Differential Allelic Expression among Long Non-Coding RNAs
Abstract
:1. Introduction
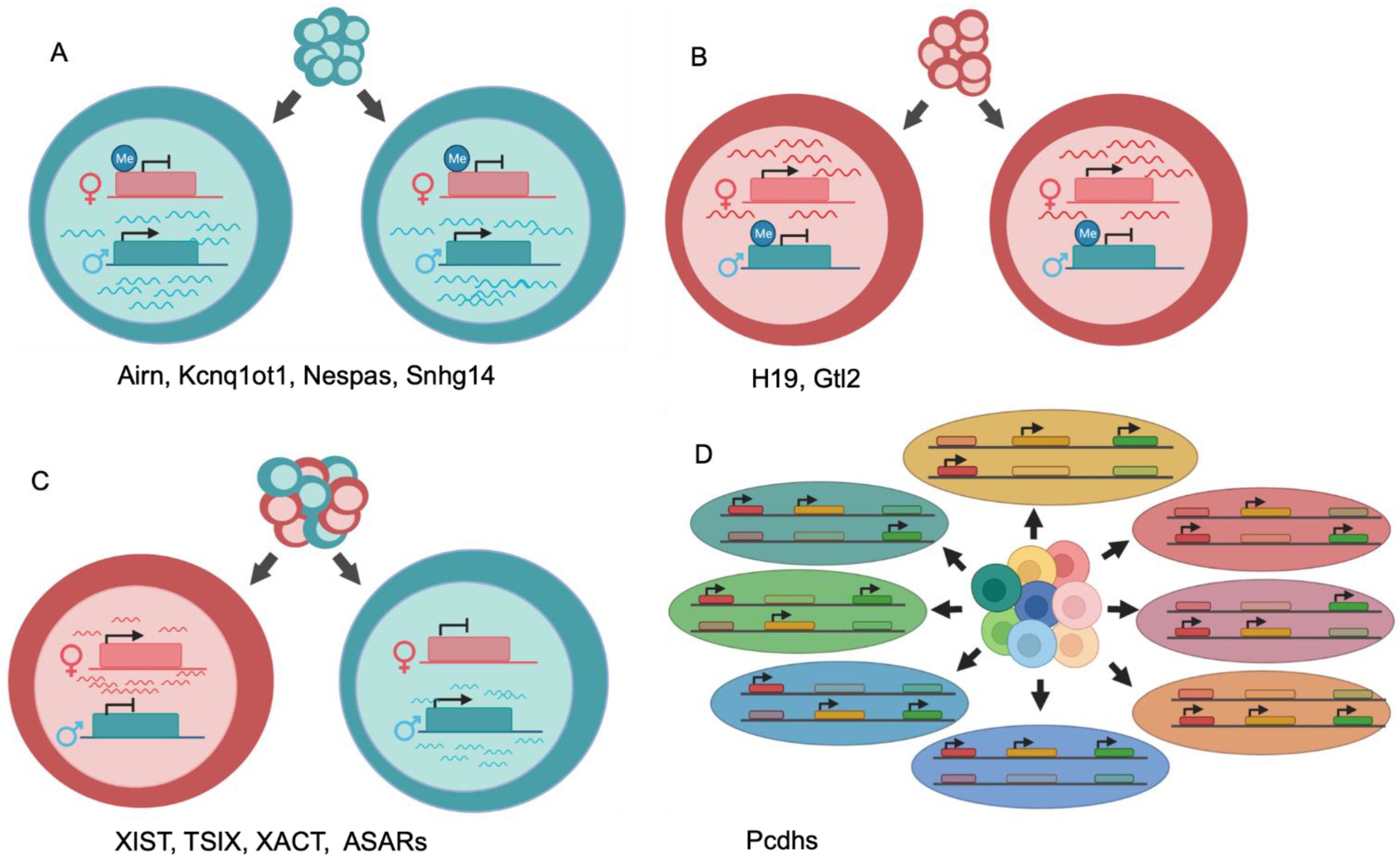
2. Differential Allelic Expression of lncRNA by Genomic Imprinting
3. Differential Allelic Expression of lncRNAs in X-Inactivation
4. Autosomal Differential Allelic Expression of Non-Canonical lncRNAs Is Required for Normal DNA Replication Timing and Chromosome Stability
5. Allele-Specific Mechanisms of Action of lncRNAs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Tay, S.-K.; Blythe, J.; Lipovich, L. Global discovery of primate-specific genes in the human genome. Proc. Natl. Acad. Sci. USA 2009, 106, 12019. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Li, Y.; Zhao, Z.; Lu, J.; Chen, H.; Ding, N.; Wang, G.; Xu, J.; Li, X. Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 2016, 7, 7120–7133. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef] [Green Version]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Reinius, B.; Sandberg, B. Random monoallelic expression of autosomal genes: Stochastic transcription and allele-level regulation. Nat. Rev. Genet. 2015, 16, 653–664. [Google Scholar] [CrossRef]
- Nica, A.C.; Dermitzakis, E.T. Expression quantitative trait loci: Present and future. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013, 368, 20120362. [Google Scholar] [CrossRef] [PubMed]
- de Goede, O.M.; Nachun, D.C.; Ferraro, N.M.; Gloudemans, M.J.; Rao, A.S.; Smail, C.; Eulalio, T.Y.; Aguet, F.; Ng, B.; Xu, J.; et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell 2021, 184, 2633–2648.e19. [Google Scholar] [CrossRef] [PubMed]
- Pernis, B.; Chiappino, G.; Kelus, A.S.; Gell, P.G. Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J. Exp. Med. 1965, 122, 853–876. [Google Scholar] [CrossRef] [PubMed]
- Mostoslavsky, R.; Alt, F.W.; Rajewsky, K. The lingering enigma of the allelic exclusion mechanism. Cell 2004, 118, 539–544. [Google Scholar] [CrossRef] [Green Version]
- Chess, A.; Simon, I.; Cedar, H.; Axel, R. Allelic inactivation regulates olfactory receptor gene expression. Cell 1994, 78, 823–834. [Google Scholar] [CrossRef]
- Ercan, S. Mechanisms of x chromosome dosage compensation. J. Genom. 2015, 3, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Kalsner, L.; Chamberlain, S.J. Prader-Willi, Angelman, and 15q11-q13 Duplication Syndromes. Pediatr. Clin. N. Am. 2015, 62, 587–606. [Google Scholar] [CrossRef] [Green Version]
- Li, S.M.; Valo, Z.; Wang, J.; Gao, H.; Bowers, C.W.; Singer-Sam, J. Transcriptome-wide survey of mouse CNS-derived cells reveals monoallelic expression within novel gene families. PLoS ONE 2012, 7, e31751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gendrel, A.V.; Attia, M.; Chen, C.J.; Diabangouaya, P.; Servant, N.; Barillot, E.; Heard, E. Developmental dynamics and disease potential of random monoallelic gene expression. Dev. Cell 2014, 28, 366–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagi, T. Genetic basis of neuronal individuality in the mammalian brain. J. Neurogenet. 2013, 27, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagi, T. Molecular codes for neuronal individuality and cell assembly in the brain. Front. Mol. Neurosci. 2012, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Zemel, S.; Bartolomei, M.S.; Tilghman, S.M. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat. Genet. 1992, 2, 61–65. [Google Scholar] [CrossRef]
- Zhang, Y.; Tycko, B. Monoallelic expression of the human H19 gene. Nat. Genet. 1992, 1, 40–44. [Google Scholar] [CrossRef]
- MacDonald, W.A.; Mann, M.R.W. Long noncoding RNA functionality in imprinted domain regulation. PLoS Genet. 2020, 16, e1008930. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.J.; Oakey, R.J. Genomic imprinting in mammals: Emerging themes and established theories. PLoS Genet. 2006, 2, e147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lottin, S.; Adriaenssens, E.; Dupressoir, T.; Berteaux, N.; Montpellier, C.; Coll, J.; Dugimont, T.; Curgy, J.J. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 2002, 23, 1885–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elias-Rizk, T.; El Hajj, J.; Segal-Bendirdjian, E.; Hilal, G. The long non coding RNA H19 as a biomarker for breast cancer diagnosis in Lebanese women. Sci. Rep. 2020, 10, 22228. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yu, B.; Li, J.; Su, L.; Yan, M.; Zhu, Z.; Liu, B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014, 5, 2318–2329. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Qin, Y.; Lv, J.; Wang, Y.; Che, H.; Chen, X.; Jiang, Y.; Li, A.; Sun, X.; Yue, E.; et al. Silencing long non-coding RNA Kcnq1ot1 alleviates pyroptosis and fibrosis in diabetic cardiomyopathy. Cell Death Dis. 2018, 9, 1000. [Google Scholar] [CrossRef]
- Shen, Y.; Xu, J.; Pan, X.; Zhang, Y.; Weng, Y.; Zhou, D.; He, S. LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis. 2020, 11, 278. [Google Scholar] [CrossRef]
- Haig, D. The Kinship Theory of Genomic Imprinting. Annu. Rev. Ecol. Syst. 2000, 31, 9–32. [Google Scholar] [CrossRef] [Green Version]
- Mozaffari, S.V.; DeCara, J.M.; Shah, S.J.; Sidore, C.; Fiorillo, E.; Cucca, F.; Lang, R.M.; Nicolae, D.L.; Ober, C. Parent-of-origin effects on quantitative phenotypes in a large Hutterite pedigree. Commun. Biol. 2019, 2, 28. [Google Scholar] [CrossRef] [Green Version]
- Loda, A.; Heard, E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019, 15, e1008333. [Google Scholar] [CrossRef] [Green Version]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef] [Green Version]
- Rodermund, L.; Coker, H.; Oldenkamp, R.; Wei, G.; Bowness, J.; Rajkumar, B.; Nesterova, T.; Pinto, D.M.S.; Schermelleh, L.; Brockdorff, N. Time-resolved structured illumination microscopy reveals key principles of Xist RNA spreading. Science 2021, 372, 6547. [Google Scholar] [CrossRef] [PubMed]
- Augui, S.; Nora, E.; Heard, E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011, 12, 429–442. [Google Scholar] [CrossRef]
- Galupa, R.; Nora, E.P.; Worsley-Hunt, R.; Picard, C.; Gard, C.; van Bemmel, J.G.; Servant, N.; Zhan, Y.; El Marjou, F.; Johanneau, C.; et al. A Conserved Noncoding Locus Regulates Random Monoallelic Xist Expression across a Topological Boundary. Mol. Cell 2020, 77, 352–367.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aeby, E.; Lee, H.G.; Lee, Y.W.; Kriz, A.; Del Rosario, B.C.; Oh, H.J.; Boukhali, M.; Haas, W.; Lee, J.T. Decapping enzyme 1A breaks X-chromosome symmetry by controlling Tsix elongation and RNA turnover. Nat. Cell Biol. 2020, 22, 1116–1129. [Google Scholar] [CrossRef]
- Ohhata, T.; Yamazawa, K.; Miura-Kamio, A.; Takahashi, S.; Sakai, S.; Tamura, Y.; Uchida, C.; Kitagawa, K.; Niida, H.; Hiratani, I.; et al. Dynamics of transcription-mediated conversion from euchromatin to facultative heterochromatin at the Xist promoter by Tsix. Cell Rep. 2021, 34, 108912. [Google Scholar] [CrossRef]
- Yang, F.; Deng, X.; Ma, W.; Berletch, J.B.; Rabaia, N.; Wei, G.; Moore, J.M.; Filippova, G.N.; Xu, J.; Liu, Y.; et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015, 16, 52. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Bonora, G.; Lewandowski, J.P.; Thakur, J.; Filippova, G.N.; Henikoff, S.; Shendure, J.; Duan, Z.; Rinn, J.L.; Deng, X.; et al. Trans- and cis-acting effects of Firre on epigenetic features of the inactive X chromosome. Nat. Commun. 2020, 11, 6053. [Google Scholar] [CrossRef] [PubMed]
- Donley, N.; Stoffregen, E.P.; Smith, L.; Montagna, C.; Thayer, M.J. Asynchronous replication, mono-allelic expression, and long range Cis-effects of ASAR6. PLoS Genet. 2013, 9, e1003423. [Google Scholar] [CrossRef] [Green Version]
- Donley, N.; Smith, L.; Thayer, M.J. ASAR15, A cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15. PLoS Genet. 2015, 11, e1004923. [Google Scholar] [CrossRef] [Green Version]
- Heskett, M.B.; Smith, L.G.; Spellman, P.; Thayer, M.J. Reciprocal monoallelic expression of ASAR lncRNA genes controls replication timing of human chromosome 6. RNA 2020, 26, 724–738. [Google Scholar] [CrossRef]
- Guenzl, P.M.; Barlow, D. Macro lncRNAs: A new layer of cis-regulatory information in the mammalian genome. RNA Biol. 2012, 9, 731–741. [Google Scholar] [CrossRef] [Green Version]
- St Laurent, G.; Shtokalo, D.; Dong, B.; Tackett, M.R.; Fan, X.; Lazorthes, S.; Nicolas, E.; Sang, N.; Triche, T.J.; McCaffrey, T.A.; et al. VlincRNAs controlled by retroviral elements are a hallmark of pluripotency and cancer. Genome Biol. 2013, 14, R73. [Google Scholar] [CrossRef] [Green Version]
- Kapranov, P.; St Laurent, G.; Raz, T.; Ozsolak, F.; Reynolds, C.P.; Sorensen, P.H.; Reaman, G.; Milos, P.; Arceci, R.J.; Thompson, J.F.; et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010, 8, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoffregen, E.P.; Donley, N.; Stauffer, D.; Smith, L.; Thayer, M.J. An autosomal locus that controls chromosome-wide replication timing and mono-allelic expression. Hum. Mol. Genet. 2011, 20, 2366–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, B.H.; Smith, L.; Huang, J.; Thayer, M. Chromosomes with delayed replication timing lead to checkpoint activation, delayed recruitment of Aurora B and chromosome instability. Oncogene 2007, 26, 1852–1861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breger, K.S.; Smith, L.; Thayer, M.J. Engineering translocations with delayed replication: Evidence for cis control of chromosome replication timing. Hum. Mol. Genet. 2005, 14, 2813–2827. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Perez, S.V.; Ferguson, D.O.; Wang, C.; Csankovszki, G.; Wang, C.; Tsai, S.C.; Dutta, D.; Perez, V.; Kim, S.; Eller, C.D.; et al. A deletion at the mouse Xist gene exposes trans-effects that alter the heterochromatin of the inactive X chromosome and the replication time and DNA stability of both X chromosomes. Genetics 2006, 174, 1115–1133. [Google Scholar] [CrossRef] [Green Version]
- Platt, E.J.; Smith, L.; Thayer, M.J. L1 retrotransposon antisense RNA within ASAR lncRNAs controls chromosome-wide replication timing. J. Cell Biol. 2018, 217, 541–553. [Google Scholar] [CrossRef] [Green Version]
- Smith, L.; Plug, A.; Thayer, M. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc. Natl. Acad. Sci. USA 2001, 98, 13300–13305. [Google Scholar] [CrossRef] [Green Version]
- Cortés-Ciriano, I.; Lee, J.J.K.; Xi, R.; Jain, D.; Jung, Y.L.; Yang, L.; Gordenin, D.; Klimczak, L.J.; Zhang, C.Z.; Pellman, D.S.; et al. Comprehensive analysis of chromothripsis in 2658 human cancers using whole-genome sequencing. Nat. Genet. 2020, 52, 331–341. [Google Scholar] [CrossRef] [Green Version]
- Forment, J.V.; Kaidi, A.; Jackson, S. Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat. Rev. Cancer 2012, 12, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Hall, L.L.; Carone, D.M.; Gomez, A.V.; Kolpa, H.J.; Byron, M.; Mehta, N.; Fackelmayer, F.O.; Lawrence, J.B. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell 2014, 156, 907–919. [Google Scholar] [CrossRef] [Green Version]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, C.; Zhang, Q.C.; Da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pullirsch, D.; Härtel, R.; Kishimoto, H.; Leeb, M.; Steiner, G.; Wutz, A. The Trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation. Development 2010, 137, 935–943. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Qu, L.; Gao, F.; Lin, J.; Liu, J.; Lin, A. LncRNAs: Architectural Scaffolds or More Potential Roles in Phase Separation. Front. Genet. 2021, 12, 369. [Google Scholar] [CrossRef]
- Strom, A.R.; Brangwynne, C. The liquid nucleome-phase transitions in the nucleus at a glance. J. Cell Sci. 2019, 132, jcs235093. [Google Scholar] [CrossRef] [Green Version]
- Pandya-Jones, A.; Markaki, Y.; Serizay, J.; Chitiashvili, T.; Leon, W.R.M.; Damianov, A.; Chronis, C.; Papp, B.; Chen, C.K.; McKee, R.; et al. A protein assembly mediates Xist localization and gene silencing. Nature 2020, 587, 145–151. [Google Scholar] [CrossRef]
- Tatavosian, R.; Kent, S.; Brown, K.; Yao, T.; Duc, H.N.; Huynh, T.N.; Zhen, C.Y.; Ma, B.; Wang, H.; Ren, X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019, 294, 1451–1463. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell 2018, 70, 1038–1053.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taiana, E.; Ronchetti, D.; Todoerti, K.; Nobili, L.; Tassone, P.; Amodio, N.; Neri, A. LncRNA NEAT1 in Paraspeckles: A Structural Scaffold for Cellular DNA Damage Response Systems? Non-Coding RNA 2020, 6, 26. [Google Scholar] [CrossRef]
- Lyon, M.F. X-chromosome inactivation: A repeat hypothesis. Cytogenet. Cell Genet. 1998, 80, 133–137. [Google Scholar] [CrossRef]
- Allen, E.; Horvath, S.; Tong, F.; Kraft, P.; Spiteri, E.; Riggs, A.D.; Marahrens, Y. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl. Acad. Sci. USA 2003, 100, 9940–9945. [Google Scholar] [CrossRef] [Green Version]
- Attig, J.; Agostini, F.; Gooding, C.; Chakrabarti, A.M.; Singh, A.; Haberman, N.; Zagalak, J.A.; Emmett, W.; Smith, C.W.; Luscombe, N.M.; et al. Heteromeric RNP Assembly at LINEs Controls Lineage-Specific RNA Processing. Cell 2018, 174, 1067–1081.e17. [Google Scholar] [CrossRef] [Green Version]
- Cavalli, M.; Baltzer, N.; Umer, H.M.; Grau, J.; Lemnian, I.; Pan, G.; Wallerman, O.; Spalinskas, R.; Sahlén, P.; Grosse, I.; et al. Allele specific chromatin signals, 3D interactions, and motif predictions for immune and B cell related diseases. Sci. Rep. 2019, 9, 2695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, H.; Zhou, Y.; Qiao, M.; Zhao, S.; Kozlova, A.; Shi, J.; Sanders, A.R.; Wang, G.; Luo, K.; et al. Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants. Science 2020, 369, 561. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heskett, M.B.; Spellman, P.T.; Thayer, M.J. Differential Allelic Expression among Long Non-Coding RNAs. Non-Coding RNA 2021, 7, 66. https://doi.org/10.3390/ncrna7040066
Heskett MB, Spellman PT, Thayer MJ. Differential Allelic Expression among Long Non-Coding RNAs. Non-Coding RNA. 2021; 7(4):66. https://doi.org/10.3390/ncrna7040066
Chicago/Turabian StyleHeskett, Michael B., Paul T. Spellman, and Mathew J. Thayer. 2021. "Differential Allelic Expression among Long Non-Coding RNAs" Non-Coding RNA 7, no. 4: 66. https://doi.org/10.3390/ncrna7040066
APA StyleHeskett, M. B., Spellman, P. T., & Thayer, M. J. (2021). Differential Allelic Expression among Long Non-Coding RNAs. Non-Coding RNA, 7(4), 66. https://doi.org/10.3390/ncrna7040066





