Long Non-Coding RNAs in Obesity-Induced Cancer
Abstract
1. Introduction
1.1. Hyperinsulinemia
1.2. Dysregulation of the Adipokine Expression
1.3. Hypoxia
1.4. Chronic Inflammation
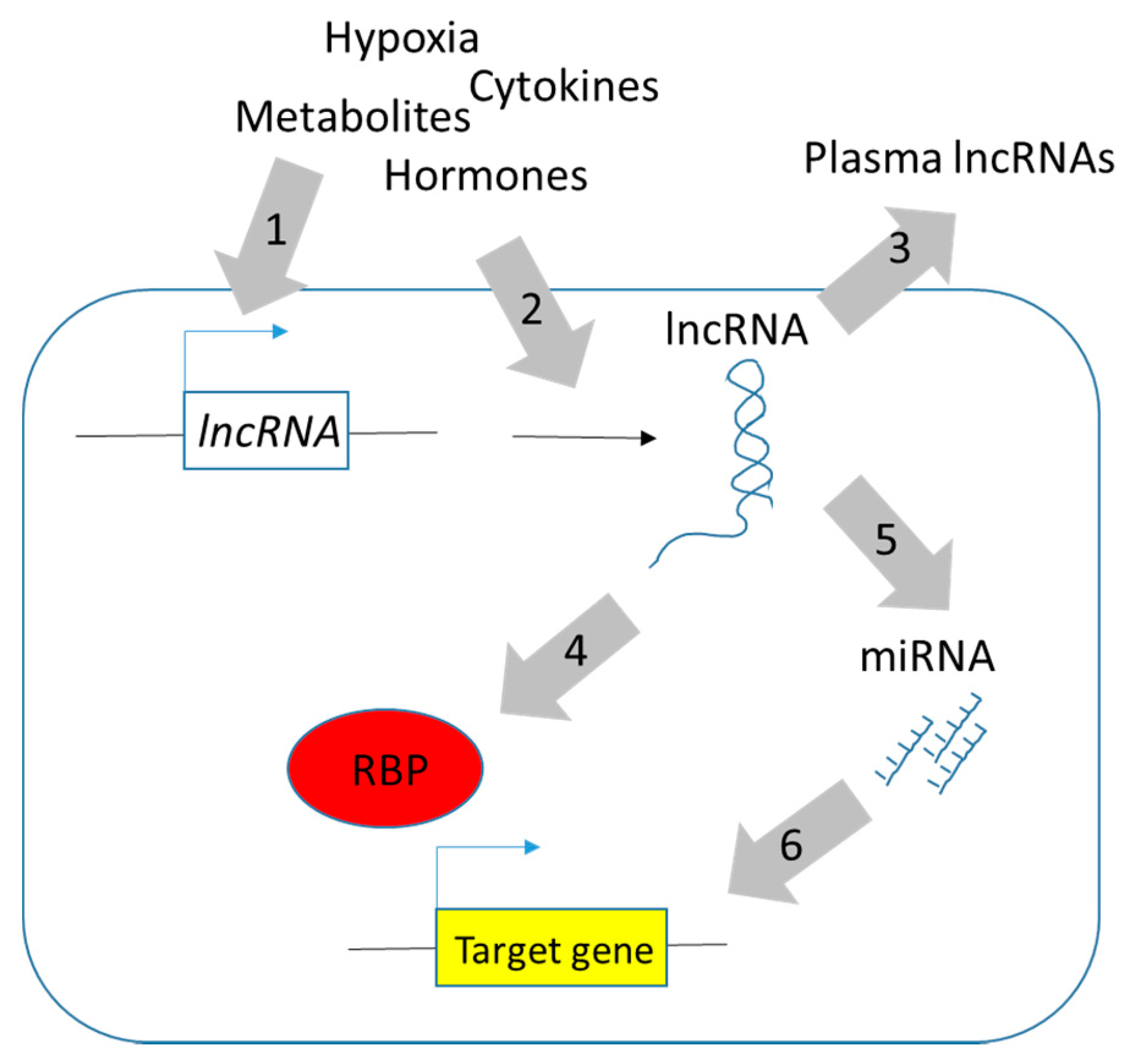
2. lncRNAs in Cancer and Energy Metabolism
2.1. Antisense Non-Coding RNA in the INK4 Locus
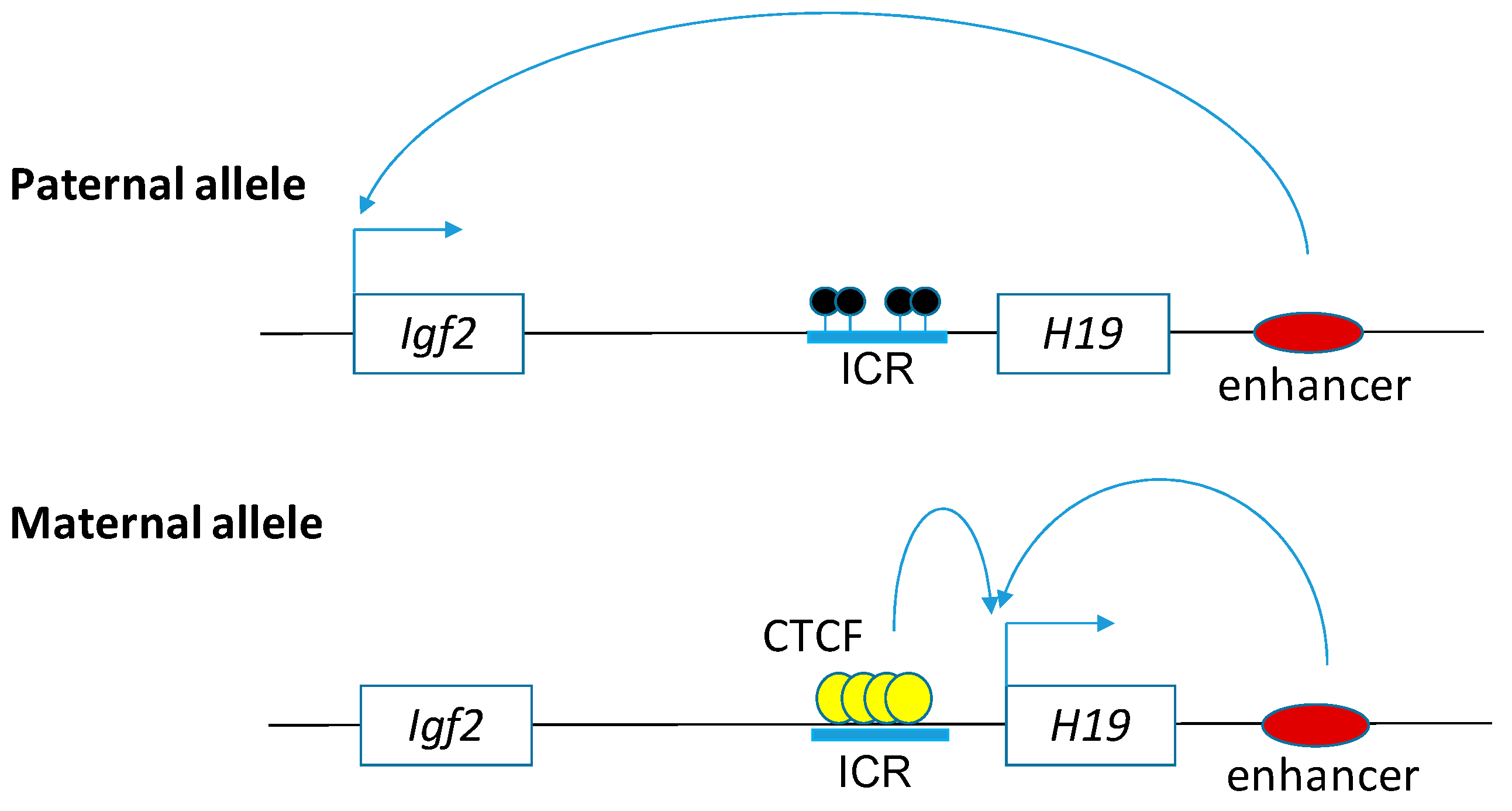
2.2. H19
2.3. HOTAIR
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3’UTR | 3’ untranslated region |
| AFP | α-fetoprotein |
| ANRIL | antisense non-coding RNA in the INK4 Locus |
| DMR | differentially methylated region |
| HOTAIR | HOX transcript antisense RNA |
| HOX | Homebox |
| IGF-1 | insulin-like growth factor-1 |
| lncRNAs | long noncoding RNAs |
| LPS | lipopolysaccharides |
| PRC2 | polycomb repressive complex-2 |
| RNA Pol II | RNA polymerase II |
References
- Tahergorabi, Z.; Khazaei, M.; Moodi, M.; Chamani, E. From obesity to cancer: A review on proposed mechanisms. Cell Biochem. Funct. 2016, 34, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.G.; Hussan, H.; Cruz-Monserrate, Z.; Conteh, L.F.; Mumtaz, K.; Conwell, D.L. A review of the impact of obesity on common gastrointestinal malignancies. Integr. Cancer Sci. Ther. 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Majumder, K.; Gupta, A.; Arora, N.; Singh, P.P.; Singh, S. Premorbid obesity and mortality in patients with pancreatic cancer: A systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2016, 14, 355–368. [Google Scholar] [CrossRef] [PubMed]
- De Pergola, G.; Silvestris, F. Obesity as a major risk factor for cancer. J. Obes. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Demark-Wahnefried, W.; Platz, E.A.; Ligibel, J.A.; Blair, C.K.; Courneya, K.S.; Meyerhardt, J.A.; Ganz, P.A.; Rock, C.L.; Schmitz, K.H.; Wadden, T.; et al. The role of obesity in cancer survival and recurrence. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1244–1259. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.L.; Dive, C.; Renehan, A.G. Biological mechanisms linking obesity and cancer risk: New perspectives. Annu. Rev. Med. 2010, 61, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Louie, S.M.; Roberts, L.S.; Nomura, D.K. Mechanisms linking obesity and cancer. Biochim. Biophys. Acta 2013, 1831, 1499–1508. [Google Scholar] [CrossRef] [PubMed]
- Stone, T.W.; McPherson, M.; Gail Darlington, L. Obesity and cancer: Existing and new hypotheses for a causal connection. EBioMedicine 2018, 30, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Conte, C.; Fabbrini, E.; Kars, M.; Mittendorfer, B.; Patterson, B.W.; Klein, S. Multiorgan insulin sensitivity in lean and obese subjects. Diabetes Care 2012, 35, 1316–1321. [Google Scholar] [CrossRef] [PubMed]
- Djiogue, S.; Nwabo Kamdje, A.H.; Vecchio, L.; Kipanyula, M.J.; Farahna, M.; Aldebasi, Y.; Seke Etet, P.F. Insulin resistance and cancer: The role of insulin and IGFs. Endocr Relat Cancer 2013, 20, R1–R17. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, E.J.; LeRoith, D. The proliferating role of insulin and insulin-like growth factors in cancer. Trends Endocrinol. Metab. 2010, 21, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Denduluri, S.K.; Idowu, O.; Wang, Z.; Liao, Z.; Yan, Z.; Mohammed, M.K.; Ye, J.; Wei, Q.; Wang, J.; Zhao, L.; et al. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis. 2015, 2, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Frasca, F.; Pandini, G.; Sciacca, L.; Pezzino, V.; Squatrito, S.; Belfiore, A.; Vigneri, R. The role of insulin receptors and IGF-I receptors in cancer and other diseases. Arch. Physiol. Biochem. 2008, 114, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose tissue remodeling: its role in energy metabolism and metabolic disorders. Front. Endocrinol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.; Oliveira, T.; Fernandes, R. Biochemistry of adipose tissue: An endocrine organ. Arch. Med. Sci. 2013, 9, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Feve, B.; Bastard, C.; Fellahi, S.; Bastard, J.P.; Capeau, J. New adipokines. Ann. D’endocrinol. 2016, 77, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Martyniak, K.; Masternak, M.M. Changes in adipose tissue cellular composition during obesity and aging as a cause of metabolic dysregulation. Exp. Gerontol. 2017, 94, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Balistreri, C.R.; Caruso, C.; Candore, G. The role of adipose tissue and adipokines in obesity-related inflammatory diseases. Mediat. Inflamm. 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Ghosh, S.; Pandit, K.; Mukhopadhyay, P.; Chowdhury, S. Leptin and cancer: Pathogenesis and modulation. Indian J. Endocrinol. Metab. 2012, 16 (Suppl. 3), S596–S600. [Google Scholar] [PubMed]
- Cleary, M.P.; Torroella-Kouri, M. Leptin in cancer: Epidemiology and mechanisms. In Adipocytokines, Energy Balance, and Cancer; Reizes, O., Berger, N.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Hosney, M.; Sabet, S.; El-Shinawi, M.; Gaafar, K.M.; Mohamed, M.M. Leptin is overexpressed in the tumor microenvironment of obese patients with estrogen receptor positive breast cancer. Exp. Ther. Med. 2017, 13, 2235–2246. [Google Scholar] [CrossRef] [PubMed]
- Candelaria, P.V.; Rampoldi, A.; Harbuzariu, A.; Gonzalez-Perez, R.R. Leptin signaling and cancer chemoresistance: Perspectives. World J. Clin. Oncol. 2017, 8, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Kusminski, C.M.; Scherer, P.E. Adipose tissue remodeling and obesity. J. Clin. Investig. 2011, 121, 2094–2101. [Google Scholar] [CrossRef] [PubMed]
- Trayhurn, P. Hypoxia and adipose tissue function and dysfunction in obesity. Physiol. Rev. 2013, 93, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, K. HIF-1 at the crossroads of hypoxia, inflammation, and cancer. Int. J. Cancer. J. Int. Cancer 2016, 138, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Masoud, G.N.; Li, W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Agani, F.; Jiang, B.H. Oxygen-independent regulation of HIF-1: Novel involvement of PI3K/AKT/mTOR pathway in cancer. Curr. Cancer Drug Targets 2013, 13, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.; Padwad, Y.S. HIF-1 in cancer therapy: Two decade long story of a transcription factor. Acta Oncol. 2017, 56, 503–515. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Gao, Z.; Yin, J.; Zhang, J.; Yun, Z.; Ye, J. Regulation of HIF-1α activity in adipose tissue by obesity-associated factors: Adipogenesis, insulin, and hypoxia. Am. J. Physiol. Endocrinol. Metab. 2011, 300, E877–E885. [Google Scholar] [CrossRef] [PubMed]
- Rausch, L.K.; Netzer, N.C.; Hoegel, J.; Pramsohler, S. The linkage between breast cancer, hypoxia, and adipose tissue. Front. Oncol. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed]
- De Marzo, A.M.; Platz, E.A.; Sutcliffe, S.; Xu, J.; Gronberg, H.; Drake, C.G.; Nakai, Y.; Isaacs, W.B.; Nelson, W.G. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 2007, 7, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, S.; Ohnishi, S.; Ma, N.; Hiraku, Y.; Murata, M. Crosstalk between DNA damage and inflammation in the multiple steps of carcinogenesis. Int. J. Mol. Sci. 2017, 18, 1808. [Google Scholar] [CrossRef] [PubMed]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Kern, P.A.; Di Gregorio, G.B.; Lu, T.; Rassouli, N.; Ranganathan, G. Adiponectin expression from human adipose tissue: Relation to obesity, insulin resistance, and tumor necrosis factor-α expression. Diabetes 2003, 52, 1779–1785. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, P. The role of adipokines in chronic inflammation. Immunotargets Ther. 2016, 5, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Boutagy, N.E.; McMillan, R.P.; Frisard, M.I.; Hulver, M.W. Metabolic endotoxemia with obesity: Is it real and is it relevant? Biochimie 2016, 124, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Henao-Mejia, J.; Flavell, R.A. Innate immune receptors: Key regulators of metabolic disease progression. Cell Metab. 2013, 17, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Seganfredo, F.B.; Blume, C.A.; Moehlecke, M.; Giongo, A.; Casagrande, D.S.; Spolidoro, J.V.N.; Padoin, A.V.; Schaan, B.D.; Mottin, C.C. Weight-loss interventions and gut microbiota changes in overweight and obese patients: A systematic review. Obes. Rev. 2017, 18, 832–851. [Google Scholar] [CrossRef] [PubMed]
- Berger, N.A.; Scacheri, P.C. Targeting epigenetics to prevent obesity promoted cancers. Cancer Prev. Res. 2018, 11, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Su, S.; Barnes, V.A.; De Miguel, C.; Pollock, J.; Ownby, D.; Shi, H.; Zhu, H.; Snieder, H.; Wang, X. A genome-wide methylation study on obesity: Differential variability and differential methylation. Epigenetics 2013, 8, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.A.; Milagro, F.I.; Claycombe, K.J.; Schalinske, K.L. Epigenetics in adipose tissue, obesity, weight loss, and diabetes. Adv. Nutr. 2014, 5, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ruan, Y.; Wang, M.; Chen, R.; Yu, N.; Sun, L.; Liu, T.; Chen, H. Differentially expressed circulating lncRNAs and mRNA identified by microarray analysis in obese patients. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Latorre, J.; Fernandez-Real, J.M. lncRNAs in adipose tissue from obese and insulin-resistant subjects: New targets for therapy? EBioMedicine 2018, 30, 10–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z. Progress and prospects of long non-coding RNAs in lipid homeostasis. Mol. Metab. 2016, 5, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Zhao, X.Y.; Li, S.; Yang, G.; Lin, J.D. Conserved function of the long non-coding RNA Blnc1 in brown adipocyte differentiation. Mol. Metab. 2017, 6, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Karapetyan, A.R.; Buiting, C.; Kuiper, R.A.; Coolen, M.W. Regulatory roles for long ncRNA and mRNA. Cancers 2013, 5, 462–490. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Crowe, M.L.; Grimmond, S.M.; Mattick, J.S. NRED: A database of long non-coding RNA expression. Nucl. Acids Res. 2009, 37, D122–D126. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Song, R.; Regev, A.; Struhl, K. Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long non-coding RNAs in cancer pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long non-coding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M.; Emanueli, C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Haider Ali, M.S.S.; Moran, M. The role of interactions of long non-coding RNAs and heterogeneous nuclear ribonucleoproteins in regulating cellular functions. Biochem. J. 2017, 474, 2925–2935. [Google Scholar] [CrossRef] [PubMed]
- Bolha, L.; Ravnik-Glavac, M.; Glavac, D. Long non-coding RNAs as biomarkers in cancer. Dis. Markers 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Goff, L.A.; Trapnell, C.; Alexander, R.; Lo, K.A.; Hacisuleyman, E.; Sauvageau, M.; Tazon-Vega, B.; Kelley, D.R.; Hendrickson, D.G.; et al. Long non-coding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3387–3392. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Gao, W.W.; Tang, H.M.; Deng, J.J.; Wong, C.M.; Chan, C.P.; Jin, D.Y. β-TrCP-mediated ubiquitination and degradation of liver-enriched transcription factor CREB-H. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, P.; Yang, W.; Ruan, X.; Kiesewetter, K.; Zhu, J.; Cao, H. Integrative transcriptome analyses of metabolic responses in mice define pivotal lncrna metabolic regulators. Cell Metab. 2016, 24, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Nakaoka, H.; Gurumurthy, A.; Hayano, T.; Ahmadloo, S.; Omer, W.H.; Yoshihara, K.; Yamamoto, A.; Kurose, K.; Enomoto, T.; Akira, S.; et al. Allelic imbalance in regulation of ANRIL through chromatin interaction at 9p21 endometriosis risk locus. PLoS Genet. 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, Z.; Chen, W.; Zhai, W.; Pan, J.; Pang, H.; Li, X. H19 promotes endometrial cancer progression by modulating epithelial-mesenchymal transition. Oncol. Lett. 2017, 13, 363–369. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Bao, W.; Li, X.; Chen, Z.; Che, Q.; Wang, H.; Wan, X.P. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int. J. Mol. Med. 2014, 33, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.X.; Wang, C.; Mao, L.W.; Wang, Y.L.; Xia, L.Q.; Zhao, W.; Shen, J.; Chen, J. Long non-coding RNA HOTAIR mediates the estrogen-induced metastasis of endometrial cancer cells via the miR-646/NPM1 axis. Am. J. Physiol. Cell Physiol. 2018, 314, C690–C701. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhang, Z.; Mao, C.; Zhou, Y.; Yu, L.; Yin, Y.; Wu, S.; Mou, X.; Zhu, Y. ANRIL inhibits p15(INK4b) through the TGFβ1 signaling pathway in human esophageal squamous cell carcinoma. Cell. Immunol. 2014, 289, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Wu, Y.; Hu, L.; He, P.; Xiong, G.; Bai, Y.; Yang, K. Long non-coding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis. Dis. Esophagus 2017, 30, 1–9. [Google Scholar] [PubMed]
- Hibi, K.; Nakamura, H.; Hirai, A.; Fujikake, Y.; Kasai, Y.; Akiyama, S.; Ito, K.; Takagi, H. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996, 56, 480–482. [Google Scholar] [PubMed]
- Wang, W.; He, X.; Zheng, Z.; Ma, X.; Hu, X.; Wu, D.; Wang, M. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol. Cancer 2017, 16. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.B.; Lian, G.Y.; Wang, H.R.; Song, E.; Yao, H.; Wang, M.H. Long non-coding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.D.; Chen, W.M.; Qi, F.Z.; Xia, R.; Sun, M.; Xu, T.P.; Yin, L.; Zhang, E.B.; De, W.; Shu, Y.Q. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J. Hematol. Oncol. 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Hua, L.; Wang, C.Y.; Yao, K.H.; Chen, J.T.; Zhang, J.J.; Ma, W.L. High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 3076–3082. [Google Scholar] [PubMed]
- Fellig, Y.; Ariel, I.; Ohana, P.; Schachter, P.; Sinelnikov, I.; Birman, T.; Ayesh, S.; Schneider, T.; de Groot, N.; Czerniak, A.; et al. H19 expression in hepatic metastases from a range of human carcinomas. J. Clin. Pathol. 2005, 58, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Lu, Y.; Xu, Q.; Tang, B.; Park, C.K.; Chen, X. HULC and H19 played different roles in overall and disease-free survival from hepatocellular carcinoma after curative hepatectomy: A preliminary analysis from gene expression omnibus. Dis. Markers 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.P.; Wang, J.P.; Wang, X.P. HOTAIR contributes to the growth of liver cancer via targeting miR-217. Oncol. Lett. 2018, 15, 7963–7972. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, J.Q.; Chen, J.Z.; Chen, H.X.; Qiu, F.N.; Yan, M.L.; Chen, Y.L.; Peng, C.H.; Tian, Y.F.; Wang, Y.D. The over expression of long non-coding RNA ANRIL promotes epithelial-mesenchymal transition by activating the ATM-E2F1 signaling pathway in pancreatic cancer: An in vivo and in vitro study. Int. J. Biol. Macromol. 2017, 102, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Lu, Y.L.; Yang, Y.; Hu, L.B.; Bai, Y.; Li, R.Q.; Zhang, G.Y.; Li, J.; Bi, C.W.; Yang, L.B.; et al. Overexpression of lncRNA ANRIL promoted the proliferation and migration of prostate cancer cells via regulating let-7a/TGF-β1/Smad signaling pathway. Cancer Biomark. 2018, 21, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Nong, K.; Zhu, H.; Wang, W.; Huang, X.; Yuan, Z.; Ai, K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014, 35, 9163–9169. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yao, J.; An, Y.; Chen, X.; Chen, W.; Wu, D.; Luo, B.; Yang, Y.; Jiang, Y.; Sun, D.; et al. lncRNA HOTAIR acts a competing endogenous RNA to control the expression of Notch3 via sponging miR-613 in pancreatic cancer. Oncotarget 2017, 8, 32905–32917. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Jutooru, I.; Chadalapaka, G.; Johnson, G.; Frank, J.; Burghardt, R.; Kim, S.; Safe, S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013, 32, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zheng, Z.P.; Li, H.; Zhang, H.Q.; Ma, F.Q. ANRIL is associated with the survival rate of patients with colorectal cancer, and affects cell migration and invasion in vitro. Mol. Med. Rep. 2016, 14, 1714–1720. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Ou, C.; Ren, W.; Xie, X.; Li, X.; Li, G. Downregulation of long non-coding RNA ANRIL suppresses lymphangiogenesis and lymphatic metastasis in colorectal cancer. Oncotarget 2016, 7, 47536–47555. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, X.; Tang, C.; Chen, X.; He, J. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int. J. Oncol. 2017, 50, 1801–1809. [Google Scholar] [CrossRef] [PubMed]
- Schultheiss, C.S.; Laggai, S.; Czepukojc, B.; Hussein, U.K.; List, M.; Barghash, A.; Tierling, S.; Hosseini, K.; Golob-Schwarzl, N.; Pokorny, J.; et al. The long non-coding RNA H19 suppresses carcinogenesis and chemoresistance in hepatocellular carcinoma. Cell Stress 2018, 1, 37–54. [Google Scholar] [CrossRef]
- Li, P.; Zhang, X.; Wang, L.; Du, L.; Yang, Y.; Liu, T.; Li, C.; Wang, C. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-κB/TS signaling in colorectal cancer. Mol. Ther. Nucl. Acids 2017, 8, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, Z.; Ning, X.; Huang, L.; Jiang, B. The long non-coding RNA HOTAIR promotes colorectal cancer progression by sponging miR-197. Oncol. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Shen, E.D.; Liao, M.M.; Hu, Y.B.; Wu, K.; Yang, P.; Zhou, L.; Chen, W.D. Expression and mechanisms of long non-coding RNA genes MEG3 and ANRIL in gallbladder cancer. Tumour Biol. 2016, 37, 9875–9886. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, X.; Liu, Y.; Yuan, J.; Yang, F.; Huang, J.; Meng, Q.; Zhou, C.; Liu, F.; Ma, J.; et al. Long non-coding RNA H19 inhibits the proliferation of fetal liver cells and the Wnt signaling pathway. FEBS Lett. 2016, 590, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Wu, X.C.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Quan, Z.W. Upregulation of H19 indicates a poor prognosis in gallbladder carcinoma and promotes epithelial-mesenchymal transition. Am. J. Cancer Res. 2016, 6, 15–26. [Google Scholar] [PubMed]
- Ma, M.Z.; Li, C.X.; Zhang, Y.; Weng, M.Z.; Zhang, M.D.; Qin, Y.Y.; Gong, W.; Quan, Z.W. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol. Cancer 2014, 13. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.T.; Xu, J.H.; Zheng, Z.R.; Zhao, Q.Q.; Zeng, X.S.; Cheng, S.X.; Liang, Y.H.; Hu, Q.F. Long non-coding RNA ANRIL promotes carcinogenesis via sponging miR-199a in triple-negative breast cancer. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 96, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.; Dumont, L.; Lottin, S.; Bolle, D.; Lepretre, A.; Delobelle, A.; Bouali, F.; Dugimont, T.; Coll, J.; Curgy, J.J. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am. J. Pathol. 1998, 153, 1597–1607. [Google Scholar] [CrossRef]
- Ding, W.; Ren, J.; Ren, H.; Wang, D. Long non-coding RNA HOTAIR modulates miR-206-mediated Bcl-w signaling to facilitate cell proliferation in breast cancer. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.J.; Lin, Y.Y.; Ding, J.X.; Feng, W.W.; Jin, H.Y.; Hua, K.Q. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int. J. Oncol. 2015, 46, 2497–2505. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Song, L.; He, J.; Sun, Y.; Liu, X.; Zou, X. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 10082–10091. [Google Scholar] [PubMed]
- Liu, S.; Lei, H.; Luo, F.; Li, Y.; Xie, L. The effect of lncRNA HOTAIR on chemoresistance of ovarian cancer through regulation of HOXA7. Biol. Chem. 2018, 399, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.J.; Hao, S.; Wang, L.L.; Hu, C.Y.; Zhang, S.; Guo, L.J.; Zhang, G.; Gao, B.; Jiang, Y.; Tian, W.G.; et al. Long non-coding RNA ANRIL promotes the invasion and metastasis of thyroid cancer cells through TGF-β/Smad signaling pathway. Oncotarget 2016, 7, 57903–57918. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Sun, W.; Dong, W.; Wang, Z.; Zhang, T.; He, L.; Zhang, H. Downregulation of long non-coding RNA H19 contributes to the proliferation and migration of papillary thyroid carcinoma. Gene 2018, 646, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, S.; Jiang, L.; Wang, X.; Song, X. HOTAIR is a promising novel biomarker in patients with thyroid cancer. Exp. Ther. Med. 2017, 13, 2274–2278. [Google Scholar] [CrossRef] [PubMed]
- Di, W.; Li, Q.; Shen, W.; Guo, H.; Zhao, S. The long non-coding RNA HOTAIR promotes thyroid cancer cell growth, invasion and migration through the miR-1-CCND2 axis. Am. J. Cancer Res. 2017, 7, 1298–1309. [Google Scholar] [PubMed]
- Congrains, A.; Kamide, K.; Ohishi, M.; Rakugi, H. ANRIL: Molecular mechanisms and implications in human health. Int. J. Mol. Sci. 2013, 14, 1278–1292. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Ink4-Arf locus in cancer and aging. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Pasmant, E.; Laurendeau, I.; Heron, D.; Vidaud, M.; Vidaud, D.; Bieche, I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007, 67, 3963–3969. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, X.; Shen, J. ANRIL: A pivotal tumor suppressor long non-coding RNA in human cancers. Tumour Biol. 2016, 37, 5657–5661. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Mathur, R.; Hu, X.; Liu, Y.; Zhang, X.; Peng, G.; Lu, X. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell. Signal. 2013, 25, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, C.; Borgel, J.; Court, F.; Cathala, G.; Forne, T.; Piette, J. CTCF is a DNA methylation-sensitive positive regulator of the INK/ARF locus. Biochem. Biophys. Res. Commun. 2010, 392, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Maurano, M.T.; Qu, H.; Varley, K.E.; Gertz, J.; Pauli, F.; Lee, K.; Canfield, T.; Weaver, M.; Sandstrom, R.; et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012, 22, 1680–1688. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.C.; Felsenfeld, G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 2000, 405, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Cunnington, M.S.; Santibanez Koref, M.; Mayosi, B.M.; Burn, J.; Keavney, B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Sharma, R.B.; Nwosu, B.U.; Alonso, L.C. Islet biology, the CDKN2A/B locus and type 2 diabetes risk. Diabetologia 2016, 59, 1579–1593. [Google Scholar] [CrossRef] [PubMed]
- Murray, R.; Bryant, J.; Titcombe, P.; Barton, S.J.; Inskip, H.; Harvey, N.C.; Cooper, C.; Lillycrop, K.; Hanson, M.; Godfrey, K.M. DNA methylation at birth within the promoter of ANRIL predicts markers of cardiovascular risk at 9 years. Clin. Epigenetics 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Lillycrop, K.; Murray, R.; Cheong, C.; Teh, A.L.; Clarke-Harris, R.; Barton, S.; Costello, P.; Garratt, E.; Cook, E.; Titcombe, P.; et al. ANRIL promoter DNA methylation: A perinatal marker for later adiposity. EBioMedicine 2017, 19, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Helgadottir, A.; Thorleifsson, G.; Manolescu, A.; Gretarsdottir, S.; Blondal, T.; Jonasdottir, A.; Jonasdottir, A.; Sigurdsson, A.; Baker, A.; Palsson, A.; et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007, 316, 1491–1493. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, H.M.; Peden, J.F.; Lorkowski, S.; Goel, A.; Ongen, H.; Green, F.; Clarke, R.; Collins, R.; Franzosi, M.G.; Tognoni, G.; et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 2008, 17, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Cai, C.; Feng, S.; Lv, J.; Li, S.; Chang, B.; Zhang, H.; Shi, W.; Han, H.; Ling, C.; et al. TOX and CDKN2A/B gene polymorphisms are associated with type 2 diabetes in Han Chinese. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Cai, M.Y.; Chen, Y.N.; Li, Z.C.; Tang, S.S.; Yang, X.L.; Chen, C.; Liu, X.; Xiong, X.D. Variants in ANRIL gene correlated with its expression contribute to myocardial infarction risk. Oncotarget 2017, 8, 12607–12619. [Google Scholar] [CrossRef] [PubMed]
- Mafi Golchin, M.; Ghaderian, S.M.H.; Akhavan-Niaki, H.; Jalalian, R.; Heidari, L.; Salami, S.A. Analysis of two CDKN2B-AS polymorphisms in relation to coronary artery disease patients in North of Iran. Int. J. Mol. Cell. Med. 2017, 6, 31–37. [Google Scholar] [PubMed]
- Arbiol-Roca, A.; Padro-Miquel, A.; Hueso, M.; Navarro, E.; Alia-Ramos, P.; Gonzalez-Alvarez, M.T.; Rama, I.; Torras, J.; Grinyo, J.M.; Cruzado, J.M.; et al. Association of ANRIL gene polymorphisms with major adverse cardiovascular events in hemodialysis patients. Clin. Chim. Acta 2017, 466, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Sharma, R.B.; Ly, S.; Stamateris, R.E.; Jesdale, W.M.; Alonso, L.C. CDKN2A/B T2D genome-wide association study risk SNPs impact locus gene expression and proliferation in human islets. Diabetes 2018, 67, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Bochenek, G.; Hasler, R.; El Mokhtari, N.E.; Konig, I.R.; Loos, B.G.; Jepsen, S.; Rosenstiel, P.; Schreiber, S.; Schaefer, A.S. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum. Mol. Genet. 2013, 22, 4516–4527. [Google Scholar] [CrossRef] [PubMed]
- Roux, B.T.; Heward, J.A.; Donnelly, L.E.; Jones, S.W.; Lindsay, M.A. Catalog of differentially expressed long non-coding RNA following activation of human and mouse innate immune response. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, I.H.; Narra, H.P.; Sahni, A.; Khanipov, K.; Schroeder, C.L.C.; Patel, J.; Fofanov, Y.; Sahni, S.K. Expression profiling of long non-coding RNA splice variants in human microvascular endothelial cells: Lipopolysaccharide effects in vitro. Med. Inflamm. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Falls, J.G.; Pulford, D.J.; Wylie, A.A.; Jirtle, R.L. Genomic imprinting: Implications for human disease. Am. J. Pathol. 1999, 154, 635–647. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Leibovitch, M.P.; Nguyen, V.C.; Gross, M.S.; Solhonne, B.; Leibovitch, S.A.; Bernheim, A. The human ASM (adult skeletal muscle) gene: Expression and chromosomal assignment to 11p15. Biochem. Biophys. Res. Commun. 1991, 180, 1241–1250. [Google Scholar] [CrossRef]
- Sasaki, H.; Ishihara, K.; Kato, R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J. Biochem. 2000, 127, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Rainier, S.; Johnson, L.A.; Dobry, C.J.; Ping, A.J.; Grundy, P.E.; Feinberg, A.P. Relaxation of imprinted genes in human cancer. Nature 1993, 362, 747–749. [Google Scholar] [CrossRef] [PubMed]
- Raveh, E.; Matouk, I.J.; Gilon, M.; Hochberg, A. The H19 long non-coding RNA in cancer initiation, progression and metastasis—A proposed unifying theory. Mol Cancer 2015, 14. [Google Scholar] [CrossRef] [PubMed]
- Bauderlique-Le Roy, H.; Vennin, C.; Brocqueville, G.; Spruyt, N.; Adriaenssens, E.; Bourette, R.P. Enrichment of human stem-like prostate cells with s-SHIP promoter activity uncovers a role in stemness for the long noncoding RNA H19. Stem. Cells Dev. 2015, 24, 1252–1262. [Google Scholar] [CrossRef] [PubMed]
- Shima, H.; Kida, K.; Adachi, S.; Yamada, A.; Sugae, S.; Narui, K.; Miyagi, Y.; Nishi, M.; Ryo, A.; Murata, S.; et al. lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res. Treat. 2018, 170, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Yuan, W.; Wu, S.; Wang, Z.; Mao, L.; Tian, T.; Lu, Y.; Zhu, B.; Yang, Y.; Wang, B.; et al. Quantitative assessment of polymorphisms in H19 lncRNA and cancer risk: A meta-analysis of 13,392 cases and 18,893 controls. Oncotarget 2016, 7, 78631–78639. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Wang, Y.F.; Zhang, X.Q.; Lv, J.M.; Li, Y.; Liu, X.X.; Xu, T.P. H19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma 2016, 63, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.T.; Pan, H.; Xia, G.F.; Qiu, C.; Zhu, Z.M. Prognostic and clinicopathological significance of long noncoding RNA H19 overexpression in human solid tumors: Evidence from a meta-analysis. Oncotarget 2016, 7, 83177–83186. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, H.; Matsuda, Y.; Yamamoto, M.; Michishita, M.; Takahashi, K.; Sasaki, N.; Ishikawa, N.; Aida, J.; Takubo, K.; Arai, T.; et al. Reduced expression of the H19 long non-coding RNA inhibits pancreatic cancer metastasis. Lab. Investig. 2018, 98, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Liu, Z.; Fang, J. H19 functions as a competing endogenous RNA to regulate human epidermal growth factor receptor expression by sequestering let7c in gastric cancer. Mol. Med. Rep. 2018, 17, 2600–2606. [Google Scholar] [PubMed]
- Sun, X.; Liu, J.; Xu, C.; Tang, S.C.; Ren, H. The insights of Let-7 miRNAs in oncogenesis and stem cell potency. J. Cell. Mol. Med. 2016, 20, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Boyerinas, B.; Park, S.M.; Hau, A.; Murmann, A.E.; Peter, M.E. The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer 2010, 17, F19–F36. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Li, T.T.; Wang, K.L.; Xiao, G.Q.; Wang, J.H.; Zhao, H.D.; Kang, Z.J.; Fan, W.J.; Zhu, L.L.; Li, M.; et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Zhu, H. Lin28 and let-7 in cell metabolism and cancer. Transl. Pediatr. 2015, 4, 4–11. [Google Scholar] [PubMed]
- Yan, J.; Zhang, Y.; She, Q.; Li, X.; Peng, L.; Wang, X.; Liu, S.; Shen, X.; Zhang, W.; Dong, Y.; et al. Long non-coding RNA H19/miR-675 axis promotes gastric cancer via FADD/Caspase 8/Caspase 3 signaling pathway. Cell. Physiol. Biochem. 2017, 42, 2364–2376. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Wang, L.; Zhang, J.; Lin, R.; Wang, L.; Sun, W.; Wu, H.; Xin, S. Long non-coding RNA H19-derived miR-675 aggravates restenosis by targeting PTEN. Biochem. Biophys. Res. Commun. 2018, 497, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Bu, D.; Ma, Y.; Zhu, J.; Chen, G.; Sun, L.; Zuo, S.; Li, T.; Pan, Y.; Wang, X.; et al. H19 overexpression induces resistance to 1, 25(OH)2D3 by targeting VDR through miR-675-5p in colon cancer cells. Neoplasia 2017, 19, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.W.; Zhang, H.; Duan, C.J.; Gao, Y.; Cheng, Y.D.; He, D.; Li, R.; Zhang, C.F. miR-675-5p enhances tumorigenesis and metastasis of esophageal squamous cell carcinoma by targeting REPS2. Oncotarget 2016, 7, 30730–30747. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, M.; Gao, W.; Xu, J.; Wang, P.; Shu, Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem. Biophys. Res. Commun. 2014, 448, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.M.; Elahi, A.; Clark, C.W.; Wang, J.; Humphries, L.A.; Centeno, B.; Bloom, G.; Fuchs, B.C.; Yeatman, T.; Shibata, D. miR-675 mediates downregulation of Twist1 and Rb in AFP-secreting hepatocellular carcinoma. Ann. Surg. Oncol. 2013, 20 (Suppl. 3), S625–S635. [Google Scholar] [CrossRef] [PubMed]
- Tsang, W.P.; Ng, E.K.; Ng, S.S.; Jin, H.; Yu, J.; Sung, J.J.; Kwok, T.T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010, 31, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yu, B.; Li, J.; Su, L.; Yan, M.; Zhu, Z.; Liu, B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014, 5, 2318–2329. [Google Scholar] [CrossRef] [PubMed]
- Vennin, C.; Spruyt, N.; Dahmani, F.; Julien, S.; Bertucci, F.; Finetti, P.; Chassat, T.; Bourette, R.P.; Le Bourhis, X.; Adriaenssens, E. H19 non-coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 2015, 6, 29209–29223. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Chen, Q.; Liu, X.; Sun, Q.; Zhao, X.; Deng, R.; Wang, Y.; Huang, J.; Xu, M.; Yan, J.; et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014, 281, 3766–3775. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, Y.; Luan, W.; Wang, P.; Tao, T.; Zhang, J.; Qian, J.; Liu, N.; You, Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, C.; Sun, L.; Huang, D.; Li, T.; He, X.; Wu, G.; Yang, Z.; Zhong, X.; Song, L.; et al. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Shyh-Chang, N.; Zhu, H.; Yvanka de Soysa, T.; Shinoda, G.; Seligson, M.T.; Tsanov, K.M.; Nguyen, L.; Asara, J.M.; Cantley, L.C.; Daley, G.Q. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 2013, 155, 778–792. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Shyh-Chang, N.; Segre, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.P.; Kharas, M.G.; et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wu, F.; Zhou, J.; Yan, L.; Jurczak, M.J.; Lee, H.Y.; Yang, L.; Mueller, M.; Zhou, X.B.; Dandolo, L.; et al. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucl. Acids Res. 2014, 42, 13799–13811. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Li, W.; Zhang, M. The function of the RNA-binding protein hnRNP in cancer metastasis. J. Cancer Res. Ther. 2013, 9, S129–S134. [Google Scholar] [PubMed]
- Bi, H.S.; Yang, X.Y.; Yuan, J.H.; Yang, F.; Xu, D.; Guo, Y.J.; Zhang, L.; Zhou, C.C.; Wang, F.; Sun, S.H. H19 inhibits RNA polymerase II-mediated transcription by disrupting the hnRNP U-actin complex. Biochim. Biophys. Acta 2013, 1830, 4899–4906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, F.; Yuan, J.H.; Yuan, S.X.; Zhou, W.P.; Huo, X.S.; Xu, D.; Bi, H.S.; Wang, F.; Sun, S.H. Epigenetic activation of the miR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 2013, 34, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Bartolomei, M.S.; Ferguson-Smith, A.C. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Kelsey, G.; Feil, R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368. [Google Scholar] [CrossRef] [PubMed]
- Rozek, L.S.; Dolinoy, D.C.; Sartor, M.A.; Omenn, G.S. Epigenetics: relevance and implications for public health. Annu. Rev. Public Health 2014, 35, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Khadilkar, A.V.; Sanwalka, N.J.; Chiplonkar, S.A.; Khadilkar, V.V.; Pandit, D. Body fat reference percentiles on healthy affluent Indian children and adolescents to screen for adiposity. Int. J. Obes. 2013, 37, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Valero, M.A.; Rother, J.; Gorlov, I.; Frazier, M.; Gorlova, O. Interplay between polymorphisms and methylation in the H19/IGF2 gene region may contribute to obesity in Mexican-American children. J. Dev. Orig. Health Dis. 2013, 4, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Perkins, E.; Murphy, S.K.; Murtha, A.P.; Schildkraut, J.; Jirtle, R.L.; Demark-Wahnefried, W.; Forman, M.R.; Kurtzberg, J.; Overcash, F.; Huang, Z.; et al. Insulin-like growth factor 2/H19 methylation at birth and risk of overweight and obesity in children. J. Pediatr. 2012, 161, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.; Lee, J.Y.; Donaldson, A.V.; Natanek, S.A.; Vaidyanathan, S.; Man, W.D.; Hopkinson, N.S.; Sayer, A.A.; Patel, H.P.; Cooper, C.; et al. Increased expression of H19/miR-675 is associated with a low fat-free mass index in patients with COPD. J. Cachexia Sarcopenia Muscle 2016, 7, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dang, Y.; Liu, J.; Ouyang, X. The function of homeobox genes and lncRNAs in cancer. Oncol. Lett. 2016, 12, 1635–1641. [Google Scholar] [CrossRef] [PubMed]
- Mallo, M.; Alonso, C.R. The regulation of Hox gene expression during animal development. Development 2013, 140, 3951–3963. [Google Scholar] [CrossRef] [PubMed]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. 2014, 92, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Hajjari, M.; Salavaty, A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 2015, 12, 1–9. [Google Scholar] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wu, X.Y.; Wu, C.L. The association between lncRNA HOTAIR and cancer lymph node metastasis and distant metastasis: A meta-analysis. Neoplasma 2018, 65, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, Y.; Xu, T.; Tian, W.; Yang, C.; Wang, X.; Zhong, S.; Ran, Q.; Yang, H.; Zhu, S. Clinical value of long noncoding RNA HOTAIR as a novel biomarker in digestive cancers: A meta-analysis. Technol. Cancer Res. Treat. 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Hruby, A.; Hu, F.B. The epidemiology of obesity: A big picture. Pharmacoeconomics 2015, 33, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Bai, D.; Liu, X.; Zhou, C.; Yang, G. Sedentary lifestyle related exosomal release of HOTAIR from gluteal-femoral fat promotes intestinal cell proliferation. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Divoux, A.; Karastergiou, K.; Xie, H.; Guo, W.; Perera, R.J.; Fried, S.K.; Smith, S.R. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity 2014, 22, 1781–1785. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.; Anderson, C.; Lippman, S.M. Physical activity, sedentary behaviour, diet, and cancer: An update and emerging new evidence. Lancet Oncol. 2017, 18, e457–e471. [Google Scholar] [CrossRef]
- Shen, D.; Mao, W.; Liu, T.; Lin, Q.; Lu, X.; Wang, Q.; Lin, F.; Ekelund, U.; Wijndaele, K. Sedentary behavior and incident cancer: A meta-analysis of prospective studies. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
| Type of Cancer | ANRIL | H19 | HOTAIR |
|---|---|---|---|
| Endometrial cancer | Upregulated [58] | Upregulated [59] | Upregulated [60,61] |
| Esophageal adenocarcinoma | Upregulated [62] | Upregulated [63,64] | Upregulated [65,66] |
| Liver cancer | Upregulated [67,68] | Upregulated [69], Downregulated [70] | Upregulated [71] |
| Pancreatic cancer | Upregulated [72,73] | Upregulated [74] | Upregulated [75,76] |
| Colorectal cancer | Upregulated [77,78] | Upregulated [64,79], Downregulated [80] | Upregulated [81,82] |
| Gallbladder cancer | Upregulated [83] | Upregulated [84,85] | Upregulated [86] |
| Breast cancer | Upregulated [87] | Upregulated [88] | Upregulated [89] |
| Ovarian cancer | Upregulated [90] | Upregulated [91] | Upregulated [92] |
| Thyroid cancer | Upregulated [93] | Downregulated [94] | Upregulated [95,96] |
| SNP-ID | Related Diseases | Remarks | References |
|---|---|---|---|
| rs10757278 | Myocardial infarction | [110] | |
| rs2891168 | Coronary artery disease | G-allele was associated with lower triglyceride level | [111] |
| rs10811661 | Type 2 diabetes | [111,112] | |
| rs10965215 and rs10738605 | Myocardial infarction | [113] | |
| rs10757274 and rs1333042 | Coronary artery disease | [114] | |
| rs10757278 | Major adverse cardio-vascular event (MACE) in patients starting on hemodialysis | [115] | |
| rs564398 | Type 2 diabetes | Reduced β-cell proliferation | [116] |
| Targeted mRNA | Targeted Region | Related Cancer or Diseases | References |
|---|---|---|---|
| FADD | 3′-UTR | Gastric cancer | [139] |
| PTEN | 3′-UTR | Restenosis | [140] |
| Vitamin D receptor | 3′-UTR | Colon cancer | [141] |
| REPS2 | 3′-UTR | Esophageal squamous cell carcinoma | [142] |
| RUNX1 | 3′-UTR | Gastric cancer | [143] |
| TWIST1 | 3′-UTR | AFP-secreting hepatocellular carcinoma | [144] |
| Retinoblastoma | 3′-UTR | AFP-secreting hepatocellular carcinoma, Colorectal cancer, glioma | [144,145] |
| CALN1 | 3′-UTR | Gastric cancer | [146] |
| c-Cbl | coding sequence | Breast cancer | [147] |
| Cbl-b | coding sequence | Breast cancer | [147] |
| TGFBI | 3′-UTR | Prostate cancer | [148] |
| Cadherin 13 | 3′-UTR | Glioma development | [149] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yau, M.Y.-C.; Xu, L.; Huang, C.-L.; Wong, C.-M. Long Non-Coding RNAs in Obesity-Induced Cancer. Non-Coding RNA 2018, 4, 19. https://doi.org/10.3390/ncrna4030019
Yau MY-C, Xu L, Huang C-L, Wong C-M. Long Non-Coding RNAs in Obesity-Induced Cancer. Non-Coding RNA. 2018; 4(3):19. https://doi.org/10.3390/ncrna4030019
Chicago/Turabian StyleYau, Mabel Yin-Chun, Lu Xu, Chien-Ling Huang, and Chi-Ming Wong. 2018. "Long Non-Coding RNAs in Obesity-Induced Cancer" Non-Coding RNA 4, no. 3: 19. https://doi.org/10.3390/ncrna4030019
APA StyleYau, M. Y.-C., Xu, L., Huang, C.-L., & Wong, C.-M. (2018). Long Non-Coding RNAs in Obesity-Induced Cancer. Non-Coding RNA, 4(3), 19. https://doi.org/10.3390/ncrna4030019





