exercise is associated with numerous protective and restorative effects on cardiovascular health [215,216,217,218,219,220], in addition to improving the prognosis of patients after cardiovascular events. Physical exercises encompass modifiable variables [218]. These include a modality (e.g., aerobic or resistance training, frequency, intensity, and duration of exercise activities), each of which affect metabolic and molecular responses. Aerobic (or resistance-based) and resisted (or strength-based) exercises represent two extremes of the diversity of existing modalities. Aerobic exercise imposes a high-frequency (repetition) and low-power (load) demand, whereas resistance exercise imposes a low-frequency and high-load demand. Therefore, the metabolic and molecular responses to different modalities are distinct and, therefore, the specificity of a given molecular response is coupled with a functional result. Aerobic physical training has been the most investigated in the literature and with more effective results than resistance training for the treatment of the main cardiovascular risk factors, as it promotes blood pressure control, improves blood lipid profile and insulin sensitivity as well as improvements in an individual’s capacity for maximum oxygen consumption [218]. More recent studies, however, have shown that combined aerobic exercise and resistance exercise training appears to be more effective in modulating CVD risk factors than just those based on a single modality [221,222], in addition to promoting compensatory physiological adaptations capable of altering the structure of the myocardium, thus improving cardiac functionality [223]. The beneficial effects of exercise to the cardiovascular system have been well described in many excellent reviews [224,225,226,227].
Aerobic exercise has been characterized as the main therapeutic alternative for hypertensive patients in view of its beneficial effects on all systems involved in blood pressure regulation [228,229]. In the cardiovascular system, for example, aerobic exercise promotes an increase in cardiac output by inducing left ventricular hypertrophy of the physiological type and a reduction in heart rate that leads to a reduction in the pressure of the coronary flow on the organ, not to mention its influence on risk factors related to the development and progression of this disease [228]. In this way, aerobic exercise promotes significant improvements in the cardiac health of hypertensive patients, delaying or even preventing adversities arising therefrom. It is not only in hypertension that exercise is successfully applied for therapeutic purposes. In order to assess the effect of exercise-based cardiac rehabilitation, a large meta-analysis involving more than 8000 patients with CHD undergoing different randomized clinical training studies observed a significant 26% decrease in cardiac mortality in these patients [230,231]. If in CHD, an important precursor of MI, if not the main one, exercise already has a significant impact on cardiac mortality, it could not be different in the context of MI. After MI, physical exercise is a great ally in preventing complications resulting from infarction and in increasing the quality of life and longevity of these individuals [232,233,234,235]. Furthermore, a variety of physical training studies in patients with HF have proven the efficacy and safety of aerobic training in these patients [236,237,238,239]. Furthermore, meta-analysis studies confirmed improvements in clinically relevant parameters for HF [240,241,242]. Although there is no doubt about the restorative role of exercise on the cardiovascular system, little is known about the molecular mechanisms behind these effects.
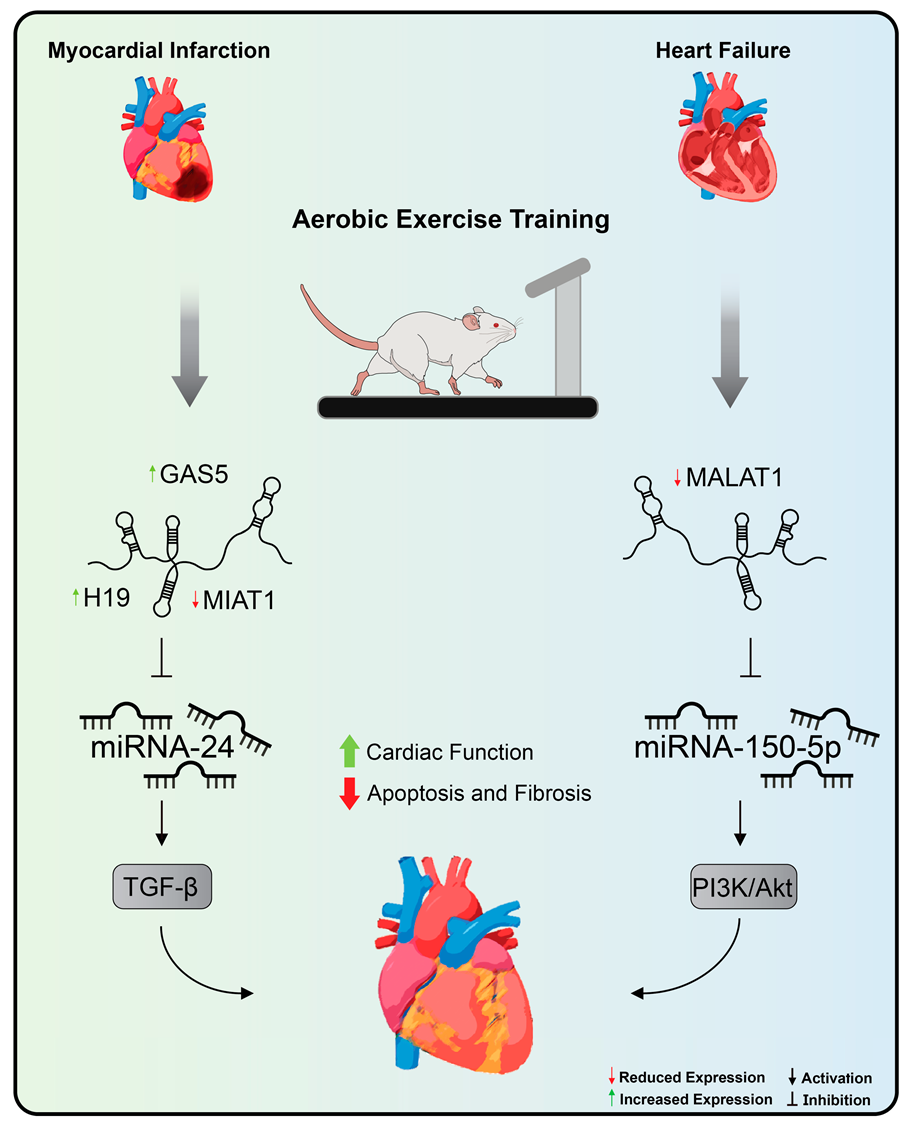
Understanding the molecular pathways involved behind exercise-induced cardiac adaptation provides potential therapeutic targets for the treatment of CVDs [2]. Widely recommended as the most effective non-pharmacological strategy for the prevention and treatment of CVDs, exercise training therefore represents another method to be applied, aiming at modulating the expression of lncRNAs [28,35]. Thus far, only two articles have investigated the role of exercise training on the modulation of lncRNAs in the context of CVDs [148,243] (Figure 4).
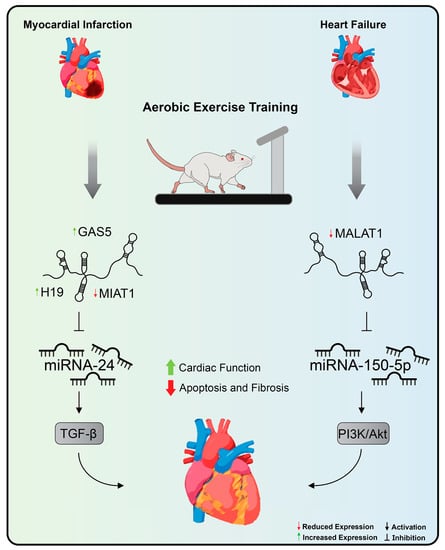
Figure 4.
Aerobic exercise training’s effects on lncRNA expression in cardiovascular diseases. Treadmill exercise promoted, by decreasing MALAT1 and increasing miRNA-150 expression levels, improvements in cardiac function and left ventricular remodeling in rats with heart failure. In addition, aerobic exercise promoted a reduction in apoptosis of cardiomyocytes and fibrotic areas in the heart of rats after myocardial infarction through the normalization of levels of expression of H19, MIAT, and GAS5. Exercise training inhibits MIAT expression, a lncRNA with pro-fibrotic action in the myocardial infarction that acts by sequestering miRNA-24 and stimulating the TGF-beta pathway. MALAT1: metastasis-associated lung adenocarcinoma transcript 1, MIAT: myocardial infarction-association transcript, and GAS5: growth arrest specific 5.
In one of these studies, it was suggested that aerobic exercise on a treadmill promoted, by decreasing MALAT1 expression levels [244], not only improvements in cardiac function and left ventricular remodeling in rats with chronic HF but also led to a reduction in the levels of reactive oxygen species and inflammatory cytokines, decreased apoptosis of cardiomyocytes, and increased autophagic proteins [148]. The overexpression of MALAT1 via plasmid vector nullified all the positive effects promoted by aerobic exercise in rats with chronic HF [148]. As a molecular mechanism, it was observed that, in vitro, MALAT1 overexpression was related to decreased miRNA-150-5p expression levels, resulting in increased cardiomyocyte apoptosis and decreased autophagy as well as suppression of the signaling pathway PI3k/Akt [148].
Aerobic exercise, also performed on the treadmill, promoted improvements in contractility and cardiac function as a pump through the improvement in left ventricular function and led to a reduction in apoptosis of cardiomyocytes and fibrotic areas in the heart of rats after MI through the normalization of levels of expression of H19, MIAT, and GAS5. H19 is an lncRNA associated with an increase in the area of infarction and fibrosis in the stages immediately after MI [130,131]. With significant expression in cardiac fibroblasts, H19 exerts its mechanism through binding to the Y-box binding protein 1 thus preventing collagen production [131]. In this sense, the positive effect of exercise on reducing fibrosis and improving cardiac function can be attributed, albeit partially, to the modulation of H19 expression levels. MIAT, in turn, is an lncRNA with pro-fibrotic action in the pathogenesis of MI that acts by sequestering miRNA-24 and, in this way, removes the inhibition of Furin, the signaling activator of the TGF-beta pathway [156]. Therefore, it is to be expected that their expression levels are high in this context. By decreasing MIAT expression levels, aerobic exercise attenuated the effects promoted by the dysregulation of this lncRNA in cardiac fibrosis. In a previous study, induction of lncRNA GAS5 expression was related to inhibition of sema3a [128]. Moreover, corroborating this finding, physical exercise promoted a significant increase in GAS and, thus, promoted beneficial effects in the rat model used in the study. Despite these findings, the researchers concluded that further investigation into the targets of these lncRNAs is needed to trace the exact molecular mechanism that links exercise to lncRNAs in MI.
15. LncRNAs in Cardiovascular Diseases: Challenges and Future Perspectives
lncRNAs have characteristics of great interest to the biomedical community. These characteristics have received attention, albeit timidly, in recent clinical trials (NCT04189029; NCT03268135; NCT02915315; NCT03279770) with the purpose of investigating the role of these transcripts as biomarkers and in the pathogenesis of some CVDs. However, one cannot fail to mention the challenges to be overcome until these transcripts finally move from research to clinical application.
The isolation, detectability, quantification, and strategy adopted for the normalization of circulating lncRNAs are key factors for the reliable identification of candidates as potential biomarkers and, in the absence of standardization, have been technical limitations of important relevance for ncRNAs in general [202,214,245]. Added to this is the fact that there may be significant variations in the expression levels of lncRNAs, including those that are significant regarding CVDs, among different body fluids such as serum, plasma, and urine or even among different compartments of the same cell [199,200]. The lack of standardization regarding the fluid to be considered as a sample for a given lncRNA may end up leading to research with wrong conclusions. In addition, cardiovascular risk factors, medication use, sex, and age are examples of some factors capable of promoting changes in the expression levels of ncRNAs such as lncRNAs [196,202]. Among the limitations found in the process until lncRNAs reach the clinic stage as therapeutic targets is the fact that lncRNAs are still in the process of characterization and annotation; even at these stages, many challenges need to be overcome. In this sense, the modulation of an lncRNA can result in opposite effects, even harmful, for the purpose in question [34], since the same lncRNA can be involved in the mechanism of different pathologies [246,247]. The availability of information on the characterization and annotation of lncRNAs, therefore, provides greater knowledge about the lncRNA in question and, consequently, of other molecules with which it may be related, which allows a broader notion about the implications involved in the modulation of one of these transcripts. Furthermore, it is well known that the intermediate step between basic research and clinical trials necessarily involves the use of animal models. At this point, the lack of conservation of the nucleotide sequence of lncRNAs among different species represents a limitation with great impact, as it makes it difficult to transpose the results obtained in preclinical studies to humans [248]. Therefore, clinical trials end up being restricted to working only with those lncRNAs that have their counterparts in humans. In addition to these challenges, there is still a need to elucidate the secondary and tertiary structures of lncRNAs, which are even more critical for the function of these transcripts than the primary structure, and these may have structural homologues in other species including those used as models of experimental animals [249,250]. Finally, there are still challenges regarding drug delivery to the target lncRNA of interest [251].
Considering the limitations mentioned here, an initiative by the scientific community is needed to reach a consensus on the methods (from the way the sample is manipulated to the chosen normalization strategy) to be used in order to ensure robust paths for identifying lncRNAs as CVD biomarkers as well as precision regarding the criteria and parameters to be adopted for the formation of groups involved in future clinical studies, whether for the identification of lncRNAs as biomarkers or for the assessment of their potential as a therapeutic target. In this regard, there is still much to be overcome regarding the challenges of the application of lncRNAs as a therapeutic approach in CVDs; the clinical trials presented here refer mostly to the use of these molecules as biomarkers, some of which also investigated the role of lncRNAs in CVDs. Although important experiments using highly sophisticated technological tools, such as RNA-seq, have been conducted to identify therapeutic candidate lncRNAs, little has been done regarding the characterization of these transcripts found in terms of regulation of the pathological process or ability to undergo regulation. It is necessary, therefore, that new information that arises about a lncRNA already known or recently discovered can be accessed by any researcher, anywhere in the world, as this ensures optimization in the field of research on lncRNAs. Even today, the lack of gene homology is an obstacle in science. Future technological advances are expected to provide solutions to overcome these and other limitations that challenge the use of lncRNAs as therapeutic targets in CVDs [34,202,214,251]. Once overcome, the benefits for patients affected by CVDs can be enormous.
16. Conclusions
This new class of non-coding transcripts playing regulatory roles in various diseases is the beginning of knowledge. Although the number of lncRNAs discovered over the years has increased, so far very little is known about the mechanisms of action and functions performed by these molecules. One of the reasons for this delay is the poor sequence conservation of interspecies lncRNAs, as variations in different animal models make the identification of biological functions and mechanisms of action of the vast majority of lncRNAs and the consequent translation of findings from animals to humans difficult. In this aspect, databases (for example, LNCipedia [252], LncTar [253], and LncRNAWiki [254,255]) have been important tools for depositing and rationalizing information about lncRNAs from different parts of the world. Despite the challenges, lncRNAs are promising candidates for therapeutic use and are characterized as a tool with great application power in personalized medicine given their specific expression pattern associated with different pathologies. It is still the beginning of this new field of study involving the modulation of the expression of lncRNAs in the context of CVDs and physical exercise. There are, therefore, great expectations regarding the application of alternative modalities to aerobic exercise to modulate the lncRNAs involved in this context, such as resistance training and also combined training. Before therapeutic application, further research is needed for a complete functional characterization of lncRNAs involved in cardiovascular pathology as well as their ability to be regulated from different physical training protocols.
Author Contributions
Conceptualization, C.C.M.C., L.F.R., B.R.d.A.P. and T.F.; methodology, C.C.M.C., L.F.R., B.R.d.A.P. and T.F.; formal analysis, E.M.O. and T.F.; data curation, C.C.M.C., L.F.R., B.R.d.A.P. and T.F.; writing—original draft preparation, C.C.M.C., L.F.R. and B.R.d.A.P.; writing—review and editing, C.C.M.C., L.F.R., B.R.d.A.P., E.M.O. and T.F.; visualization, C.C.M.C., L.F.R., B.R.d.A.P., E.M.O. and T.F.; supervision, T.F. All authors have read and agreed to the published version of the manuscript.
Funding
The researchers were supported by the Sao Paulo Research Foundation (FAPESP: #2015/22814-5 and #2015/17275-8), the MicroRNA Research Center (NAPmiR, University of Sao Paulo), the National Council for Scientific and Technological Development (CNPq: #313479/2017-8, #100870/2021-0, and #143938/2021-5), and the Coordination for the Improvement of Higher Education Personnel (CAPES-PROEX: #88887.484856/2020-00 and #88887.640701/2021-00).
Acknowledgments
The authors thank the laboratory team for their technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Cardiovascular Diseases; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Sharma, S.; Merghani, A.; Mont, L. Exercise and the Heart: The Good, the Bad, and the Ugly. Eur. Hear. J. 2015, 36, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.K.; Saltin, B. Exercise as Medicine-Evidence for Prescribing Exercise as Therapy in 26 Different Chronic Diseases. Scand. J. Med. Sci. Sports 2015, 25, 1–72. [Google Scholar] [CrossRef] [PubMed]
- Dunstan, D.W.; Dogra, S.; Carter, S.E.; Owen, N. Sit Less and Move More for Cardiovascular Health: Emerging Insights and Opportunities. Nat. Rev. Cardiol. 2021, 18, 637–648. [Google Scholar] [CrossRef]
- ENCODE: Encyclopedia of DNA Elements. Available online: https://www.encodeproject.org/ (accessed on 28 August 2021).
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-Throughput Sequencing Technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Delihas, N. Discovery and Characterization of the First Non-Coding RNA That Regulates Gene Expression, micFRNA: A Historical Perspective. World J. Biol. Chem. 2015, 6, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M. Non-Coding DNA Evolution: Junk DNA Revisited. Encycl. Evol. Biol. 2016, 6, 124–129. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or Transcriptional Noise? Evidence for Selection within Long Noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef]
- Mattick, J.; Makunin, I.V. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Churchman, L.S. Not Just Noise: Genomics and Genetics Bring Long Noncoding RNAs into Focus. Mol. Cell 2017, 65, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Chen, R.; Qu, L.; Cao, X. Noncoding RNA: From Dark Matter to Bright Star. Sci. China Life Sci. 2020, 63, 463–468. [Google Scholar] [CrossRef]
- Mongelli, A.; Martelli, F.; Farsetti, A.; Gaetano, C. The Dark That Matters: Long Non-Coding RNAs as Master Regulators of Cellular Metabolism in Non-communicable Diseases. Front. Physiol. 2019, 10, 369. [Google Scholar] [CrossRef]
- Kaikkonen, M.U.; Lam, M.T.; Glass, C.K. Non-Coding RNAs as Regulators of Gene Expression and Epigenetics. Cardiovasc. Res. 2011, 90, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-Coding RNAs as Regulators in Epigenetics. Oncol. Rep. 2016, 37, 3–9. [Google Scholar] [CrossRef]
- Chen, Y.-C.A.; Aravin, A.A. Non-Coding RNAs in Transcriptional Regulation. Curr. Mol. Biol. Rep. 2015, 1, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.S.; Zhou, R.; Rana, T.M. Gene Regulation by Non-Coding RNAs. Crit. Rev. Biochem. Mol. Biol. 2013, 49, 16–32. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.N. MicroRNAs as Therapeutic Targets and Biomarkers of Cardiovascular Disease. Sci. Transl. Med. 2014, 6, 239ps3. [Google Scholar] [CrossRef] [PubMed]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol. Biol. 2017, 1617, 57–67. [Google Scholar] [CrossRef]
- Gomes, C.P.; de Gonzalo-Calvo, D.; Toro, R.; Fernandes, T.; Theisen, D.; Wang, D.-Z.; Devaux, Y. Non-Coding RNAs and Exercise: Pathophysiological Role and Clinical Application in the Cardiovascular System. Clin. Sci. 2018, 132, 925–942. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.J.; Bye, A.; el Azzouzi, H.; Wisløff, U. MicroRNAs as Important Regulators of Exercise Adaptation. Prog. Cardiovasc. Dis. 2017, 60, 130–151. [Google Scholar] [CrossRef]
- Altana, V.; Geretto, M.; Pulliero, A. MicroRNAs and Physical Activity. MicroRNA 2015, 4, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Meurer, S.; Krüger, K.; Mooren, F. MicroRNAs unter Einfluss Körperlicher Belastung. Ger. J. Sports Med. 2016, 2016, 27–34. [Google Scholar] [CrossRef]
- Yao, R.; Wang, Y.; Chen, L.-L. Cellular Functions of Long Noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, A.; Feschotte, C. Volatile Evolution of Long Noncoding RNA Repertoires: Mechanisms and Biological Implications. Trends Genet. 2014, 30, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Kraus, W.L. From Discovery to Function: The Expanding Roles of Long Non-Coding RNAs in Physiology and Disease. Endocr. Rev. 2015, 36, 25–64. [Google Scholar] [CrossRef] [PubMed]
- Dhanoa, J.K.; Sethi, R.S.; Verma, R.; Arora, J.S.; Mukhopadhyay, C.S. Long Non-Coding RNA: Its Evolutionary Relics and Biological Implications in Mammals: A Review. J. Anim. Sci. Technol. 2018, 60, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.M. On the Origin of lncRNAs: Missing Link Found. Trends Genet. 2017, 33, 660–662. [Google Scholar] [CrossRef]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef]
- McMullen, J.R.; Drew, B.G. Long Non-Coding RNAs (lncRNAs) in Skeletal and Cardiac Muscle: Potential Therapeutic and Diagnostic Targets? Clin. Sci. 2016, 130, 2245–2256. [Google Scholar] [CrossRef]
- Gomes, C.P.C.; Spencer, H.; Ford, K.L.; Michel, L.Y.M.; Baker, A.H.; Emanueli, C.; Balligand, J.-L.; Devaux, Y. The Function and Therapeutic Potential of Long Non-Coding RNAs in Cardiovascular Development and Disease. Mol. Ther.-Nucleic Acids 2017, 8, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Bonilauri, B.; Dallagiovanna, B. Long Non-Coding RNAs Are Differentially Expressed after Different Exercise Training Programs. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J. Non-Coding RNAs: The Architects of Eukaryotic Complexity. EMBO Rep. 2001, 2, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Pheasant, M.; Mattick, J. The Relationship between Non-Protein-Coding DNA and Eukaryotic Complexity. BioEssays 2007, 29, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.A. The Genetic Organization of Chromosomes. Annu. Rev. Genet. 1971, 5, 237–256. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E.; Crick, F.H.C. Selfish DNA: The Ultimate Parasite. Nature 1980, 284, 604–607. [Google Scholar] [CrossRef]
- Britten, R.J.; Davidson, E.H. Repetitive and Non-Repetitive DNA Sequences and a Speculation on the Origins of Evolutionary Novelty. Q. Rev. Biol. 1971, 46, 111–138. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- NONCODE. NONCODE Outline. Available online: http://www.noncode.org/analysis.php (accessed on 28 August 2021).
- GENCODE. GENCODE General Stats. Available online: https://www.gencodegenes.org/human/stats.html (accessed on 28 August 2021).
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.; et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Bánfai, B.; Jia, H.; Khatun, J.; Wood, E.; Risk, B.; Gundling, W.E.; Kundaje, A.; Gunawardena, H.P.; Yu, Y.; Xie, L.; et al. Long Noncoding RNAs Are Rarely Translated in Two Human Cell Lines. Genome Res. 2012, 22, 1646–1657. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.-H.; Guo, J.U. Coding Functions of “Noncoding” RNAs. Science 2020, 367, 1074–1075. [Google Scholar] [CrossRef] [PubMed]
- Bunch, H. Gene Regulation of Mammalian Long Non-Coding RNA. Mol. Genet. Genom. 2017, 293, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat. Rev. Genet. 2015, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.; Kjems, J. Natural RNA Circles Function as Efficient microRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2012, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nat. Cell Biol. 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.-F.; Yang, L.; Zhang, Y.; Xiang, J.-F.; Wu, Y.-W.; Carmichael, G.G.; Chen, L.-L. Long Noncoding RNAs with snoRNA Ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the Classification of Long Non-Coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A Class of Long Noncoding RNAs Synthesized at Enhancers: Figure 1. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Zangrando, J.; Schroen, B.; Creemers, E.E.; Pedrazzinik, T.; Chang, C.-P.; Dorn, G.W., 2nd; Thum, T.; Heymans, S.; Cardiolinc Network. Long Noncoding RNAs in Cardiac Development and Ageing. Nat. Rev. Cardiol. 2015, 12, 415–425. [Google Scholar] [CrossRef]
- Pang, K.C.; Frith, M.C.; Mattick, J.S. Rapid Evolution of Noncoding RNAs: Lack of Conservation Does Not Mean Lack of Function. Trends Genet. 2006, 22, 1–5. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczyk, M.; Kasprowicz, M.K.; Kasprzyk, M.E.; Wrzesinski, J. Human Long Noncoding RNA Interactome: Detection, Characterization and Function. Int. J. Mol. Sci. 2020, 21, 1027. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.A.; Chang, H.Y. Long Noncoding RNAs in Cell-Fate Programming and Reprogramming. Cell Stem Cell 2014, 14, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Chang, H.Y. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.B.; Uchida, S. Functional Characterization of Long Noncoding RNAs. Curr. Opin. Cardiol. 2020, 35, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Pontier, D.B.; Gribnau, J. Xist Regulation and Function eXplored. Qual. Life Res. 2011, 130, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin Signature Reveals over a Thousand Highly Conserved Large Non-Coding RNAs in Mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long Noncoding RNAs with Enhancer-Like Function in Human Cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs Transactivate STAU1-Mediated mRNA Decay by Duplexing with 3′ UTRs via Alu Elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-DiNardo, D.; Kanduri, C. Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Transcriptional Silencing through Chromatin-Level Regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-F.; Chang, Y.-C.E.; Lu, C.-Y.; Hsuan, C.-F.; Chang, W.-T.; Yang, K.-C. Expedition to the Missing Link: Long Noncoding RNAs in Cardiovascular Diseases. J. Biomed. Sci. 2020, 27, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Dimmeler, S. Long Noncoding RNAs in Cardiovascular Diseases. Circ. Res. 2015, 116, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA Gas5 Is a Growth Arrest- and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Redon, S.; Reichenbach, P.; Lingner, J. The Non-Coding RNA TERRA Is a Natural Ligand and Direct Inhibitor of Human Telomerase. Nucleic Acids Res. 2010, 38, 5797–5806. [Google Scholar] [CrossRef] [PubMed]
- Guenther, M.G.; Levine, S.S.; Boyer, L.A.; Jaenisch, R.; Young, R.A. A Chromatin Landmark and Transcription Initiation at Most Promoters in Human Cells. Cell 2007, 130, 77–88. [Google Scholar] [CrossRef]
- Huang, Z.-W.; Tian, L.-H.; Yang, B.; Guo, R.-M. Long Noncoding RNA H19 Acts as a Competing Endogenous RNA to Mediate CTGF Expression by Sponging miR-455 in Cardiac Fibrosis. DNA Cell Biol. 2017, 36, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.M.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.; Gupta, R.A.; et al. A Long Noncoding RNA Maintains Active Chromatin to Coordinate Homeotic Gene Expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.R.; Morales, D.R.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many Human Large Intergenic Noncoding RNAs Associate with Chromatin-Modifying Complexes and Affect Gene Expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T. The X as Model for RNA’s Niche in Epigenomic Regulation. Cold Spring Harb. Perspect. Biol. 2010, 2, a003749. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T. Lessons from X-chromosome Inactivation: Long ncRNA As Guides and Tethers to the Epigenome. Genes Dev. 2009, 23, 1831–1842. [Google Scholar] [CrossRef]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular Signals of Epigenetic States. Science 2010, 330, 612–616. [Google Scholar] [CrossRef]
- Dekker, J. Faculty Opinions Recommendation of Long Noncoding RNA in Genome Regulation: Prospects and Mechanisms. RNA Biol. 2010, 7, 582–585. [Google Scholar] [CrossRef]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Xiong, Y. Long Non-Coding RNA ANRIL Is Required for the PRC2 Recruitment to and Silencing of p15INK4B Tumor Suppressor Gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Spitale, R.C.; Tsai, M.-C.; Chang, H.Y. RNA Templating the Epigenome: Long Noncoding RNAs as Molecular Scaffolds. Epigenetics 2011, 6, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tang, Y.; Sun, H.; Lin, X.; Jiang, B. The Roles of Long Noncoding RNAs in Myocardial Pathophysiology. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ning, Q. The Emerging Roles of Long Noncoding RNAs in Common Cardiovascular Diseases. Hypertens. Res. 2015, 38, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Somoza, A.S.; Devaux, Y.; Martelli, F. Long Noncoding RNAs and Cardiac Disease. Antioxidants Redox Signal. 2018, 29, 880–901. [Google Scholar] [CrossRef] [PubMed]
- Hobuß, L.; Bär, C.; Thum, T. Long Non-Coding RNAs: At the Heart of Cardiac Dysfunction? Front. Physiol. 2019, 10, 30. [Google Scholar] [CrossRef]
- Ounzain, S.; Micheletti, R.; Arnan, C.; Plaisance, I.; Cecchi, D.; Schroen, B.; Reverter, F.; Alexanian, M.; Gonzales, C.; Ng, S.-Y.; et al. CARMEN, A Human Super Enhancer-Associated Long Noncoding RNA Controlling Cardiac Specification, Differentiation and Homeostasis. J. Mol. Cell. Cardiol. 2015, 89, 98–112. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The Tissue-Specific lncRNA Fendrr Is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Werber, M.; Wittler, L.; Timmermann, B.; Grote, P.; Herrmann, B.G. The Tissue-Specific Transcriptomic Landscape of the Mid-Gestational Mouse Embryo. Development 2014, 141, 2325–2330. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple Knockout Mouse Models Reveal lincRNAs Are Required for Life and Brain Development. eLife 2013, 2, e01749. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.; Soltani, B.M. LncRNAs in Cardiomyocyte Maturation: New Window for Cardiac Regenerative Medicine. Non-Coding RNA 2021, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Ding, N.; Li, J.; Jin, X.; Li, L.; Pan, T.; Huo, C.; Li, Y.; Xu, J.; Li, X. Landscape of the Long Non-Coding RNA Transcriptome in Human Heart. Brief. Bioinform. 2018, 20, 1812–1825. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, X. The Functions of LncRNA in the Heart. Diabetes Res. Clin. Pr. 2020, 168, 108249. [Google Scholar] [CrossRef]
- DiStefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. Methods Mol. Biol. 2018, 1706, 91–110. [Google Scholar] [CrossRef]
- Ismail, N.; Abdullah, N.; Murad, N.A.; Jamal, R.; Sulaiman, S. Long Non-Coding RNAs (lncRNAs) in Cardiovascular Disease Complication of Type 2 Diabetes. Diagnostics 2021, 11, 145. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long Noncoding RNAs in Cancer Metastasis. Nat. Rev. Cancer 2021, 21, 446–460. [Google Scholar] [CrossRef]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a Novel Non-Coding RNA, MIAT, That Confers Risk of Myocardial Infarction. J. Hum. Genet. 2006, 51, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hypoxia-Inducible Factor 1 and Cardiovascular Disease. Annu. Rev. Physiol. 2014, 76, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Wang, Y.; Ding, H.; Xue, S.; Yu, H.; Hu, L.; Qi, H.; Wang, Y.; Zhu, W.; et al. KCNQ 1 OT 1, HIF 1A-AS 2 and APOA 1-AS Are Promising Novel Biomarkers for Diagnosis of Coronary Artery Disease. Clin. Exp. Pharmacol. Physiol. 2019, 46, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Lin, X.; Yang, L.; Fan, X.; Wang, W.; Li, S.; Li, J.; Liu, X.; Bao, M.; Cui, X.; et al. AK098656, a Novel Vascular Smooth Muscle Cell–Dominant Long Noncoding RNA, Promotes Hypertension. Hypertension 2018, 71, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, H.M.; Peden, J.F.; Lorkowski, S.; Goel, A.; Ongen, H.; Green, F.; Clarke, R.; Collins, R.; Franzosi, M.G.; Tognoni, G.; et al. Susceptibility to Coronary Artery Disease and Diabetes Is Encoded by Distinct, Tightly Linked SNPs in the ANRIL Locus on Chromosome 9p. Hum. Mol. Genet. 2007, 17, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Bayoglu, B.; Yuksel, H.; Cakmak, H.A.; Dirican, A.; Cengiz, M. Polymorphisms in the Long Non-Coding RNA CDKN2B-AS1 May Contribute to Higher Systolic Blood Pressure Levels in Hyper-Tensive Patients. Clin. Biochem. 2016, 49, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Vausort, M.; Wagner, D.R.; Devaux, Y. Long Noncoding RNAs in Patients with Acute Myocardial Infarction. Circ. Res. 2014, 115, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Ali, I.S.; Riaz, M.; Younas, A.; Sadeque, A.; Niazi, A.K.; Niazi, S.H.; Ali, S.H.B.; Azam, M.; Qamar, R. Association of ANRIL Polymorphism (rs1333049:C>G) with Myocardial Infarction and Its Pharmacogenomic Role in Hypercholesterolemia. Gene 2013, 515, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Shu, L.; Zhang, W.; Huang, C.; Huang, G.; Su, G.; Xu, J. lncRNA ANRIL protects H9c2 cells against hypoxia-induced injury through targeting the miR-7-5p/SIRT1 axis. J. Cell. Physiol. 2019, 235, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Zaccagnini, G.; Perfetti, A.; Fuschi, P.; Valaperta, R.; Voellenkle, C.; Castelvecchio, S.; Gaetano, C.; Finato, N.; Beltrami, A.P.; et al. Long Noncoding Rna Dysregulation in Ischemic Heart Failure. J. Transl. Med. 2016, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, C.-Y.; Zhou, L.-Y.; Wang, J.; Wang, M.; Zhao, B.; Zhao, W.-K.; Jian-Xun, W.; Yan-Fang, Z.; Zhang, X.-J.; et al. APF lncRNA Regulates Autophagy and Myocardial Infarction by Targeting miR-188-3p. Nat. Commun. 2015, 6, 6779. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.-T.; Wang, B.-Q. Clinical Significance of lncRNA-AWPPH in Coronary Artery Diseases. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11747–11751. [Google Scholar] [CrossRef]
- Greco, S.; Zaccagnini, G.; Fuschi, P.; Voellenkle, C.; Carrara, M.; Sadeghi, I.; Bearzi, C.; Maimone, B.; Castelvecchio, S.; Stellos, K.; et al. Increased BACE1-AS Long Noncoding RNA and β-Amyloid Levels in Heart Failure. Cardiovasc. Res. 2017, 113, 453–463. [Google Scholar] [CrossRef]
- Li, H.; Liu, X.; Zhang, L.; Li, X. LncRNA BANCR Facilitates Vascular Smooth Muscle Cell Proliferation and Migration through JNK Pathway. Oncotarget 2017, 8, 114568–114575. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Zhou, L.-Y.; Liu, F.; Zhou, Q.-Y.; Liu, C.-Y.; Fan, Y.-Y.; Li, P.-F. CARL lncRNA Inhibits Anoxia-Induced Mitochondrial Fission and Apoptosis in Cardiomyocytes by Impairing miR-539-Dependent PHB2 Downregulation. Nat. Commun. 2014, 5, 3596. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, L.; Xuan, L.; Pan, Z.; Li, K.; Liu, S.; Huang, Y.; Zhao, X.; Huang, L.; Wang, Z.; et al. Reciprocal Changes of Circulating Long Non-Coding RNAs ZFAS1 and CDR1AS Predict Acute Myocardial Infarction. Sci. Rep. 2016, 6, 22384. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.-J.; Ji, Y.-X.; Zhang, P.; Deng, K.-Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The Long Noncoding RNA CHAER Defines an Epigenetic Checkpoint in Cardiac Hypertrophy. Nat. Med. 2016, 22, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Viereck, J.; Kumarswamy, R.; Foinquinos, A.; Xiao, K.; Avramopoulos, P.; Kunz, M.; Dittrich, M.; Maetzig, T.; Zimmer, K.; Remke, J.; et al. Long Noncoding RNA CHAST Promotes Cardiac Remodeling. Sci. Transl. Med. 2016, 8, 326ra22. [Google Scholar] [CrossRef]
- Wang, K.; Liu, F.; Zhou, L.-Y.; Long, B.; Yuan, S.-M.; Wang, Y.; Liu, C.-Y.; Sun, T.; Zhang, X.-J.; Li, P.-F. The Long Noncoding RNA CHRF Regulates Cardiac Hypertrophy by Targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.J.; Van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The Long Noncoding RNA CHROME Regulates Cholesterol Homeostasis in Primates. Nat. Metab. 2018, 1, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cai, Y.; Wu, G.; Chen, X.; Liu, Y.; Wang, X.; Yu, J.; Li, C.; Chen, X.; Jose, P.A.; et al. Plasma Long Non-Coding RNA, CoroMarker, a Novel Biomarker for Diagnosis of Coronary Artery Disease. Clin. Sci. 2015, 129, 675–685. [Google Scholar] [CrossRef]
- Long, B.; Li, N.; Xu, X.-X.; Li, X.-X.; Xu, X.-J.; Guo, D.; Zhang, D.; Wu, Z.-H.; Zhang, S.-Y. Long Noncoding RNA FTX Regulates Cardiomyocyte Apoptosis by Targeting miR-29b-1-5p and Bcl2l2. Biochem. Biophys. Res. Commun. 2018, 495, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-N.; Shan, K.; Yao, M.-D.; Yao, J.; Wang, J.-J.; Li, X.; Liu, B.; Zhang, Y.-Y.; Ji, Y.; Jiang, Q.; et al. Long Noncoding RNA-GAS5: A Novel Regulator of Hypertension-Induced Vascular Remodeling. Hypertension 2016, 68, 736–748. [Google Scholar] [CrossRef]
- Liu, K.; Liu, C.; Zhang, Z. lncRNA GAS5 Acts as a ceRNA for miR-21 in Suppressing PDGF-BB-Induced Proliferation and Migration in Vascular Smooth Muscle Cells. J. Cell. Biochem. 2019, 120, 15233–15240. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yang, S.-T.; Liu, J.; Zhang, K.-X.; Leng, J.-Y. Silence of LncRNA GAS5 Protects Cardiomyocytes H9c2 against Hypoxic Injury via Sponging miR-142-5p. Mol. Cells 2019, 42, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hou, Y.-M.; Gao, F.; Xiao, J.-W.; Li, C.-C.; Tang, Y. lncRNA GAS5 Regulates Myocardial Infarction by Targeting the miR-525-5p/CALM2 Axis. J. Cell. Biochem. 2019, 120, 18678–18688. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Liu, X.; Sui, X.; Pei, Y.; Liang, Z.; Zhou, N. Long Non-Coding RNA GAS5 Reduces Cardiomyocyte Apoptosis Induced by MI through sema3a. Int. J. Biol. Macromol. 2018, 120, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Zhang, E.; Senapati, P.; Amaram, V.; Reddy, M.A.; Stapleton, K.; Leung, A.; Lanting, L.; Wang, M.; Chen, Z.; et al. A Novel Angiotensin II–Induced Long Noncoding RNA Giver Regulates Oxidative Stress, Inflammation, and Proliferation in Vascular Smooth Muscle Cells. Circ. Res. 2018, 123, 1298–1312. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zou, Y.-G.; Xue, Y.-Z.; Wang, X.-H.; Gao, H.; Dong, H.-W.; Zhang, Q. Long Non-Coding RNA H19 Protects Acute Myocardial Infarction through Activating Autophagy in Mice. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5647–5651. [Google Scholar] [PubMed]
- Choong, O.K.; Chen, C.-Y.; Zhang, J.; Lin, J.-H.; Lin, P.-J.; Ruan, S.-C.; Kamp, T.J.; Hsieh, P.C. Hypoxia-Induced H19/YB-1 Cascade Modulates Cardiac Remodeling after Infarction. Theranostics 2019, 9, 6550–6567. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-M.; Li, X.-M.; Song, N.; Zhai, H.; Gao, X.-M.; Yang, Y.-N. Long Non-Coding RNAs H19, MALAT1 and MIAT as Potential Novel Biomarkers for Diagnosis of Acute Myocardial Infarction. Biomed. Pharmacother. 2019, 118, 109208. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Xiong, G.; Jiang, X.; Song, T. The Overexpression of lncRNA H19 as a Diagnostic Marker for Coronary Artery Disease. Rev. Assoc. Med. Bras. 2019, 65, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Zhu, M.; Wang, H.; Zhao, S.; Zhao, D.; Yang, Y.; Wang, Z.-M.; Wang, F.; Yang, Z.-J.; Lu, X.; et al. Association of Polymorphisms in Long Non-Coding RNA H19 with Coronary Artery Disease Risk in a Chinese Population. Mutat. Res. Mol. Mech. Mutagen. 2015, 772, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; An, X.; Li, Z.; Song, Y.; Li, L.; Zuo, S.; Liu, N.; Yang, G.; Wang, H.; Cheng, X.; et al. The H19 Long Noncoding RNA Is a Novel Negative Regulator of Cardiomyocyte Hypertrophy. Cardiovasc. Res. 2016, 111, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Boeckel, J.-N.; Perret, M.F.; Glaser, S.F.; Seeger, T.; Heumüller, A.W.; Chen, W.; John, D.; Kokot, K.E.; Katus, H.A.; Haas, J.; et al. Identification and Regulation of the Long Non-Coding RNA Heat2 in Heart Failure. J. Mol. Cell. Cardiol. 2019, 126, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Liu, Y.; Guo, S.; Yao, R.; Wu, L.; Xiao, L.; Wang, Z.; Liu, Y.; Zhang, Y. Circulating Long Noncoding RNA HOTAIR is an Essential Mediator of Acute Myocardial Infarction. Cell. Physiol. Biochem. 2017, 44, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.; Chen, R.; Lin, F.; Mai, A.; Chen, J.; Li, H.; Xu, Z.; Dong, S. Long Noncoding RNA HOTTIP Promotes Endothelial Cell Proliferation and Migration via Activation of the Wnt/β-Catenin Pathway. J. Cell. Biochem. 2017, 119, 2797–2805. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Creemers, E.E.; Boon, R.A.; Werfel, S.; Thum, T.; Engelhardt, S.; Dimmeler, S.; Squire, I. Circular RNAs in Heart Failure. Eur. J. Hear. Fail. 2017, 19, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; de Groote, P.; Pinet, F.; Thum, T. Circulating Long Noncoding RNA, LIPCAR, Predicts Survival in Patients with Heart Failure. Circ. Res. 2014, 114, 1569–1575. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, W.; Long, Q.-Q.; Zhang, J.; Lian-Sheng, W.; Liu, D.-C.; Yan, J.-J.; Yang, Z.-J.; Wang, L.-S. Increased Plasma Levels of lncRNA H19 and LIPCAR Are Associated with Increased Risk of Coronary Artery Disease in a Chinese Population. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.-S.; Cheng, J.; Cai, M.-Y.; Yang, X.-L.; Liu, X.G.; Zheng, B.-Y.; Xiong, X.-D. Association of lincRNA-p21Haplotype with Coronary Artery Disease in a Chinese Han Population. Dis. Markers 2016, 2016, 1–7. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Z.; Zhang, W.; Wang, Y. Long Noncoding RNA LINC00968 Promotes Endothelial Cell Proliferation and Migration via Regulating miR-9-3p Expression. J. Cell. Biochem. 2019, 120, 8214–8221. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhou, X.; Huang, J. Long Non-Coding RNA-ROR Mediates the Reprogramming in Cardiac Hypertrophy. PLoS ONE 2016, 11, e0152767. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.; Trac, C.; Jin, W.; Lanting, L.; Akbany, A.; Sætrom, P.; Schones, D.E.; Natarajan, R. Novel Long Noncoding RNAs Are Regulated by Angiotensin II in Vascular Smooth Muscle Cells. Circ. Res. 2013, 113, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, S.; Song, F.; Zhou, Y.; He, X. Lnc-Ang362 Is a Pro-Fibrotic Long Non-Coding RNA Promoting Cardiac Fibrosis after Myocardial Infarction by Suppressing Smad7. Arch. Biochem. Biophys. 2020, 685, 108354. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wu, X.; Han, Y.; Tian, E.; Cheng, J. LncRNA MALAT1 Protects Cardiomyocytes from Isoproterenol-Induced Apoptosis through Sponging miR-558 to Enhance ULK1-Mediated Protective Autophagy. J. Cell. Physiol. 2018, 234, 10842–10854. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Xu, Y.-N.; Wang, Q.; Liu, M.-J.; Zhang, P.; Zhao, L.-T.; Liu, F.; Zhao, D.-Y.; Pei, H.-N.; Yao, X.-B.; et al. Aerobic Exercise Improves Cardiac Function in Rats with Chronic Heart Failure through Inhibition of the Long Non-Coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1). Ann. Transl. Med. 2021, 9, 340. [Google Scholar] [CrossRef] [PubMed]
- Toraih, E.; El-Wazir, A.; Alghamdi, S.A.; Alhazmi, A.S.; El-Wazir, M.; Abdel-Daim, M.; Fawzy, M.S. Association of Long Non-Coding RNA MIAT and MALAT1 Expression Profiles in Peripheral Blood of Coronary Artery Disease Patients with Previous Cardiac Events. Genet. Mol. Biol. 2019, 42, 509–518. [Google Scholar] [CrossRef]
- Wang, K.; Sun, T.; Li, N.; Wang, Y.; Wang, J.; Zhou, L.-Y.; Long, B.; Liu, C.-Y.; Liu, F.; Li, P.-F. MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361. PLoS Genet. 2014, 10, e1004467. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhao, Z.-A.; Liu, J.; Hao, K.; Yu, Y.; Han, X.; Li, J.; Wang, Y.; Lei, W.; Dong, N.; et al. Long Noncoding RNA Meg3 Regulates Cardiomyocyte Apoptosis in Myocardial Infarction. Gene Ther. 2018, 25, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, M.-T.; Gupta, S.K.; Viereck, J.; Foinquinos, A.; Samolovac, S.; Kramer, F.L.; Garg, A.; Remke, J.; Zimmer, K.; Batkai, S.; et al. Inhibition of the Cardiac Fibroblast–Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction. Circ. Res. 2017, 121, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, C.; Meng, M.; Tang, H. Long Noncoding RNA MHRT Protects Cardiomyocytes against H2O2-Induced Apoptosis. Biomol. Ther. 2016, 24, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Li, W.; Lin, C.-H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.-Y.; Lin, C.-J.; et al. A Long Noncoding Rna Protects the Heart from Pathological Hypertrophy. Nat. Cell Biol. 2014, 514, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Xuan, L.; Sun, L.; Zhang, Y.; Huang, Y.; Hou, Y.; Li, Q.; Guo, Y.; Feng, B.; Cui, L.; Wang, X.; et al. Circulating Long Non-Coding Rnas Nron and MHRT as Novel Predictive Biomarkers of Heart Failure. J. Cell. Mol. Med. 2017, 21, 1803–1814. [Google Scholar] [CrossRef]
- Qu, X.; Du, Y.; Shu, Y.; Gao, M.; Sun, F.; Luo, S.; Yang, T.; Zhan, L.; Yuan, Y.; Chu, W.; et al. MIAT Is a Pro-Fibrotic Long Non-Coding RNA Governing Cardiac Fibrosis in Post-Infarct Myocardium. Sci. Rep. 2017, 7, srep42657. [Google Scholar] [CrossRef]
- Zhu, X.-H.; Yuan, Y.-X.; Rao, S.-L.; Wang, P. LncRNA MIAT Enhances Cardiac Hypertrophy Partly through Sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3653. [Google Scholar]
- Zangrando, J.; Zhang, L.; Vausort, M.; Maskali, F.; Marie, P.-Y.; Wagner, D.R.; Devaux, Y. Identification of Candidate Long Non-Coding RNAs in Response to Myocardial Infarction. BMC Genom. 2014, 15, 460. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yong, C.; Yu, K.; Yu, R.; Zhang, R.; Yu, L.; Li, S.; Cai, S. Long Noncoding RNA (lncRNA) n379519 Promotes Cardiac Fibrosis in Post-Infarct Myocardium by Targeting miR-30. Med. Sci. Monit. 2018, 24, 3958–3965. [Google Scholar] [CrossRef]
- Hu, Y.-W.; Guo, F.-X.; Xu, Y.-J.; Li, P.; Lu, Z.-F.; McVey, D.G.; Zheng, L.; Wang, Q.; Ye, J.; Kang, C.-M.; et al. Long Noncoding RNA NEXN-AS1 Mitigates Atherosclerosis by Regulating the Actin-Binding Protein NEXN. J. Clin. Investig. 2019, 129, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Tu, G.; Zou, L.; Liu, S.; Wu, B.; Lv, Q.; Wang, S.; Xue, Y.; Zhang, C.; Yi, Z.; Zhang, X.; et al. Long Noncoding NONRATT021972 siRNA Normalized Abnormal Sympathetic Activity Mediated by the Upregulation of P2X7 Receptor in Superior Cervical Ganglia after Myocardial Ischemia. Purinergic Signal. 2016, 12, 521–535. [Google Scholar] [CrossRef]
- Chen, S.; Chen, R.; Zhang, T.; Lin, S.; Chen, Z.; Zhao, B.; Li, H.; Wu, S. Relationship of Cardiovascular Disease Risk Factors and Noncoding RNAs with Hypertension: A Case-Control Study. BMC Cardiovasc. Disord. 2018, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Liu, C.-Y.; An, T.; Zhang, J.; Zhou, L.-Y.; Wang, M.; Dong, Y.-H.; Li, N.; Gao, J.-N.; et al. The Long Noncoding RNA NRF Regulates Programmed Necrosis and Myocardial Injury during Ischemia and Reperfusion by Targeting miR-873. Cell Death Differ. 2016, 23, 1394–1405. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhuang, Y.; Zhu, H.; Wu, H.; Li, D.; Zhan, L.; Yang, W.; Yuan, Y.; Xie, Y.; Yang, S.; et al. LncRNA PCFL Promotes Cardiac Fibrosis via miR-378/GRB2 Pathway Following Myocardial Infarction. J. Mol. Cell. Cardiol. 2019, 133, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Ballantyne, M.D.; Pinel, K.; Dakin, R.S.; Vesey, A.T.; Diver, L.; MacKenzie, R.M.; Garcia, R.; Welsh, P.; Sattar, N.A.; Hamilton, G.; et al. Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation. Circulation 2016, 133, 2050–2065. [Google Scholar] [CrossRef]
- Huang, S.; Tao, W.; Guo, Z.; Cao, J.; Huang, X. Suppression of Long Noncoding RNA TTTY15 Attenuates Hypoxia-Induced Cardiomyocytes Injury by Targeting miR-455-5p. Gene 2019, 701, 1–8. [Google Scholar] [CrossRef]
- Chen, J.; Hu, Q.; Zhang, B.-F.; Liu, X.-P.; Yang, S.; Jiang, H. Long Noncoding RNA UCA1 Inhibits Ischaemia/Reperfusion Injury Induced Cardiomyocytes Apoptosis via Suppression of Endoplasmic Reticulum Stress. Genes Genom. 2019, 41, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, B.; Liu, N.; Qi, C.; Xiao, Y.; Tian, X.; Li, T.; Liu, B. Circulating Long Noncoding RNA UCA1 as a Novel Biomarker of Acute Myocardial Infarction. BioMed Res. Int. 2016, 2016, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, L.; Xu, Y.; Liu, Y.; Li, Z.; Xue, X.; Wan, S.; Wang, H. Long Noncoding RNA Upregulated in Hypothermia Treated Cardiomyocytes Protects against Myocardial Infarction through Improving Mitochondrial Function. Int. J. Cardiol. 2018, 266, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.-C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The Long Noncoding RNA Wisper Controls Cardiac Fibrosis and Remodeling. Sci. Transl. Med. 2017, 9, eaai9118. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Li, M.; Shao, Y.; Zhang, Y.; Gong, M.; Yang, X.; Wang, Y.; Tan, Z.; Sun, L.; Xuan, L.; et al. lncRNA-ZFAS1 Induces Mitochondria-Mediated Apoptosis by Causing Cytosolic Ca2+ Overload in Myocardial Infarction Mice Model. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Fang, Y.; Xu, Y.; Wang, R.; Hu, L.; Guo, D.; Xue, F.; Guo, W.; Zhang, D.; Hu, J.; Li, Y.; et al. Recent Advances on the Roles of LncRNAs in Cardiovascular Disease. J. Cell. Mol. Med. 2020, 24, 12246–12257. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.; Binder, P.; Chen, H.; Wang, X. Regulation of Long Non-Coding RNAs and MicroRNAs in Heart Disease: Insight into Mechanisms and Therapeutic Approaches. Front. Physiol. 2020, 11, 798. [Google Scholar] [CrossRef] [PubMed]
- Brozovich, F.V.; Nicholson, C.J.; Degen, C.; Gao, Y.Z.; Aggarwal, M.; Morgan, K.G. Mechanisms of Vascular Smooth Muscle Contraction and the Basis for Pharmacologic Treatment of Smooth Muscle Disorders. Pharmacol. Rev. 2016, 68, 476–532. [Google Scholar] [CrossRef] [PubMed]
- Bátkai, S.; Thum, T. MicroRNAs in Hypertension: Mechanisms and Therapeutic Targets. Curr. Hypertens. Rep. 2012, 14, 79–87. [Google Scholar] [CrossRef]
- Jusic, A.; Devaux, Y. Noncoding RNAs in Hypertension. Hypertension 2019, 74, 477–492. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ning, Q. Long Noncoding RNAs as Novel Players in the Pathogenesis of Hypertension. Hypertens. Res. 2020, 43, 597–608. [Google Scholar] [CrossRef]
- Libby, P.; Buring, J.E.; Badimon, L.; Hansson, G.K.; Deanfield, J.; Bittencourt, M.S.; Tokgözoğlu, L.; Lewis, E.F. Atherosclerosis. Nat. Rev. Dis. Prim. 2019, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, L.; Li, H.; Han, X.; Chen, S.; Yang, B.; Hu, Z.; Zhu, H.; Cai, C.; Chen, J.; et al. Characterization of LncRNA Expression Profile and Identification of Novel LncRNA Biomarkers to Diagnose Coronary Artery Disease. Atherosclerosis 2018, 275, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Devlin, A.M.; Bottiglieri, T.; Domann, F.E.; Lentz, S.R. Tissue-specific Changes in H19 Methylation and Expression inMice with Hyperhomocysteinemia. J. Biol. Chem. 2005, 280, 25506–25511. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Cai, J.; Han, Y.; Chen, J.; Huang, Z.-P.; Chen, C.; Cai, Y.; Huang, H.; Yang, Y.; Liu, Y.; et al. LincRNA-p21 Regulates Neointima Formation, Vascular Smooth Muscle Cell Proliferation, Apoptosis, and Atherosclerosis by Enhancing p53 Activity. Circulation 2014, 130, 1452–1465. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Sun, L.; Zhu, S. LncRNA Myocardial Infarction-Associated Transcript (MIAT) Contributed to Cardiac Hypertrophy by Regulating TLR4 via miR-93. Eur. J. Pharmacol. 2018, 818, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, J.; Huang, K. Inhibition of the lncRNA Mirt1 Attenuates Acute Myocardial Infarction by Suppressing NF-κB Activation. Cell. Physiol. Biochem. 2017, 42, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiao, L.; Sun, L.-H.; Li, Y.; Gao, Y.; Xu, C.; Shao, Y.; Li, M.; Li, C.; Lu, Y.; et al. LncRNA ZFAS1 as a SERCA2a Inhibitor to Cause Intracellular Ca 2+ Overload and Contractile Dysfunction in a Mouse Model of Myocardial Infarction. Circ. Res. 2018, 122, 1354–1368. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, M.; Jessup, M.; Mullens, W.; Reza, N.; Shah, A.M.; Sliwa, K.; Mebazaa, A. Acute Heart Failure. Nat. Rev. Dis. Prim. 2020, 6, 1–15. [Google Scholar] [CrossRef]
- Malik, A.; Brito, D.; Chhabra, L. Congestive Heart Failure; StatPearls Publishing: Treausure Island, FL, USA, 2021. [Google Scholar]
- Gomes, C.P.C.; Schroen, B.; Kuster, G.M.; Robinson, E.L.; Ford, K.; Squire, I.B.; Heymans, S.; Martelli, F.; Emanueli, C.; Devaux, Y. Regulatory RNAs in Heart Failure. Circulation 2020, 141, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Haddad, F.; Bodell, P.W.; Qin, A.X.; Giger, J.M.; Baldwin, K.M. Role of Antisense RNA in Coordinating Cardiac Myosin Heavy Chain Gene Switching. J. Biol. Chem. 2003, 278, 37132–37138. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.; Minobe, W.; Bristow, M.R.; Leinwand, L.A. Myosin Heavy Chain Isoform Expression in the Failing and Nonfailing Human Heart. Circ. Res. 2000, 86, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-S.; Flammer, A.J.; Lerman, L.O.; Lerman, A. Personalized Medicine in Cardiovascular Diseases. Korean Circ. J. 2012, 42, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Califf, R.M. Future of Personalized Cardiovascular Medicine: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2018, 72, 3301–3309. [Google Scholar] [CrossRef] [PubMed]
- Kirchhof, P.; Sipido, K.R.; Cowie, M.R.; Eschenhagen, T.; Fox, K.A.A.; Katus, H.; Schroeder, S.; Schunkert, H.; Priori, S.G.; Alonso, A.; et al. The Continuum of Personalized Cardiovascular Medicine: A Position Paper of the European Society of Cardiology. Eur. Hear. J. 2014, 35, 3250–3257. [Google Scholar] [CrossRef] [PubMed]
- Expert Group. Biomarkers Biomarkers in Cardiology-Part 1-In Heart Failure and Specific Cardiomyopathies. Arq. Bras. Cardiol. 2014, 103, 451–459. [Google Scholar] [CrossRef]
- Bazzell, B.G.; Rainey, W.E.; Auchus, R.J.; Zocco, D.; Bruttini, M.; Hummel, S.L.; Byrd, J.B. Human Urinary mRNA as a Biomarker of Cardiovascular Disease. Circ. Genom. Precis. Med. 2018, 11, e002213. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Goretti, E.; Wagner, D.R.; Devaux, Y. Mirnas as Biomarkers of Myocardial Infarction: A Step Forward towards Personalized Medicine? Trends Mol. Med. 2014, 20, 716–725. [Google Scholar] [CrossRef] [PubMed]
- De Gonzalo-Calvo, D.; Kenneweg, F.; Bang, C.; Toro, R.; Van Der Meer, R.W.; Rijzewijk, L.J.; Smit, J.W.; Lamb, H.J.; Llorente-Cortes, V.; Thum, T. Circulating Long Non-Coding RNAs as Biomarkers of Left Ventricular Diastolic Function and Remodelling in Patients with Well-Controlled Type 2 Diabetes. Sci. Rep. 2016, 6, 37354. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.-X. LncRNA H19 Promotes Atherosclerosis by Regulating MAPK and NF-kB Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 322–328. [Google Scholar]
- Kitow, J.; Derda, A.A.; Beermann, J.; Kumarswarmy, R.; Pfanne, A.; Fendrich, J.; Lorenzen, J.M.; Xiao, K.; Bavendiek, U.; Bauersachs, J.; et al. Mitochondrial Long Noncoding RNAs as Blood Based Biomarkers for Cardiac Remodeling in Patients with Hypertrophic Cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Chen, G.; Yang, J.; Fan, X.; Gong, Y.; Xu, G.; Cui, Q.; Geng, B. Transcriptome Analysis Reveals Distinct Patterns of Long Noncoding RNAs in Heart and Plasma of Mice with Heart Failure. PLoS ONE 2013, 8, e77938. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in Extracellular Vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef] [PubMed]
- Viereck, J.; Thum, T. Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury. Circ. Res. 2017, 120, 381–399. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Z.; Chen, X.; Zhang, S. Long Non-Coding RNAs: From Disease Code to Drug Role. Acta Pharm. Sin. B 2020, 11, 340–354. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Jaé, N.; Holdt, L.; Dimmeler, S. Long Noncoding RNAs. J. Am. Coll. Cardiol. 2016, 67, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J. Targeting Noncoding Rnas in Disease. J. Clin. Investig. 2017, 127, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Lennox, K.A.; Behlke, M.A. Cellular Localization of Long Non-Coding RNAs Affects Silencing by RNAi More Than by Antisense Oligonucleotides. Nucleic Acids Res. 2015, 44, 863–877. [Google Scholar] [CrossRef]
- Zamore, P.D. RNA Interference: Listening to the Sound of Silence. Nat. Genet. 2001, 8, 746–750. [Google Scholar] [CrossRef]
- Kurreck, J. Design of Antisense Oligonucleotides Stabilized by Locked Nucleic Acids. Nucleic Acids Res. 2002, 30, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- Lennox, K.A.; Behlke, M.A. Tips for Successful lncRNA Knockdown Using Gapmers. Methods Mol. Biol. 2020, 2176, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.F.; Baker, B.F.; Pham, N.; Swayze, E.; Geary, R.S. Pharmacology of Antisense Drugs. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 81–105. [Google Scholar] [CrossRef] [PubMed]
- Prelich, G. Gene Overexpression: Uses, Mechanisms, and Interpretation. Genetics 2012, 190, 841–854. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-Based Genome-Scale Identification of Functional Long Noncoding RNA Loci in Human Cells. Science 2016, 355, eaah7111. [Google Scholar] [CrossRef] [PubMed]
- Aparicio-Prat, E.; Arnan, C.; Sala, I.; Bosch, N.; Guigó, R.; Johnson, R. DECKO: Single-Oligo, Dual-CRISPR Deletion of Genomic Elements Including Long Non-Coding RNAs. BMC Genom. 2015, 16, 846. [Google Scholar] [CrossRef]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-Coding RNAs in Cardiovascular Diseases: Diagnostic and Therapeutic Perspectives. Eur. Hear. J. 2017, 39, 2704–2716. [Google Scholar] [CrossRef]
- Tao, L.; Bei, Y.; Lin, S.; Zhang, H.; Zhou, Y.; Jiang, J.; Chen, P.; Shen, S.; Xiao, J.; Li, X. Exercise Training Protects against Acute Myocardial Infarction via Improving Myocardial Energy Metabolism and Mitochondrial Biogenesis. Cell. Physiol. Biochem. 2015, 37, 162–175. [Google Scholar] [CrossRef]
- Tian, D.; Meng, J. Exercise for Prevention and Relief of Cardiovascular Disease: Prognoses, Mechanisms, and Approaches. Oxidative Med. Cell. Longev. 2019, 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Sadoshima, J. Mechanisms of Physiological and Pathological Cardiac Hypertrophy. Nat. Rev. Cardiol. 2018, 15, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, J.; Gao, F. Exercise and Cardiovascular Protection; Springer: Singapore, 2020; pp. 205–216. [Google Scholar]
- Nystoriak, M.A.; Bhatnagar, A. Cardiovascular Effects and Benefits of Exercise. Front. Cardiovasc. Med. 2018, 5, 135. [Google Scholar] [CrossRef]
- Pinckard, K.; Baskin, K.K.; Stanford, K.I. Effects of Exercise to Improve Cardiovascular Health. Front. Cardiovasc. Med. 2019, 6, 69. [Google Scholar] [CrossRef]
- Schroeder, E.C.; Franke, W.D.; Sharp, R.L.; Lee, D.-C. Comparative Effectiveness of Aerobic, Resistance, and Combined Training on Cardiovascular Disease Risk Factors: A Randomized Controlled Trial. PLoS ONE 2019, 14, e0210292. [Google Scholar] [CrossRef]
- Marzolini, S.; Oh, P.I.; Brooks, D. Effect of Combined Aerobic and Resistance Training versus Aerobic Training Alone in Individuals with Coronary Artery Disease: A Meta-Analysis. Eur. J. Prev. Cardiol. 2012, 19, 81–94. [Google Scholar] [CrossRef]
- Weiner, R.B.; Baggish, A.L. Exercise-Induced Cardiac Remodeling. Prog. Cardiovasc. Dis. 2012, 54, 380–386. [Google Scholar] [CrossRef]
- Vega, R.B.; Konhilas, J.; Kelly, D.P.; Leinwand, L.A. Molecular Mechanisms Underlying Cardiac Adaptation to Exercise. Cell Metab. 2017, 25, 1012–1026. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, B.C.; Ooi, J.Y.Y.; Weeks, K.L.; Patterson, N.L.; McMullen, J.R. Understanding Key Mechanisms of Exercise-Induced Cardiac Protection to Mitigate Disease: Current Knowledge and Emerging Concepts. Physiol. Rev. 2018, 98, 419–475. [Google Scholar] [CrossRef] [PubMed]
- Fiuza-Luces, C.; Santos-Lozano, A.; Joyner, M.; Carrera-Bastos, P.; Picazo, O.; Zugaza, J.L.; Izquierdo, M.; Ruilope, L.M.; Lucia, A. Exercise Benefits in Cardiovascular Disease: Beyond Attenuation of Traditional Risk Factors. Nat. Rev. Cardiol. 2018, 15, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Moreira, J.B.N.; Wohlwend, M.; Wisløff, U. Exercise and Cardiac Health: Physiological and Molecular Insights. Nat. Metab. 2020, 2, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Whelton, S.P.; Chin, A.; Xin, X.; He, J. Effect of Aerobic Exercise on Blood Pressure. Ann. Int. Med. 2002, 136, 493–503. [Google Scholar] [CrossRef]
- Moraes-Silva, I.C.; Mostarda, C.T.; Silva-Filho, A.C.; Irigoyen, M.C. Hypertension and Exercise Training: Evidence from Clinical Studies. Adv. Exp. Med. Biol. 2017, 1000, 65–84. [Google Scholar] [CrossRef] [PubMed]
- Gielen, S.; Laughlin, M.H.; O’Conner, C.; Duncker, D.J. Exercise Training in Patients with Heart Disease: Review of Beneficial Effects and Clinical Recommendations. Prog. Cardiovasc. Dis. 2015, 57, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.S.; Brown, A.; Ebrahim, S.; Jolliffe, J.; Noorani, H.; Rees, K.; Skidmore, B.; Stone, J.A.; Thompson, D.; Oldridge, N. Exercise-Based Rehabilitation for Patients with Coronary Heart Disease: Systematic Review and Meta-Analysis of Randomized Controlled Trials. Am. J. Med. 2004, 116, 682–692. [Google Scholar] [CrossRef]
- Ribeiro, F.; Alves, A.J.; Teixeira, M.; Miranda, F.; Azevedo, C.; Duarte, J.A.; Oliveira, J. Exercise Training Enhances Autonomic Function after Acute Myocardial Infarction: A Randomized Controlled Study. Rev. Port. Cardiol. 2012, 31, 135–141. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, S.A.; Claudio, E.R.G.; Mengal, V.F.; De Oliveira, S.G.; Merlo, E.; Podratz, P.L.; Gouvêa, S.A.; Graceli, J.B.; De Abreu, G.R. Exercise Training Reduces Cardiac Dysfunction and Remodeling in Ovariectomized Rats Submitted to Myocardial Infarction. PLoS ONE 2014, 9, e115970. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Yang, S.-D.; Wang, M.-M.; Feng, Y.-S.; Dong, F.; Zhang, F. The Beneficial Role of Exercise Training for Myocardial Infarction Treatment in Elderly. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef]
- Moraes-Silva, I.C.; Rodrigues, B.; Coelho-Junior, H.J.; Feriani, D.J.; Irigoyen, M.-C. Myocardial Infarction and Exercise Training: Evidence from Basic Science. Adv. Exp. Med. Biol. 2017, 999, 139–153. [Google Scholar] [CrossRef]
- Hambrecht, R.; Gielen, S.; Linke, A.; Fiehn, E.; Yu, J.; Walther, C.; Schoene, N.; Schuler, G. Effects of Exercise Training on Left Ventricular Function and Peripheral Resistance in Patients with Chronic Heart Failure. JAMA 2000, 283, 3095–3101. [Google Scholar] [CrossRef]
- Belardinelli, R.; Georgiou, D.; Cianci, G.; Purcaro, A. Randomized, Controlled Trial of Long-Term Moderate Exercise Training in Chronic Heart Failure. Circulation 1999, 99, 1173–1182. [Google Scholar] [CrossRef]
- Coats, A.S.; Adamopoulos, S.; Radaelli, A.; McCance, A.; Meyer, T.E.; Bernardi, L.; Solda, P.L.; Davey, P.; Ormerod, O.; Forfar, C. Controlled Trial of Physical Training in Chronic Heart Failure. Exercise Performance, Hemodynamics, Ventilation, and Autonomic Function. Circulation 1992, 85, 2119–2131. [Google Scholar] [CrossRef] [PubMed]
- ExTraMATCH Collaborative. Exercise Training Meta-Analysis of Trials in Patients with Chronic Heart Failure. BMJ 2004, 328, 189. [Google Scholar] [CrossRef]
- Davies, E.J.; Moxham, T.; Rees, K.; Singh, S.; Coats, A.J.; Ebrahim, S.; Lough, F.; Taylor, R.S. Exercise Training for Systolic Heart Failure: Cochrane Systematic Review and Meta-Analysis. Eur. J. Hear. Fail. 2010, 12, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Van Tol, B.A.F.; Huijsmans, R.J.; Kroon, D.W.; Schothorst, M.; Kwakkel, G. Effects of Exercise Training on Cardiac Performance, Exercise Capacity and Quality of Life in Patients with Heart Failure: A Meta-Analysis. Eur. J. Hear. Fail. 2006, 8, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Haykowsky, M.J.; Liang, Y.; Pechter, D.; Jones, L.W.; McAlister, F.A.; Clark, A. A Meta-Analysis of the Effect of Exercise Training on Left Ventricular Remodeling in Heart Failure Patients: The Benefit Depends on the Type of Training Performed. J. Am. Coll. Cardiol. 2007, 49, 2329–2336. [Google Scholar] [CrossRef] [PubMed]
- Farsangi, S.J.; Rostamzadeh, F.; Sheikholeslami, M.; Jafari, E.; Karimzadeh, M.R. Modulation of the Expression of Long Non-Coding RNAs H19, GAS5, and MIAT by Endurance Exercise in the Hearts of Rats with Myocardial Infarction. Cardiovasc. Toxicol. 2020, 21, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Song, D.; Song, X.; Song, C. The Role of lncRNA MALAT1 in Cardiovascular Disease. IUBMB Life 2020, 72, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, L.; Batte, K.E.; Trgovcich, J.; Wisler, J.; Marsh, C.B.; Piper, M. Methodological Challenges in Utilizing Mi RNAs as Circulating Biomarkers. J. Cell. Mol. Med. 2014, 18, 371–390. [Google Scholar] [CrossRef]
- Congrains, A.; Kamide, K.; Oguro, R.; Yasuda, O.; Miyata, K.; Yamamoto, E.; Kawai, T.; Kusunoki, H.; Yamamoto, H.; Takeya, Y.; et al. Genetic Variants at the 9p21 Locus Contribute to Atherosclerosis through Modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012, 220, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Sazzini, M.; Garagnani, P.; Ferrari, A.; Boattini, A.; Lonetti, A.; Papayannidis, C.; Mantovani, V.; Marasco, E.; Ottaviani, E.; et al. A Polymorphism in the Chromosome 9p21 ANRIL Locus Is Associated to Philadelphia Positive Acute Lymphoblastic Leukemia. Leuk. Res. 2011, 35, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Diederichs, S. The Four Dimensions of Noncoding RNA Conservation. Trends Genet. 2014, 30, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Spitale, R.C.; Crisalli, P.; Flynn, R.A.; Torre, E.A.; Kool, E.T.; Chang, H.Y. RNA SHAPE Analysis in Living Cells. Nat. Chem. Biol. 2013, 9, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Tang, Y.; Kwok, C.K.; Zhang, Y.; Bevilacqua, P.C.; Assmann, S.M. In Vivo Genome-Wide Profiling of RNA Secondary Structure Reveals Novel Regulatory Features. Nature 2014, 505, 696–700. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Shah, R.; Dimmeler, S.; Freedman, J.E.; Holley, C.; Lee, J.-M.; Moore, K.; Musunuru, K.; Wang, D.-Z.; Xiao, J.; et al. Noncoding RNAs in Cardiovascular Disease: Current Knowledge, Tools and Technologies for Investigation, and Future Directions: A Scientific Statement from the American Heart Association. Circ. Genom. Precis. Med. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a Reference Set of Human Long Non-Coding RNAs. Nucleic Acids Res. 2019, 47, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A Tool for Predicting the RNA Targets of Long Noncoding RNAs. Brief. Bioinform. 2014, 16, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, A.; Zou, D.; Xu, X.; Xia, L.; Yu, J.; Bajic, V.B.; Zhang, Z. LncRNAWiki: Harnessing Community Knowledge in Collaborative Curation of Human Long Non-Coding RNAs. Nucleic Acids Res. 2014, 43, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Cao, J.; Liu, L.; Li, Z.; Shireen, H.; Pervaiz, N.; Batool, F.; Raza, R.Z.; Zou, D.; Bao, Y.; et al. Community Curation and Expert Curation of Human Long Noncoding RNAs with LncRNAWiki and LncBook. Curr. Protoc. Bioinform. 2019, 67, e82. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).