The Role of lncRNAs in Gene Expression Regulation through mRNA Stabilization
Abstract
1. Introduction
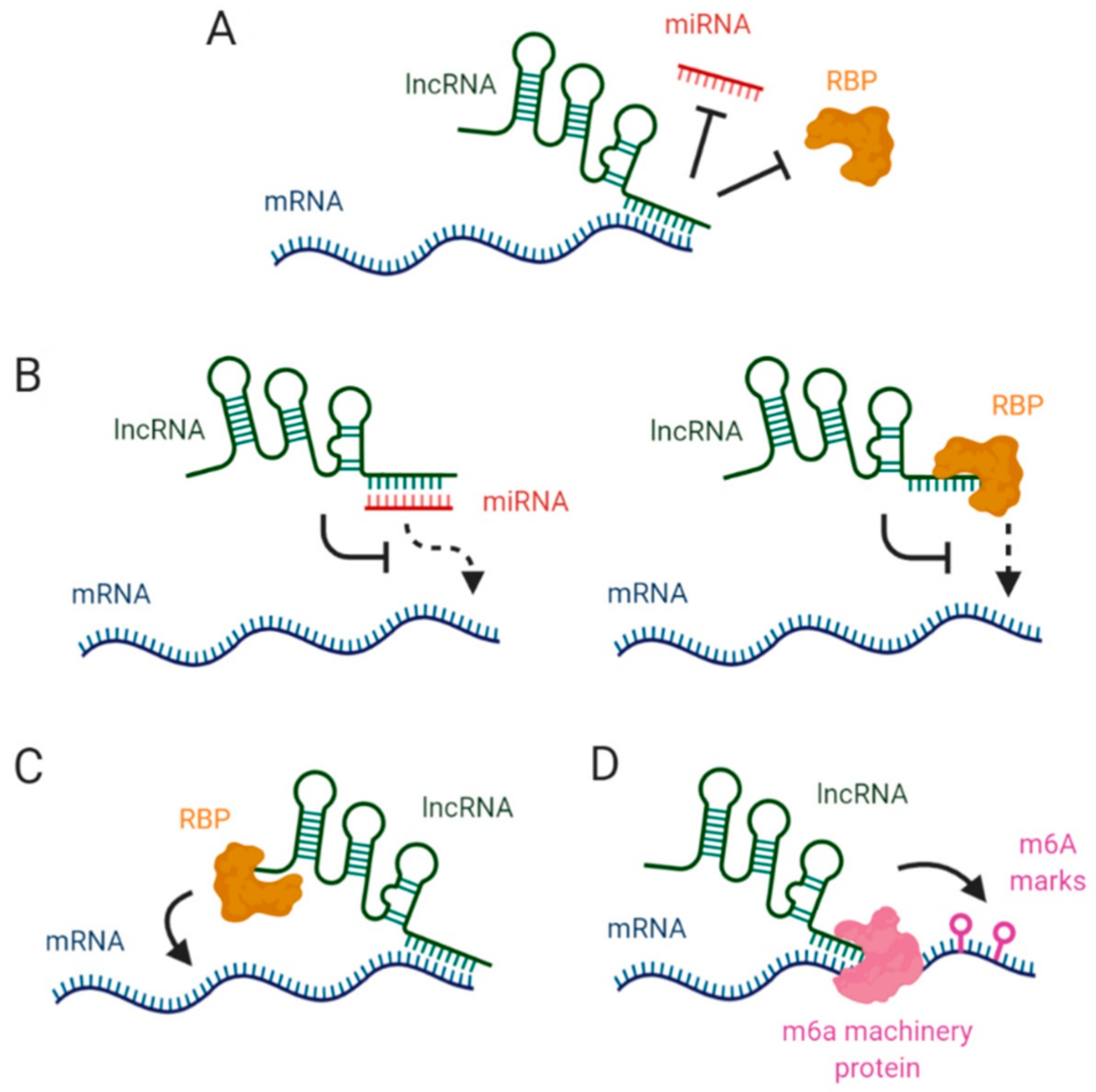
2. LncRNAs Affecting mRNA Stability via miRNA Blockage
3. Interaction between lncRNAs and RBPs in mRNA Stabilization
4. LncRNAs, Epitranscriptomic Changes and mRNA Stability
5. Concluding Remarks and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bicknell, A.A.; Ricci, E.P. When mRNA translation meets decay. Biochem. Soc. Trans. 2017, 45, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Laalami, S.; Zig, L.; Putzer, H. Initiation of mRNA decay in bacteria. Cell. Mol. Life Sci. 2014, 71, 1799–1828. [Google Scholar] [CrossRef] [PubMed]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [PubMed]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Ross, J. mRNA Stability in Mammalian Cells. Microbiol. Mol. Biol. Rev. 1995, 59, 423–450. [Google Scholar] [CrossRef]
- Pérez-Ortín, J.E.; Alepuz, P.; Chávez, S.; Choder, M. Eukaryotic mRNA decay: Methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 2013, 425, 3750–3775. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Green, R. Connections Underlying Translation and mRNA Stability. J. Mol. Biol. 2016, 428, 3558–3564. [Google Scholar] [CrossRef]
- Nilsen, T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007, 23, 243–249. [Google Scholar] [CrossRef]
- Eulalio, A.; Huntzinger, E.; Nishihara, T.; Rehwinkel, J.; Fauser, M.; Izaurralde, E. Deadenylation is a widespread effect of miRNA regulation. RNA 2009, 15, 21–32. [Google Scholar] [CrossRef]
- Karousis, E.D.; Mühlemann, O. Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb. Perspect. Biol. 2019, 11, a032862. [Google Scholar] [CrossRef]
- Schoenberg, D.R.; Maquat, L.E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012, 13, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Łabno, A.; Tomecki, R.; Dziembowski, A. Cytoplasmic RNA decay pathways—Enzymes and mechanisms. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 3125–3147. [Google Scholar] [CrossRef] [PubMed]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Jensen, T.H. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015, 16, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Moretti, F.; Kaiser, C.; Zdanowicz-Specht, A.; Hentze, M.W. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat. Struct. Mol. Biol. 2012, 19, 603–608. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef]
- Grudzien-Nogalska, E.; Kiledjian, M. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip. Rev. RNA. 2017, 8, e1379. [Google Scholar] [CrossRef]
- Kondo, Y.; Shinjo, K.; Katsushima, K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017, 108, 1927–1933. [Google Scholar] [CrossRef]
- Akhade, V.S.; Pal, D.; Kanduri, C. Long Noncoding RNA: Genome organization and mechanism of action. In Advances in Experimental Medicine and Biology; Springer New York LLC: New York, NY, USA, 2017; Volume 1008, pp. 47–74. [Google Scholar]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 19, 861–874. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, I.H.; Narra, H.P.; Sahni, A.; Khanipov, K.; Schroeder, C.L.C.; Patel, J.; Fofanov, Y.; Sahni, S.K. Expression Profiling of Long Noncoding RNA Splice Variants in Human Microvascular Endothelial Cells: Lipopolysaccharide Effects In Vitro. Mediat. Inflamm. 2017, 2017, 3427461. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long non-coding RNAs in the regulation of gene expression: Physiology and disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. MicroRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef]
- Fan, M.; Li, X.; Jiang, W.; Huang, Y.; Li, J.; Wang, Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp. Ther. Med. 2013, 5, 1143–1146. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.H.; Deng, Q.; Li, Y.; Huang, T.; Zhou, S.; Cai, Y.D. Cancer-Related Triplets of mRNA-lncRNA-miRNA Revealed by Integrative Network in Uterine Corpus Endometrial Carcinoma. BioMed Res. Int. 2017, 2017, 3859582. [Google Scholar] [CrossRef]
- Sun, J.; Wang, X.; Fu, C.; Wang, X.; Zou, J.; Hua, H.; Bi, Z. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol. Biol. Rep. 2016, 43, 427–436. [Google Scholar] [CrossRef]
- Valencia-Sanchez, M.A.; Liu, J.; Hannon, G.J.; Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006, 20, 515–524. [Google Scholar] [CrossRef]
- Chen, C.Y.; Zheng, D.; Xia, Z.; Shyu, A.B. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009, 16, 1160–1166. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, S.; Henriksson, S.; Corcoran, M.; Méndez-Vidal, C.; Wiman, K.G.; Farnebo, M. Wrap53, a Natural p53 Antisense Transcript Required for p53 Induction upon DNA Damage. Mol. Cell 2009, 33, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Su, W.Y.; Li, J.T.; Cui, Y.; Hong, J.; Du, W.; Wang, Y.C.; Lin, Y.W.; Xiong, H.; Wang, J.L.; Kong, X.; et al. Bidirectional regulation between WDR83 and its natural antisense transcript DHPS in gastric cancer. Cell Res. 2012, 22, 1374–1389. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; Yap, C.C.; Suzuki, M.; Kawai, J.; et al. Molecular biology: Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566. [Google Scholar] [PubMed]
- Nishizawa, M.; Ikeya, Y.; Okumura, T.; Kimura, T. Post-transcriptional inducible gene regulation by natural antisense RNA. Front. Biosci. 2015, 20, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Wanowska, E.; Kubiak, M.R.; Rosikiewicz, W.; Makałowska, I.; Szcześniak, M.W. Natural antisense transcripts in diseases: From modes of action to targeted therapies. Wiley Interdiscip. Rev. RNA 2018, 9, e1461. [Google Scholar] [CrossRef]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013, 14, 880–893. [Google Scholar] [CrossRef]
- Zinad, H.S.; Natasya, I.; Werner, A. Natural antisense transcripts at the interface between host genome and mobile genetic elements. Front. Microbiol. 2017, 8, 2292. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Roberson, E.D.; Mucke, L. 100 Years and counting: Prospects for defeating Alzheimer’s disease. Science 2006, 314, 781–784. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Zhang, M.; Huang, J.; Modarresi, F.; van der Brug, M.P.; Nalls, M.A.; Cookson, M.R.; St-Laurent, G.; Wahlestedt, C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010, 11, R56. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wei, Q.; Qi, Y.; Ruan, X.; Wu, F.; Li, L.; Zhou, J.; Liu, W.; Jiang, T.; Zhang, J.; et al. PTB-AS, a Novel Natural Antisense Transcript, Promotes Glioma Progression by Improving PTBP1 mRNA Stability with SND1. Mol. Ther. 2019, 27, 1621–1637. [Google Scholar] [CrossRef] [PubMed]
- Izaguirre, D.I.; Zhu, W.; Hai, T.; Cheung, H.C.; Krahe, R.; Cote, G.J. PTBP1-dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol. Carcinog. 2012, 51, 895–906. [Google Scholar] [CrossRef]
- Xue, Y.; Ouyang, K.; Huang, J.; Zhou, Y.; Ouyang, H.; Li, H.; Wang, G.; Wu, Q.; Wei, C.; Bi, Y.; et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated MicroRNA circuits. Cell 2013, 152, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Jing, Y.; Cao, Y. Overexpression of miR-100 inhibits growth of osteosarcoma through FGFR3. Tumor Biol. 2015, 36, 8405–8411. [Google Scholar] [CrossRef]
- Yuan, J.H.; Liu, X.N.; Wang, T.T.; Pan, W.; Tao, Q.F.; Zhou, W.P.; Wang, F.; Sun, S.H. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol. 2017, 19, 820–832. [Google Scholar] [CrossRef]
- Wang, G.Q.; Wang, Y.; Xiong, Y.; Chen, X.C.; Ma, M.L.; Cai, R.; Gao, Y.; Sun, Y.M.; Yang, G.S.; Pang, W.J. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci. Rep. 2016, 6, 21865. [Google Scholar] [CrossRef]
- Pardo, P.S.; Boriek, A.M. The physiological roles of Sirt1 in skeletal muscle. Aging 2011, 3, 430–437. [Google Scholar] [CrossRef]
- Barbagallo, C.; Brex, D.; Caponnetto, A.; Cirnigliaro, M.; Scalia, M.; Magnano, A.; Caltabiano, R.; Barbagallo, D.; Biondi, A.; Cappellani, A.; et al. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol. Ther. Nucleic Acids 2018, 12, 229–241. [Google Scholar] [CrossRef]
- Ulitsky, I.; Shkumatava, A.; Jan, C.H.; Sive, H.; Bartel, D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011, 147, 1537–1550. [Google Scholar] [CrossRef]
- Yang, J.; Jiang, B.; Hai, J.; Duan, S.; Dong, X.; Chen, C. Long noncoding RNA opa-interacting protein 5 antisense transcript 1 promotes proliferation and invasion through elevating integrin α6 expression by sponging miR-143-3p in cervical cancer. J. Cell. Biochem. 2019, 120, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xiong, D.; Yang, H.; Ye, L.; Mei, S.; Wu, J.; Chen, S.; Shang, X.; Wang, K.; Huang, L. Long noncoding RNA OPA-interacting protein 5 antisense transcript 1 upregulated SMAD3 expression to contribute to metastasis of cervical cancer by sponging miR-143-3p. J. Cell. Physiol. 2019, 234, 5264–5275. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, H.; Walther, W.; Zincke, F.; Kobelt, D.; Imbastari, F.; Erdem, M.; Kortüm, B.; Dahlmann, M.; Stein, U. MACC1—the first decade of a key metastasis molecule from gene discovery to clinical translation. Cancer Metastasis Rev. 2018, 37, 805–820. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Y.; Lin, L.; Huang, Q.; He, W.; Zhang, S.; Dong, S.; Wen, Z.; Rao, J.; Liao, W.; et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol. Cancer 2018, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, Y.; Chen, S.; Li, W.; Chen, W.; Gu, W. LncRNA MACC1-AS1 sponges multiple miRNAs and RNA-binding protein PTBP1. Oncogenesis 2019, 8, 73. [Google Scholar] [CrossRef]
- Johnsson, P.; Ackley, A.; Vidarsdottir, L.; Lui, W.O.; Corcoran, M.; Grandér, D.; Morris, K.V. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013, 20, 440–446. [Google Scholar] [CrossRef]
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultraconserved elements in the human genome. Science 2004, 304, 1321–1325. [Google Scholar] [CrossRef]
- Xiao, L.; Wu, J.; Wang, J.Y.; Chung, H.K.; Kalakonda, S.; Rao, J.N.; Gorospe, M.; Wang, J.Y. Long Noncoding RNA uc.173 Promotes Renewal of the Intestinal Mucosa by Inducing Degradation of MicroRNA 195. Gastroenterology 2018, 154, 599–611. [Google Scholar] [CrossRef]
- Franklin, J.L.; Rankin, C.R.; Levy, S.; Snoddy, J.R.; Zhang, B.; Washington, M.K.; Thomson, J.M.; Whitehead, R.H.; Coffey, R.J. Malignant transformation of colonic epithelial cells by a colon-derived long noncoding RNA. Biochem. Biophys. Res. Commun. 2013, 440, 99–104. [Google Scholar] [CrossRef]
- Chen, C.Y.; Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem Sci. 1995, 20, 465–470. [Google Scholar] [CrossRef]
- Vlasova, I.A.; Bohjanen, P.R. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008, 5, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Brewer, G. The regulation of mRNA stability in mammalian cells: 2.0. Gene 2012, 500, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef]
- Licatalosi, D.D.; Darnell, R.B. Resolving RNA complexity to decipher regulatory rules governing biological networks. Nat. Rev. Genet. 2010, 11, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Ferrè, F.; Colantoni, A.; Helmer-Citterich, M. Revealing protein-lncRNA interaction. Brief. Bioinform. 2016, 17, 106–116. [Google Scholar] [CrossRef]
- Zhu, J.J.; Fu, H.J.; Wu, Y.G.; Zheng, X.F. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013, 56, 876–885. [Google Scholar] [CrossRef]
- Wang, X.; Arai, S.; Song, X.; Reichart, D.; Du, K.; Pascual, G.; Tempst, P.; Rosenfeld, M.G.; Glass, C.K.; Kurokawa, R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008, 454, 126–130. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1471. [Google Scholar] [CrossRef]
- He, R.Z.; Luo, D.X.; Mo, Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019, 6, 6–15. [Google Scholar] [CrossRef]
- Briata, P.; Gherzi, R. Long Non-Coding RNA-Ribonucleoprotein Networks in the Post-Transcriptional Control of Gene Expression. Non-Coding RNA 2020, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Zhang, P.; Li, J.; Wu, M. Last, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 MRNA stability in human cells. eLife 2017, 6, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jiang, Y.; Xu, X.; Su, X.; Liu, Y.; Ma, Y.; Zhao, Y.; Shen, Z.; Huang, B.; Cao, X. Inducible degradation of lncRNA Sros1 promotes IFN-γ-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat. Immunol. 2019, 20, 1621–1630. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Panda, A.C.; Kang, M.J.; Guo, R.; Kim, J.; Grammatikakis, I.; Yoon, J.H.; Dudekula, D.B.; Noh, J.H.; Yang, X.; et al. NAR Breakthrough Article 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014, 42, 10099–10111. [Google Scholar] [CrossRef]
- Cmarik, J.L.; Min, H.; Hegamyer, G.; Zhan, S.; Kulesz-Martin, M.; Yoshinaga, H.; Matsuhashi, S.; Colburn, N.H. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc. Natl. Acad. Sci. USA 1999, 96, 14037–14042. [Google Scholar] [CrossRef]
- Jadaliha, M.; Gholamalamdari, O.; Tang, W.; Zhang, Y.; Petracovici, A.; Hao, Q.; Tariq, A.; Kim, T.G.; Holton, S.E.; Singh, D.K.; et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 2018, 14, e1007802. [Google Scholar] [CrossRef]
- Cammas, A.; Sanchez, B.J.; Lian, X.J.; Dormoy-Raclet, V.; van derGiessen, K.; de Silanes, I.L.; Ma, J.; Wilusz, C.; Richardson, J.; Gorospe, M.; et al. Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation. Nat. Commun. 2014, 5, 4190. [Google Scholar] [CrossRef]
- Zou, Z.; Ma, T.; He, X.; Zhou, J.; Ma, H.; Xie, M.; Liu, Y.; Lu, D.; Di, S.; Zhang, Z. Long intergenic non-coding RNA 00324 promotes gastric cancer cell proliferation via binding with HuR and stabilizing FAM83B expression article. Cell Death Dis. 2018, 9, 717. [Google Scholar] [CrossRef]
- Pan, L.; Li, Y.; Jin, L.; Li, J.; Xu, A. TRPM2-AS promotes cancer cell proliferation through control of TAF15. Int. J. Biochem. Cell Biol. 2020, 120, 105683. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Komiya, M.; Matsumura, K.; Negishi, L.; Suda, S.; Okuno, M.; Yokota, N.; Osada, T.; Nagashima, T.; Hiyoshi, M.; et al. MYU, a Target lncRNA for Wnt/c-Myc Signaling, Mediates Induction of CDK6 to Promote Cell Cycle Progression. Cell Rep. 2016, 16, 2554–2564. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, Z.; Trottier, J.; Barbier, O.; Wang, L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology 2017, 65, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, D.; Zhang, W.; Guo, M.; Zhan, Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012, 31, 4415–4427. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, X.; Xie, M.; Liu, M.; Ye, M.; Li, M.; Chen, X.-M.; Li, X.; Zhou, R. The NF-κB–Responsive Long Noncoding RNA FIRRE Regulates Posttranscriptional Regulation of Inflammatory Gene Expression through Interacting with hnRNPU. J. Immunol. 2017, 199, 3571–3582. [Google Scholar] [CrossRef]
- Giovarelli, M.; Bucci, G.; Ramos, A.; Bordo, D.; Wilusz, C.J.; Chen, C.Y.; Puppo, M.; Briata, P.; Gherzi, R. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc. Natl. Acad. Sci. USA 2014, 111, E5023–E5028. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef]
- Lan, Y.; Xiao, X.; He, Z.; Luo, Y.; Wu, C.; Li, L.; Song, X. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018, 46, 5809–5821. [Google Scholar] [CrossRef]
- Kim, J.; Abdelmohsen, K.; Yang, X.; De, S.; Grammatikakis, I.; Noh, J.H.; Gorospe, M. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016, 44, 2378–2392. [Google Scholar] [CrossRef]
- Gumireddy, K.; Li, A.; Yan, J.; Setoyama, T.; Johannes, G.J.; Ørom, U.A.; Tchou, J.; Liu, Q.; Zhang, L.; Speicher, D.W.; et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 2013, 32, 2672–2684. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 Suppresses Target mRNA Translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef]
- Kang, M.J.; Abdelmohsen, K.; Hutchison, E.R.; Mitchell, S.J.; Grammatikakis, I.; Guo, R.; Noh, J.H.; Martindale, J.L.; Yang, X.; Lee, E.K.; et al. HuD regulates coding and noncoding RNA to induce APP→Aβ processing. Cell Rep. 2014, 7, 1401–1409. [Google Scholar] [CrossRef]
- Kim, Y.K.; Furic, L.; Parisien, M.; Major, F.; DesGroseillers, L.; Maquat, L.E. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007, 26, 2670–2681. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. LncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 39 UTRs via Alu eleme. Nature 2011, 470, 284–290. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.P.; Liu, X.X.; Xia, R.; Yin, L.; Kong, R.; Chen, W.M.; Huang, M.D.; Shu, Y.Q. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene 2015, 34, 5648–5661. [Google Scholar] [CrossRef] [PubMed]
- Uchida, T.; Rossignol, F.; Matthay, M.A.; Mounier, R.; Couette, S.; Clottes, E.; Clerici, C. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1α and HIF-2α expression in lung epithelial cells: Implication of natural antisense HIF-1α. J. Biol. Chem. 2004, 279, 14871–14878. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, F.; Vaché, C.; Clottes, E. Natural antisense transcripts of hypoxia-inducible factor 1alpha are detected in different normal and tumour human tissues. Gene 2002, 299, 135–140. [Google Scholar] [CrossRef]
- Bhartiya, D.; Scaria, V. Genomic variations in non-coding RNAs: Structure, function and regulation. Genomics 2016, 107, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Ding, Y.; Bai, Y.; Li, J.; Zhang, G.; Wang, S.; Gao, W.; Xu, L.; Wang, H. Detection of Allosteric Effects of lncRNA Secondary Structures Altered by SNPs in Human Diseases. Front. Cell Dev. Biol. 2020, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Rubio, A.; Ghosh, S. Disease-associated SNPs in inflammation-related lncRNAs. Front. Immunol. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Ricaño-Ponce, I.; Zhernakova, D.v.; Deelen, P.; Luo, O.; Li, X.; Isaacs, A.; Karjalainen, J.; di Tommaso, J.; Borek, Z.A.; Zorro, M.M.; et al. Refined mapping of autoimmune disease associated genetic variants with gene expression suggests an important role for non-coding RNAs. J. Autoimmun. 2016, 68, 62–74. [Google Scholar] [CrossRef]
- Gonzalez-Moro, I.; Olazagoitia-Garmendia, A.; Colli, M.L.; Cobo-Vuilleumier, N.; Postler, T.S.; Marselli, L.; Marchetti, P.; Ghosh, S.; Gauthier, B.R.; Eizirik, D.L.; et al. The T1D-associated lncRNA Lnc13 modulates human pancreatic β cell inflammation by allele-specific stabilization of STAT1 mRNA. Proc. Natl. Acad. Sci. USA 2020, 117, 8661–8663. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Rubio, A.; Fernandez-Jimenez, N.; Kratchmarov, R.; Luo, X.; Bhagat, G.; Green, P.H.R.; Schneider, R.; Kiledjian, M.; Bilbao, J.R.; Ghosh, S. A long noncoding RNA associated with susceptibility to celiac disease. Science 2016, 352, 91–95. [Google Scholar] [CrossRef]
- De Beeck, A.O.; Eizirik, D.L. Viral infections in type 1 diabetes mellitus-why the β cells? Nat. Rev. Endocrinol. 2016, 12, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, A.; Ho, T.T.; Zhang, Z.; Zhou, N.; Ding, X.; Zhang, X.; Xu, M.; Mo, Y.Y. Linc-RoR promotes c-Myc expression through hnRNPi and AUF1. Nucleic Acids Res. 2015, 44, 3059–3069. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Bucci, G.; Rizzotto, D.; Bordo, D.; Marzi, M.J.; Puppo, M.; Flinois, A.; Spadaro, D.; Citi, S.; Emionite, L.; et al. LncRNA EPR controls epithelial proliferation by coordinating Cdkn1a transcription and mRNA decay response to TGF-β. Nat. Commun. 2019, 10, 1969. [Google Scholar] [CrossRef]
- Zapparoli, E.; Briata, P.; Rossi, M.; Brondolo, L.; Bucci, G.; Gherzi, R. Comprehensive multi-omics analysis uncovers a group of TGF-β-regulated genes among lncRNA EPR direct transcriptional targets. Nucleic Acids Res. 2020, 48, 9053–9066. [Google Scholar] [CrossRef]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N 6-methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation contributed reagents/analytic tools and/or grant support; HHS Public Access. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Wang, Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am. J. Transl. Res. 2019, 11, 4909–4921. [Google Scholar] [PubMed]
- Yan, J.; Huang, X.; Zhang, X.; Chen, Z.; Ye, C.; Xiang, W.; Huang, Z. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem. Biophys. Res. Commun. 2019, 521, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gu, Y.; Zhang, E.; Zhang, K.; Qin, N.; Dai, J.; Zhu, M.; Liu, J.; Xie, K.; Jiang, Y.; et al. A cancer-testis non-coding RNA LIN28B-AS1 activates driver gene LIN28B by interacting with IGF2BP1 in lung adenocarcinoma. Oncogene 2019, 38, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wu, Z.; Wang, X.; Wang, Y.; Hu, X.; Qin, W.; Lu, S.; Xu, D.; Wu, Y.; Chen, Q.; et al. LNC942 promoting METTL14-mediated m6A methylation in breast cancer cell proliferation and progression. Oncogene 2020, 39, 5358–5372. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sebastian-delaCruz, M.; Gonzalez-Moro, I.; Olazagoitia-Garmendia, A.; Castellanos-Rubio, A.; Santin, I. The Role of lncRNAs in Gene Expression Regulation through mRNA Stabilization. Non-Coding RNA 2021, 7, 3. https://doi.org/10.3390/ncrna7010003
Sebastian-delaCruz M, Gonzalez-Moro I, Olazagoitia-Garmendia A, Castellanos-Rubio A, Santin I. The Role of lncRNAs in Gene Expression Regulation through mRNA Stabilization. Non-Coding RNA. 2021; 7(1):3. https://doi.org/10.3390/ncrna7010003
Chicago/Turabian StyleSebastian-delaCruz, Maialen, Itziar Gonzalez-Moro, Ane Olazagoitia-Garmendia, Ainara Castellanos-Rubio, and Izortze Santin. 2021. "The Role of lncRNAs in Gene Expression Regulation through mRNA Stabilization" Non-Coding RNA 7, no. 1: 3. https://doi.org/10.3390/ncrna7010003
APA StyleSebastian-delaCruz, M., Gonzalez-Moro, I., Olazagoitia-Garmendia, A., Castellanos-Rubio, A., & Santin, I. (2021). The Role of lncRNAs in Gene Expression Regulation through mRNA Stabilization. Non-Coding RNA, 7(1), 3. https://doi.org/10.3390/ncrna7010003




