Abstract
There is increasing evidence that long non-coding RNAs (lncRNAs) are important for normal reproductive development, yet very few lncRNAs have been identified in phalluses so far. Unlike eutherians, phallus development in the marsupial tammar wallaby occurs post-natally, enabling manipulation not possible in eutherians in which differentiation occurs in utero. We treated with sex steroids to determine the effects of androgen and oestrogen on lncRNA expression during phallus development. Hormonal manipulations altered the coding and non-coding gene expression profile of phalluses. We identified several predicted co-regulatory lncRNAs that appear to be co-expressed with the hormone-responsive candidate genes regulating urethral closure and phallus growth, namely IGF1, AR and ESR1. Interestingly, more than 50% of AR-associated coding genes and lncRNAs were also associated with ESR1. In addition, we identified and validated three novel co-regulatory and hormone-responsive lncRNAs: lnc-BMP5, lnc-ZBTB16 and lncRSPO4. Lnc-BMP5 was detected in the urethral epithelium of male phalluses and was downregulated by oestrogen in males. Lnc-ZBTB16 was downregulated by oestrogen treatment in male phalluses at day 50 post-partum (pp). LncRSPO4 was downregulated by adiol treatment in female phalluses but increased in male phalluses after castration. Thus, the expression pattern and hormone responsiveness of these lncRNAs suggests a physiological role in the development of the phallus.
Keywords:
hypospadias; lncRNA; RNA-Seq; WGCNA; marsupial; androstanediol; phallus; androgen receptor; oestrogen receptor; castration 1. Introduction
The 20,000 protein coding genes only account for about 1% of the human genome [1,2]. However, non-protein coding RNAs (ncRNAs) comprise at least three times more DNA than protein coding RNAs in the human genome [3,4,5,6]. Many ncRNAs participate in regulating genome function [7] and gene regulation [8] during all biological processes including sex determination and differentiation [9,10]. Long non-coding RNAs (lncRNAs) form a major class of non-coding transcripts [4,11] characterised by a transcript longer than 200 nucleotides with either no significant open reading frames (ORF) or with ORF shorter than 30–60 amino acids [12]. However, little is known about the interactions between protein coding genes and lncRNAs due to the low conservation of lncRNA sequences, lack of understanding of their regulation or what they regulate. Even well-identified mouse lncRNAs are generally not sequence conserved, with only 14% of lncRNAs that have a conserved orthologue in the human genome [13,14].
Long ncRNAs play an important role in gene regulation during appendage development, sexual differentiation and reproductive development (reviewed in [9,10,15,16]). A recent study suggests that anti-sense lncRNAs have an important role in regulating appendage development in mouse [16]. Many lncRNAs in the reproductive tract are androgen or oestrogen sensitive. For instance, CTBP1-AS, a lncRNA that promotes prostate cancer, is androgen responsive [17], whereas NEAT1, a lncRNA that also promotes prostate cancer, is oestrogen responsive [18]. The transcriptional regulation of lncRNAs can be either cis or trans (reviewed in [19]) so their targets are not always easily defined.
Unlike eutherian mammals, in which sexual differentiation takes place in utero, sexual differentiation occurs largely post-natally in marsupials. For the phallus, the initiation of genital tubercle (GT) outgrowth in the tammar begins 2 days before birth in both sexes [20]. From the day of birth to day 50 post-partum (pp), there are no morphological differences between male and female phalluses [21]. After day 50 pp, the distance between the urethral meatus and the anus becomes greater in males than in females [20,21,22] and the male GT elongates [23]. By day 150 pp, urethral closure is complete, with the urethral meatus in the glans penis. In contrast, the urethral groove does not fuse in females, as seen in other mammals [21,23].
Phallus development in the male tammar is androgen dependent [24] and can be affected by sex hormones [23,24,25]. Interestingly, the earliest sexually dimorphic differences occur at a time when circulating androgen levels are similar in both males and females [21,23,26,27,28], suggesting that hormonal-responsive pathways are activated before phallus differentiation. This hormone-sensitive window (also known as the androgen imprinting window or male programming window) [23,27,29] was the first of several now described in eutherian mammals and occurs between day 25 to 30 pp [25]. Altering androgen levels during this window of sensitivity in the tammar wallaby significantly altered gene expression in multiple signalling pathways, including SHH signalling, IGF1 signalling and AP-1 signalling, and also leads to hypospadias or sex-reversed phalluses at later stages [23,24,30]. The postnatal phallus development and androgen imprinting mechanism in the tammar makes it possible to administer androgen or oestrogen directly to pouch young without interference from the maternal physiology or placental transfer.
It is clear that altering sex steroids during the androgen imprinting window changes multiple signalling pathways and causes hypospadias in the tammar. Although several studies suggest that hormone-responsive lncRNAs regulate reproductive tract development by affecting coding genes [10,15,17,18], there are few studies on the regulation of hormone-responsive lncRNAs during phallus development. In this study, we used the marsupial tammar wallaby Macropus eugenii as a research model to study the co-expression network between hormone-responsive coding genes and lncRNAs during phallus development. We investigated the regulation of multiple hormone-responsive lncRNAs that are co-expressed with coding genes in the developing tammar phallus. We treated female tammars with androgen and males with oestrogen or castration during the androgen imprinting window of sensitivity to identify hormone-responsive lncRNAs. We identified novel putative co-regulatory hormone-responsive lncRNAs of three genes regulating phallus development: insulin growth factor (IGF1), androgen receptor (AR) and estrogen receptor alpha (ESR1). IGF1, one of many hormone-responsive coding genes [31], may induce urethral closure by activating cell proliferation in the urorectal septum of male tammar phalluses and enhance growth [31]. In the mouse, the balance between AR and ESR1 activity is important to maintain normal penile development [32]. We used co-expression analysis to identify novel co-regulatory hormone-responsive lncRNAs during phallus development. Our data demonstrate that hormonal exposures have a significant impact on the network of coding and lncRNAs during phallus development, which extends our understanding on the aetiology of abnormal phallus development, including hypospadias.
2. Results
2.1. Principal Component Analysis
The 3-dimensional (3-D) principal component analysis (PCA) of RNA-Seq data separated the response of each treatment from the control groups (Figure 1). There was a sexually dimorphic expression pattern between day 50 pp male and female phalluses. In males treated with oestrogen, the phallus gene expression pattern diverged from that of normal day 50 pp males. Similarly, in females treated with adiol, the phallus expression pattern differed from normal day 50 pp female phalluses. Castration resulted in a pattern more like females than males. About 14% of these differentially expressed genes presented above were predicted lncRNAs (Table S1, see Supplementary Materials).
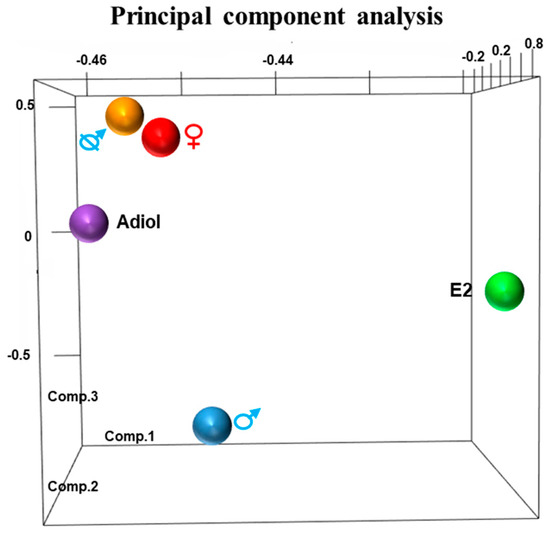
Figure 1.
Principal component analysis (PCA) demonstrates the variance of expression profile between treated phalluses and normal phalluses. Principal component 1–3 represent for 94.3%, 4.5% and 0.73% of the variance. Adiol and oestrogen treatment altered the gene expression pattern away from that in normal day 50 post-partum (pp) female and male phalluses, respectively. The gene expression pattern in oestrogen treatment group was different from that in adiol treatment group. Castration has mostly reversed the gene expression pattern in the phallus from males to females. E2: oestrogen.
2.2. Predicted Hormone-Responsive long non-coding RNAs
Co-expression networks of IGF1 (Figure 2), AR and ESR1 (Figure 3) were built to identify their potential co-regulatory lncRNAs. IGF1 was the hub gene in one of 16 modules produced by co-expression analysis. The expression correlation between identified lncRNAs and IGF1 was >0.8, and between identified lncRNAs and AR and ESR1 was >0.7. More than 50% of AR-associated coding genes and lncRNAs were also associated with ESR1 (Figure 3). However, none of these lncRNAs were located within 100 kilobases (kb) up-or down-stream of IGF1, AR and ESR1.
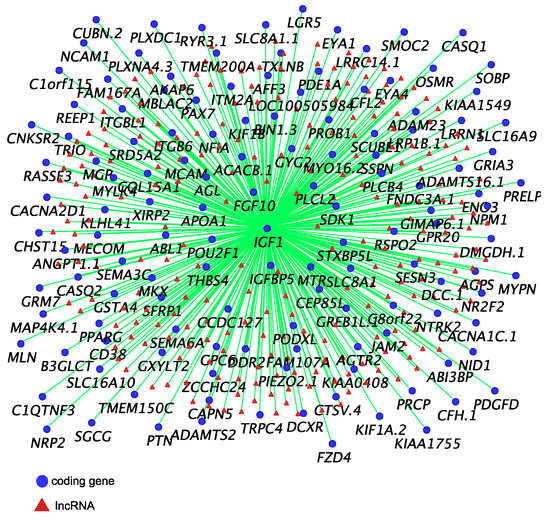
Figure 2.
IGF1 co-expressed coding genes and predicted co-regulatory long non-coding RNAs (lncRNAs) (R ≥ 0.8).
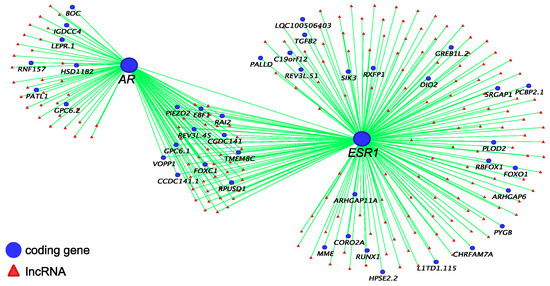
Figure 3.
AR and ESR1 co-expressed coding genes and predicted co-regulatory lncRNAs (R ≥ 0.7).
Three predicted co-regulatory lncRNAs, lnc-BMP5, lnc-ZNTB16 and lnc-RSPO4, were identified by mapping co-expressed coding genes and lncRNAs in the tammar genome. The expression correlation between lnc-BMP5, lnc-ZNTB16 and lnc-RSPO4 and their neighbouring coding genes, BMP5, ZBTB16, and RSPO4, respectively, was >0.7 and the correlation coefficient was significant between those coding genes and lncRNAs. Lnc-BMP5 was localized around 1 kb downstream of Bone Morphogenetic Protein 5 (BMP5) (Figure S1). It was detected in the urethral epithelium of male phalluses at day 20 pp and in the epithelium of the male phallus at day 90 pp, suggesting it may be involved in phallus development (Figure 4). BMP5 and lnc-BMP5 were downregulated by oestrogen in males (Figure 5). Lnc-ZBTB16 was within 1 kb downstream of zinc finger and BTB domain containing 16 (ZBTB16) (Figure S1). Both lnc-ZBTB16 and ZBTB16 had similar transcriptome and quantitative PCR (qPCR) expression patterns (Figure 6).
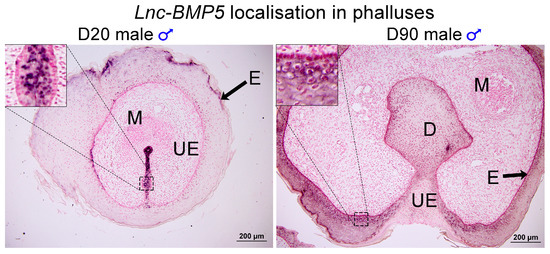
Figure 4.
Lnc-BMP5 mRNA localisation in day 20 pp and day 90 pp male phalluses. Lnc-BMP5 was detected in the nucleus of urethral epithelial cells of male phalluses at day 20 pp and in male phallus epithelium at 90 pp. D: diverticulum, E: epithelium, M: mesenchyme, UE: urethral epithelium. Scale bar: 200 µm.
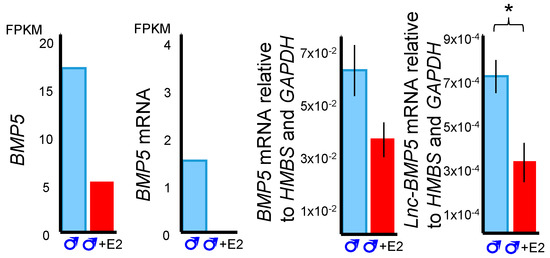
Figure 5.
BMP5 and lnc-BMP5 expression from RNA-Seq (left) and validated in quantitative PCR (qPCR) (right). BMP5 and lnc-BMP5 was downregulated by oestrogen in males. E2: oestrogen. FPKM: Fragments Per Kilobase of transcript per Million mapped reads. *: p < 0.05, ***: p < 0.001. Error bar: SEM.
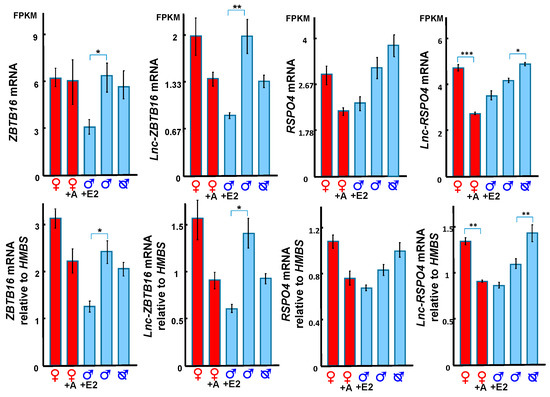
Figure 6.
ZBTB16, lnc-ZBTB16, RSPO4 and lnc-RSPO4 transcript expression from RNA-Seq (top) and gene expression validated by qPCR (bottom). ZBTB16 and lnc-ZBTB16 were both downregulated by oestrogen in males. Lnc-RSPO4 was downregulated in females by adiol treatment, but upregulated in males after castration. A: androgen, E2: oestrogen. FPKM: Fragments Per Kilobase of transcript per Million mapped reads. *: p < 0.05, **: p < 0.005, ***: p < 0.001. Error bar: SEM.
Lnc-ZBTB16 and ZBTB16 were significantly downregulated after oestrogen treatment (Figure 6). Lnc-RSPO4 was located within 1 kb upstream of R-Spondin 4 (RSPO4) (Figure S1). Lnc-RSPO4 and RSPO4 had similar qPCR and transcriptome expression patterns (Figure 6). Lnc-RSPO4 was significantly down-regulated after adiol treatment (Figure 6). Although RSPO4 expression was not as significantly changed as that of lnc-RSPO4, the expression correlation between RSPO4 and lnc-RSPO4 was significant.
3. Discussion
This study in the tammar suggests that phallus differentiation is under the control of complex network interactions between hormone-responsive coding genes and lncRNAs. Multiple predicted lncRNAs of important phallus regulating genes, including IGF1, AR and ESR1, were identified from co-expression analysis. We predicted and validated 3 novel co-regulatory lncRNAs (lnc-BMP5, lnc-ZBTB16 and lnc-RSPO4). Those lncRNAs were all hormone-responsive and were located near their co-expressed and hormone-responsive coding genes in the tammar genome.
3.1. A Global View of Differentially Expressed Genes and Phallus Phenotype
Three-dimensional PCA grouped the transcriptomic responses to treatments by phenotype showing a similar relationship to the morphological changes of phalluses as measured by qPCR in our previous study in the tammar [30]. Both adiol treatment in females and oestrogen treatment in males induce urethral closure in a micro-penis [23,30]. Similarly, in the 3-D PCA results, both adiol treatment of females and oestrogen treatment of males had a gene expression pattern distinct from that of normal female and male phalluses, respectively. Although both treatments resulted in a micro-penis with a closed urethra [30], the 3-D PCA highlighted differences between the adiol and oestrogen treatment groups. It is possible that the presence of their intact testes in the oestrogen-treated males counteracted some of the oestrogenic effects.
3.2. Co-Regulatory lncRNAs of IGF1, AR and ESR1
IGF1 plays an important role in regulating phallus development in humans [33,34] and its sexually dimorphic expression suggests it may be equally important in the tammar phallus [31]. The balance between local expression of IGF1 and insulin growth factor binding proteins (IGFBPs) is required to maintain the IGF1 signalling pathway during normal tissue growth [31,35,36]. Our co-expression analysis showed that IGF1 was highly correlated (R > 0.8) with IGFBP5 and Fibroblast Growth Factor (FGF10), another important growth factor in regulating phallus development in mice [37,38,39,40] and implicated in the tammar [31]. Similarly, a large group of predicted lncRNAs were also highly correlated (R > 0.8) with IGF1 expression. However, little is known about the functional role of those lncRNAs during reproductive tract development. AR was correlated with ESR1 and more than 50% of AR-correlated coding genes and lncRNAs were also correlated with ESR1. These data further confirm a potential interaction between AR and ESR1 networks during phallus development, as seen in mice [32]. Interestingly, none of those lncRNAs were located within 100 kb up-or down-stream of IGF1, AR and ESR1, suggesting that those lncRNAs may have other gene targets.
3.3. BMP5 and Lnc-BMP5 in Phallus Development
Bone morphogenetic proteins (BMPs), especially Bmp7, play an important role in regulating urethra formation in mice [41,42,43,44,45]. Loss of Bmp7 signalling causes hypospadias [41,43]. In contrast, some BMPs, such as Bmp4, suppress urethral outgrowth, increase apoptosis and inhibit cell proliferation of the genital tubercle mesenchyme in mice [45]. Several BMPs are hormone-responsive during mouse phallus development [32]. In male mice treated with flutamide, an androgen receptor inhibitor, expression of Bmp5 decreases but Bmp2 expression increases [32]. Male mice exposed to oestradiol benzoate, a synthetic oestrogen, upregulated Bmp8b [32]. Interestingly, BMP5, and its neighbouring lnc-BMP5 were downregulated by oestrogen treatment during tammar phallus sexual differentiation. In addition, lnc-BMP5 was localised in the urethral epithelium of tammar phalluses. Our data suggested that BMP5 and lnc-BMP5 were susceptible to oestrogen treatment, suggesting these might be another pair of genes that regulate phallus development, similar to the other Bmps mentioned above.
3.4. Hormonal Regulation of ZBTB16 and its Correlated lncRNA
We identified a new lncRNA, lnc-ZBTB16 within 1kb upstream of ZBTB16, which had the highest correlation with ZBTB16 expression. ZBTB16 gene encodes a zinc finger transcription factor and the missense mutation of ZBTB16 induces micro-penis in boys [46,47], indicating an important role for this gene during genital development. With co-expression analysis, we showed that ZBTB16 and lnc-ZBTB16 were both downregulated by oestrogen, suggesting a functional relationship between the two. Interestingly, oestrogen treatment in the tammar from day 20–40 induces a micro-penis by day 150 pp [30], similar to boys with the missense mutation of ZBTB16 [46,47]. We suggest that ZBTB16 and its co-expressed lnc-ZBTB16 might be the primary target of oestrogen that induces micropenis.
3.5. RSPO4 and lnc-RSPO4 May Regulate Phallus Development Through Activating WNT5A
RSPO4, a ligand of leucine rich repeat containing G protein-coupled (LGR) 4–6 receptors, potentiates the Wingless-Type MMTV Integration Site Family Member (WNT)/β-catenin signalling pathway [48,49,50,51]. The WNT/β-catenin signalling pathway interacts with androgens and hedgehog signalling to regulate normal phallus development in mammals [30,32,52,53,54,55,56,57]. In the tammar, RSPO4 and lnc-RSPO4 were downregulated in adiol treated female phalluses at day 50 pp, but upregulated in phalluses of earlier castrated males at day 50 pp. The expression pattern of RSPO4 and lnc-RSPO4 was similar to that of WNT5A and SHH [25,30]. RSPO4 and lnc-RSPO4 may also be involved in the interaction between androgen, SHH and WNT5A driving masculinization of the phallus, as occurs in knockout mice [52,53]. Our data demonstrate that there is a complex network of hormonally-regulated coding and lncRNA regulatory transcripts during phallus development, extending our understanding of the aetiology of abnormal phallus development, including hypospadias and potential effects of environmental steroids on the reproductive system.
This study confirms that exogenous hormonal manipulation altered the expression pattern of genes, and highlighted the importance of both androgen and oestrogen in phallus development. We identified multiple co-regulatory lncRNAs with their associated coding genes, such as IGF1, AR and ESR1. There appears to be an interaction between AR and ERα signalling during phallus development in the tammar, as seen in mice. Furthermore, we validated three novel co-regulatory hormonal responsive lncRNAs co-expressed with BMP5, ZBTB16 and RSPO4 that may also regulate phallus differentiation, as seen in mice. The results demonstrate that tammar phallus differentiation is under a complex regulation network of protein-coding genes and their co-expressed lncRNAs, all of which are susceptible to exogenous sex steroids.
4. Materials & Methods
4.1. Animals
Tammar wallabies (Macropus eugenii) of Kangaroo Island (South Australia) origin were kept in our breeding colony in Melbourne. The sex of the young was identified by the presence of scrotal or mammary primordia [58]. The age of the pouch young was estimated from measurements of head length and weight from published growth curves [59]. Phalluses were collected from pouch young tammar wallaby after anaesthesia with Zoletil 100 (Tiletamine HCl 50 mg/mL, Zolazepam HCl 50 mg/mL, Virbac, Cat# 1p6080-3, 1 mL/kg,) and killing by an overdose of sodium pentobarbitone (60 mg/mL, to effect). Samples were snap-frozen and stored at −80 °C for RNA-Seq analysis or fixed in 4% (w/v) paraformaldehyde, embedded in paraffin, and cut into 7 µm sections for histological analysis and section in situ hybridization. All experimental procedures complied with the Australian National Health and Medical Research Council (2013) guidelines and approved by the University of Melbourne Animal Experimentation Ethics Committees. Phalluses for section in situ hybridisation (n = 3) were collected as above from day 20 pp and day 90 pp males. Male pouch young were castrated (n = 5) as previously described [23,25,60] or treated with oestradiol benzoate (2.5 mg/kg/day, dissolved in triolein oil, Cat#50-50-0, Sigma, St. Louis, MO, USA) orally from day 20 pp to day 40 pp (n = 5) as previously described [61]. Female pouch young (n = 5) were injected with 5α-androstane-3α,17β-diol (adiol, 10 mg/kg/week, dissolved in triolein oil, Cat#A1170-000, Steraloids, Newport, USA) from day 20 pp to day 40 pp as previously described [23]. The treated pouch young and control phalluses were collected at day 50 pp (n = 5) for RNA-Seq analysis and qPCR (n = 5).
4.2. Transcriptome Analysis
Two RNA-seq data-sets were used: one, standard 100bpx2 RNA-seq (no replicates but a single pool of 5 samples for each tissue) and the other, strand-specific (5 replicates of each tissue). RNA samples were further tested using Bioanalyzer (Cat#G2939BA, Agilent, Santa Clara, CA, USA). Appropriate amounts of RNA were subjected to multiplex indexed-RNA-Seq analyses using TruSeq RNA Sample prep Kit (Cat#15008136A, Illumina, San Diego, CA, USA) or TruSeq Stranded Total RNA Prep Kit (Cat#20020596, Illumina, San Diego, CA, USA), and HiSeq2000 analyser (Cat#15011190, Illumina, San Diego, CA, USA). Roughly 130–150 × 106 reads (100 bp each from the both ends) were obtained for each index-tag. Reads were obtained from each sample after quality control (Q.C.) > 30 filtering. The average quality score was ~39. The RNA-Seq data were assembled with Trimmomatic [62] and Cufflinks-Cuffdiff [63] pipeline and analysed with CummeRbund R package [64]. The average of mapping rate from all samples was above 80% and the quality was good for differential expression analysis. The original (Renfree et al., 2011) and the updated tammar wallaby genome 3.0 [65] were used as a guide for transcriptome assembly. The annotation was obtained by blasting against the UNIPROT protein database [1]. Differentially expressed genes were extracted with CummeRbund R package and basic R scripts [66]. 3-D PCA analysis used the R package rgl [67]. Principal components 1–3 represent 94.3%, 4.5% and 0.73% of the variance. Long ncRNAs were predicted by using protein potential calculator [68]. BMP5 and lnc-BMP5 were extracted from the standard 100bpx2 RNA-seq data-set (no replicate single pool), whereas RSPO4, lncRSPO4, ZBTB16 and lnc-ZBTB16 were extracted from the strand-specific RNA-seq data-set (5 replicates).
4.3. Weighted Gene Co-Expression Network Analysis
Co-expression analysis between protein coding genes and lncRNAs were conducted by Weighted Gene Co-Expression Network Analysis (WGCNA), R package [69]. RNA-Seq data was normalised (log-transformed) before entering WGCNA pipeline [70]. WGCNA produced 16 different modules named with different colours. The co-expression figure was produced by using VisANT [71]. The target genes of the co-expression network were selected based on their correlation coefficient in each colour-coded module and their importance in phallus development from all relevant publications cited in this paper. IGF1 was analysed in a different module (Figure 2) from AR and ESR1 (Figure 3). The co-regulatory lncRNAs were selected based on their correlation coefficient (R ≥ 0.7) with IGF1, AR and ESR1, if they were not located on the same scaffold of those coding genes. The co-regulatory and hormone-responsive lncRNAs were selected by firstly filtering with differentially expressed lncRNAs extracted from RNA-Seq into each co-expressed module (Figure S2), then selected based on their correlation coefficient (R ≥ 0.7) with the coding genes and whether they are located within 100 kb upstream or downstream of respective coding genes.
4.4. RNA Extraction and cDNA Preparation
RNA was extracted from developing phalluses with the RNeasy Mini kit (Cat# 74804, QIAgen, Hilden, Germany) following on the manufacturer’s recommendations. The RNA was calculated with a Qubit 2.0 fluorometer (Cat#Q32866, ThermoFisher Scientific, Waltham, MA, USA). All RNA samples were treated with DNA-free enzyme (Cat#1906, ThermoFisher Scientific) to prevent genomic DNA contamination. 200 ng of total RNA was used for complementary DNA (cDNA) template synthesis by using transcription high fidelity cDNA Synthesis kit (Cat# 05081963001, Invitrogen, Waltham, MA, USA).
4.5. Quantitative PCR
FastStart Universal SYBR mix (Cat#04913914001, Roche, Basel, Switzerland) was used to detect gene expression level. PCR primers (Table S2) were designed with program of Primer 3 online (http://primer3.ut.ee/). The estimated efficiency of each set of primers was within 95%~105%. The amplification conditions for qPCR were: 1 cycle of 95 °C for 10 min; 45 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min; and 1 cycle of 72 °C for 7 min. All genes were run with 5 replicates. Two house-keeping genes (HMBS and GAPDH) were used to normalise the expression level. The method that we used to calculate the expression level was described in the MIQE guidelines [72,73]).
4.6. Section in situ Hybridization
Phalluses for section in situ hybridization were collected from day 20 pp and day 90 pp pouch young (n = 3 each stage). The paraffin embedded phalluses were sectioned at 7 µm. Primers were designed with online program of Primer 3 online (http://primer3.ut.ee/) (lnc-BMP5: forward: 5′-GCCAGTTTCCAGACTTTGTGA-3′, reverse: 5′-GGAGCTTTGACTTTGTTTTCTTC-3′). The probes for in situ hybridization (ISH) were labelled with DIG RNA labelling Mix (Roche, Cat#11277073910) and generated with T7/SP6 polymerase synthase kit (Promega, Cat #P1460). The sections were pre-hybridized for 2 h at 42 °C and hybridized for 16–18 h at 42 °C, sections washed and incubated with Anti-Digoxigenin-AP (1:300 dilution, Roche, Cat #11093274910) for 16–18 h at 4 °C and colour developed with NBT/BCIP (1:50 dilution, Roche, Cat #11681451001). The sections were counter-stained with Nuclear Fast Red solution (Cat #N3020 Sigma Aldrich, St. Louis, MO, USA). Negative controls were incubated with sense probe (Figure S3).
4.7. Statistics
Three replicates were used for section in situ hybridisation. Five replicates were used for qPCR and RNA-Seq analysis. FPKM (fragments per kilobase of exon model per million reads mapped) is a normalised estimation of gene expression based on RNA-Seq data. FPKM are calculated from the number of reads that mapped to each particular gene sequence taking into account the gene length and the sequencing depth. Student’s t-test was used to calculate the significance (p < 0.05) of normalized RNA-Seq data and qPCR data. The correlation of IGF1 in black module is 0.944. The correlation of AR and ESR1 in green module are 0.655 and 0.766. The p-values of the correlation coefficient between coding genes and lncRNAs are lower than 0.0001 in their respective module. All error bars represent standard error of the mean (SEM).
Supplementary Materials
The following are available online at http://www.mdpi.com/2311-553X/5/1/3/s1. Figure S1: Genomic location of lnc-BMP5, lnc-ZBTB16 and lnc-RSPO4 in the tammar wallaby. Figure S2. Differentially expressed lncRNAs co-expression network (R ≥ 0.7). Figure S3. Negative control of lnc-BMP5 in situ hybridisation result in day 20 pp and day 90 pp male phalluses. CC: corpus cavernosa, E: epithelium, M: mesenchyme, UE: urethral epithelium. Scale bar: 500 μm. Table S1: List of gene expression in RNA-Seq data. Table S2: List of primers used for qPCR.
Author Contributions
Conceptualization, Y.C., G.S., A.P. and M.B.R.; Data curation, Y.K., A.T. and A.F.; Formal analysis, Y.C. and A.F.; Funding acquisition, G.S., A.J.P. and M.R.; Investigation, Y.C., H.Y. and A.T.; Methodology, Y.C., Y.K., H.Y., A.T. and A.F.; Project administration, G.S., A.J.P. and M.B.R.; Resources, H.Y. and M.B.R.; Supervision, G.S., A.J.P. and M.R.; Writing—original draft, Y.C.; Writing—review & editing, Y.C., G.S., A.J.P., H.Y., A.F. and M.B.R.
Funding
This study was supported by a grant from the Australian National Health and Medical Research Council to MBR, AJP, GS, HY and JDW and was supported, in part, by the Transdisciplinary Research Integration Center (TRIC) Project of the Research Organization of Information and Systems (ROIS), Japan.
Acknowledgments
We thank members of the Wallaby research group (Walgroup) for assistance when handling animals. We also thank members of RIKEN and NIG for the technical support on NGS.
Conflicts of interest
The authors declare that they have no conflicts of interest.
References
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigó, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; Thurman, R.E.; et al. Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kapranov, P.; Drenkow, J.; Dike, S.; Brubaker, S.; Patel, S.; Long, J.; Stern, D.; Tammana, H.; Helt, G. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005, 308, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Non–coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Managadze, D.; Lobkovsky, A.E.; Wolf, Y.I.; Shabalina, S.A.; Rogozin, I.B.; Koonin, E.V. The vast, conserved mammalian lincRNome. PLoS Comput. Boil. 2013, 9, e1002917. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Boil. 2016, 17, 756–770. [Google Scholar] [CrossRef]
- Rastetter, R.H.; Smith, C.A.; Wilhelm, D. The role of non-coding RNA in male sex determination and differentiation. Reproduction 2015, 150, R93–R107. [Google Scholar] [CrossRef]
- Wilhelm, D.; Bernard, P. Non-Coding RNA and the Reproductive System; Springer: New York, NY, USA, 2016. [Google Scholar]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P. Constructing the landscape of the mammalian transcriptome. J. Exp. Boil. 2007, 210, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.H.; Chu, E.T.J.; Spektor, R.; Soloway, P.D. Long non-coding RNA regulation of reproduction and development. Mol. Reprod. Dev. 2015, 82, 932–956. [Google Scholar] [CrossRef] [PubMed]
- Kherdjemil, Y.; Lalonde, R.L.; Sheth, R.; Dumouchel, A.; de Martino, G.; Pineault, K.M.; Wellik, D.M.; Stadler, H.S.; Akimenko, M.A.; Kmita, M. Evolution of Hoxa11 regulation in vertebrates is linked to the pentadactyl state. Nature 2016, 539, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Mori, A.; Lehmann, S.; O’Kelly, J.; Kumagai, T.; Desmond, J.C.; Pervan, M.; McBride, W.H.; Kizaki, M.; Koeffler, H.P. Capsaicin, a component of red peppers, inhibits the growth of androgen-independent, p53 mutant prostate cancer cells. Cancer Res. 2006, 66, 3222–3229. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Sboner, A.; Nair, S.S.; Giannopoulou, E.; Li, R.; Hennig, S.; Mosquera, J.M.; Pauwels, J.; Park, K.; Kossai, M.; et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014, 5, 5383. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Boil. 2013, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Renfree, M.B.; Short, R.; Shaw, G. Sexual differentiation of the urogenital system of the fetal and neonatal tammar wallaby, Macropus eugenii. Anat. Embryol. 1996, 194, 111–134. [Google Scholar] [CrossRef]
- Leihy, M.; Shaw, G.; Wilson, J.; Renfree, M. Development of the penile urethra in the tammar wallaby. Sex. Dev. 2011, 5, 241–249. [Google Scholar] [CrossRef]
- Butler, C.; Shaw, G.; Renfree, M. Development of the penis and clitoris in the tammar wallaby, Macropus Eugenii. Anat. Embryol. 1999, 199, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Leihy, M.W.; Shaw, G.; Wilson, J.D.; Renfree, M.B. Penile development is initiated in the tammar wallaby pouch young during the period when 5α-androstane-3α, 17β-diol is secreted by the testes. Endocrinology 2004, 145, 3346–3352. [Google Scholar] [CrossRef] [PubMed]
- Renfree, M.B.; Chew, K.Y.; Shaw, G. Inducing sex reversal of the urogenital system of marsupials. Differentiation 2014, 87, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Chew, K.; Pask, A.; Hickford, D.; Shaw, G.; Renfree, M. A dual role for SHH during phallus development in a marsupial. Sex. Dev. 2014, 8, 166–177. [Google Scholar] [CrossRef]
- Wilson, J.D.; Auchus, R.J.; Leihy, M.W.; Guryev, O.L.; Estabrook, R.W.; Osborn, S.M.; Shaw, G.; Renfree, M.B. 5α-androstane-3α, 17β-diol is formed in tammar wallaby pouch young testes by a pathway involving 5α-pregnane-3α, 17α-diol-20-one as a key intermediate. Endocrinology 2003, 144, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.D.; George, F.W.; Shaw, G.; Renfree, M.B. Virilization of the male pouch young of the tammar wallaby does not appear to be mediated by plasma testosterone or dihydrotestosterone. Boil. Reprod. 1999, 61, 471–475. [Google Scholar] [CrossRef]
- Shaw, G.; Renfree, M.B.; Leihy, M.W.; Shackleton, C.H.; Roitman, E.; Wilson, J.D. Prostate formation in a marsupial is mediated by the testicular androgen 5α-androstane-3α, 17β-diol. Proc. Natl. Acad. Sci. USA 2000, 97, 12256–12259. [Google Scholar] [CrossRef]
- Wilson, J.D.; Shaw, G.; Leihy, M.L.; Renfree, M.B. The marsupial model for male phenotypic development. Trends Endocrinol. Metab. 2002, 13, 78–83. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, H.; Pask, A.J.; Fujiyama, A.; Suzuki, Y.; Sugano, S.; Shaw, G.; Renfree, M.B. Hormone-responsive genes in the SHH and WNT/β-catenin signaling pathways influence urethral closure and phallus growth. Boil. Reprod. 2018, 99, 806–816. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, H.; Pask, A.J.; Fujiyama, A.; Suzuki, Y.; Sugano, S.; Shaw, G.; Renfree, M. Effects of androgen and oestrogen on the IGF pathways controlling phallus growth. Reproduction 2019, 157, 1–12. [Google Scholar] [CrossRef]
- Zheng, Z.; Armfield, B.A.; Cohn, M.J. Timing of androgen receptor disruption and estrogen exposure underlies a spectrum of congenital penile anomalies. Proc. Natl. Acad. Sci. USA 2015, 112, E7194–E7203. [Google Scholar] [CrossRef] [PubMed]
- Laron, Z.; Pertzelan, A.; Mannheimer, S. Genetic pituitary dwarfism with high serum concentation of growth hormone—A new inborn error of metabolism? Isr. J. Med. Sci. 1966, 2, 152–155. [Google Scholar] [PubMed]
- Laron, Z.; Klinger, B. Effect of insulin-like growth factor-1treatment on serum androgens and testicular and penile size in males with laron syndrome (primary growth hormone resistance). Eur. J. Endocrinol. 1998, 138, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Xu, Q. Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen. Comp. Endocrinol. 2005, 142, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Clemmons, D.R. Role of insulin-like growth factor binding proteins in controlling IGF actions. Mol. Cell. Endocrinol. 1998, 140, 19–24. [Google Scholar] [CrossRef]
- Satoh, Y.; Haraguchi, R.; Wright, T.J.; Mansour, S.L.; Partanen, J.; Hajihosseini, M.K.; Eswarakumar, V.P.; Lonai, P.; Yamada, G. Regulation of external genitalia development by concerted actions of FGF ligands and FGF receptors. Anat. Embryol. 2004, 208, 479–486. [Google Scholar] [CrossRef]
- Harada, M.; Omori, A.; Nakahara, C.; Nakagata, N.; Akita, K.; Yamada, G. Tissue-specific roles of FGF signaling in external genitalia development. Dev. Dyn. 2015, 244, 759–773. [Google Scholar] [CrossRef]
- Gredler, M.L.; Seifert, A.W.; Cohn, M.J. Tissue-specific roles of Fgfr2 in development of the external genitalia. Development 2015, 142, 2203–2212. [Google Scholar] [CrossRef]
- Petiot, A.; Perriton, C.L.; Dickson, C.; Cohn, M.J. Development of the mammalian urethra is controlled by Fgfr2-IIIb. Development 2005, 132, 2441–2450. [Google Scholar] [CrossRef]
- Wu, X.; Ferrara, C.; Shapiro, E.; Grishina, I. Bmp7 expression and null phenotype in the urogenital system suggest a role in re-organization of the urethral epithelium. Gene Expr. Patterns 2009, 9, 224–230. [Google Scholar] [CrossRef]
- Beleza-Meireles, A.; Lundberg, F.; Lagerstedt, K.; Zhou, X.; Omrani, D.; Frisén, L.; Nordenskjöld, A. FGFR2, FGF8, FGF10 and BMP7 as candidate genes for hypospadias. Eur. J. Hum. Genet. 2007, 15, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.A.; Nguyen, S.B.; Scott, V.; Stadler, H.S. Loss of Bmp7 and Fgf8 signaling in Hoxa13-mutant mice causes hypospadias. Development 2003, 130, 3095–3109. [Google Scholar] [CrossRef] [PubMed]
- Bouty, A.; Ayers, K.L.; Pask, A.; Heloury, Y.; Sinclair, A.H. The genetic and environmental factors underlying hypospadias. Sex. Dev. 2015, 9, 239–259. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Bachiller, D.; Chen, Y.P.; Kamikawa, M.; Ogi, H.; Haraguchi, R.; Ogino, Y.; Minami, Y.; Mishina, Y.; Ahn, K.; et al. Regulation of outgrowth and apoptosis for the terminal appendage: External genitalia development by concerted actions of BMP signaling. Development 2003, 130, 6209–6220. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, D.; Köster, B.; Gillessen-Kaesbach, G. Absence of thumbs, A/hypoplasia of radius, hypoplasia of ulnae, retarded bone age, short stature, microcephaly, hypoplastic genitalia, and mental retardation. Am. J. Med. Genet. Part A 2002, 108, 209–213. [Google Scholar] [CrossRef]
- Fischer, S.; Kohlhase, J.; Böhm, D.; Schweiger, B.; Hoffmann, D.; Heitmann, M.; Horsthemke, B.; Wieczorek, D. Biallelic loss of function of the promyelocytic leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J. Med. Genet. 2008, 45, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Carmon, K.S.; Gong, X.; Lin, Q.; Thomas, A.; Liu, Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 11452–11457. [Google Scholar] [CrossRef]
- Carmon, K.S.; Gong, X.; Yi, J.; Thomas, A.; Liu, Q. RSPO–LGR4 functions via IQGAP1 to potentiate Wnt signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 1221–1229. [Google Scholar] [CrossRef]
- Chen, P.-H.; Chen, X.; Lin, Z.; Fang, D.; He, X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013, 27, 1345–1350. [Google Scholar] [CrossRef]
- Kim, K.-A.; Zhao, J.; Andarmani, S.; Kakitani, M.; Oshima, T.; Binnerts, M.E.; Abo, A.; Tomizuka, K.; Funk, W.D. R-Spondin proteins: A novel link to β-catenin activation. Cell Cycle 2006, 5, 23–26. [Google Scholar] [CrossRef]
- Miyagawa, S.; Moon, A.; Haraguchi, R.; Inoue, C.; Harada, M.; Nakahara, C.; Suzuki, K.; Matsumaru, D.; Kaneko, T.; Matsuo, I.; et al. Dosage-dependent hedgehog signals integrated with Wnt/β-catenin signaling regulate external genitalia formation as an appendicular program. Development 2009, 136, 3969–3978. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, S.; Satoh, Y.; Haraguchi, R.; Suzuki, K.; Iguchi, T.; Taketo, M.M.; Nakagata, N.; Matsumoto, T.; Takeyama, K.-I.; Kato, S.; et al. Genetic interactions of the androgen and Wnt/β-catenin pathways for the masculinization of external genitalia. Mol. Endocrinol. 2009, 23, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.C.; Matsumaru, D.; Ho, A.S.; Garcia-Barceló, M.; Yuan, Z.; Smith, D.; Kodjabachian, L.; Tam, P.K.; Yamada, G.; Lui, V.C. Dysregulation of wnt inhibitory factor 1 (Wif1) expression resulted in aberrant Wnt-β-catenin signaling and cell death of the cloaca endoderm, and anorectal malformations. Cell Death Differ. 2014, 21, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yin, Y.; Long, F.; Ma, L. Tissue-specific requirements of β-catenin in external genitalia development. Development 2008, 135, 2815–2825. [Google Scholar] [CrossRef] [PubMed]
- Seifert, A.W.; Zheng, Z.; Ormerod, B.K.; Cohn, M.J. Sonic hedgehog controls growth of external genitalia by regulating cell cycle kinetics. Nat. Commun. 2010, 1, 23. [Google Scholar] [CrossRef]
- Perriton, C.L.; Powles, N.; Chiang, C.; Maconochie, M.K.; Cohn, M.J. Sonic hedgehog signaling from the urethral epithelium controls external genital development. Dev. Boil. 2002, 247, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Wai-Sum, O.; Short, R.V.; Renfree, M.B.; Shaw, G. Primary genetic control of somatic sexual differentiation in a mammal. Nature 1988, 331, 716–717. [Google Scholar] [CrossRef]
- Poole, W.; Simms, N.; Wood, J.; Lubulwa, M. Tables for Age Determination of the Kangaroo Island Wallaby (Tammar), Macropus Eugenii, from Body Measurements; Wildlife and Ecology Technical Memorandum #32; CSIRO: Lyneham, ACT, Australia, 1991; ISBN 0-643-05235-6. [Google Scholar]
- Renfree, M.B.; Fenelon, J.; Wijiyanti, G.; Wilson, J.D.; Shaw, G. Wolffian duct differentiation by physiological concentrations of androgen delivered systemically. Dev. Boil. 2009, 334, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Coveney, D.; Shaw, G.; Renfree, M.B. Estrogen-induced gonadal sex reversal in the tammar wallaby. Boil. Reprod. 2001, 65, 613–621. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Goff, L.; Trapnell, C.; Kelley, D. cummeRbund: Analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. R Packag. Version 2018, 2. [Google Scholar] [CrossRef]
- O’Neill, R. (University of Connecticut, Storrs-Mansfield, Connecticut, USA); Heider, T.N. (University of Connecticut, Storrs-Mansfield, Connecticut, USA); Fujiyama, A. (Advanced Genomics Center, National Institute of Genetics, Mishima, Shizuoka, Japan); Shaw, G. (University of Melbourne, Melbourne, Victoria, Australia); Renfree, M.B. (University of Melbourne, Melbourne, Victoria, Australia); Pask, A.J. (University of Melbourne, Melbourne, Victoria, Australia). Personal communication, 2018.
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar] [CrossRef]
- Adler, D.; Murdoch, D.; Nenadic, O.; Urbanek, S. rgl: 3D Visualization Device System (OpenGL). R Package Version 0.75. 2007. Available online: https://CRAN.R-project.org/package=rgl (accessed on 31 May 2018).
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. Wgcna: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Parikshak, N.N.; Luo, R.; Zhang, A.; Won, H.; Lowe, J.K.; Chandran, V.; Horvath, S.; Geschwind, D.H. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 2013, 155, 1008–1021. [Google Scholar] [CrossRef]
- Hu, Z.; Chang, Y.-C.; Wang, Y.; Huang, C.-L.; Liu, Y.; Tian, F.; Granger, B.; DeLisi, C. Visant 4.0: Integrative network platform to connect genes, drugs, diseases and therapies. Nucleic Acids Res. 2013, 41, 225–231. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; Vandesompele, J. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).