Z Probe, An Efficient Tool for Characterizing Long Non-Coding RNA in FFPE Tissues
Abstract
1. Introduction
2. Results
2.1. Schematic and Controls
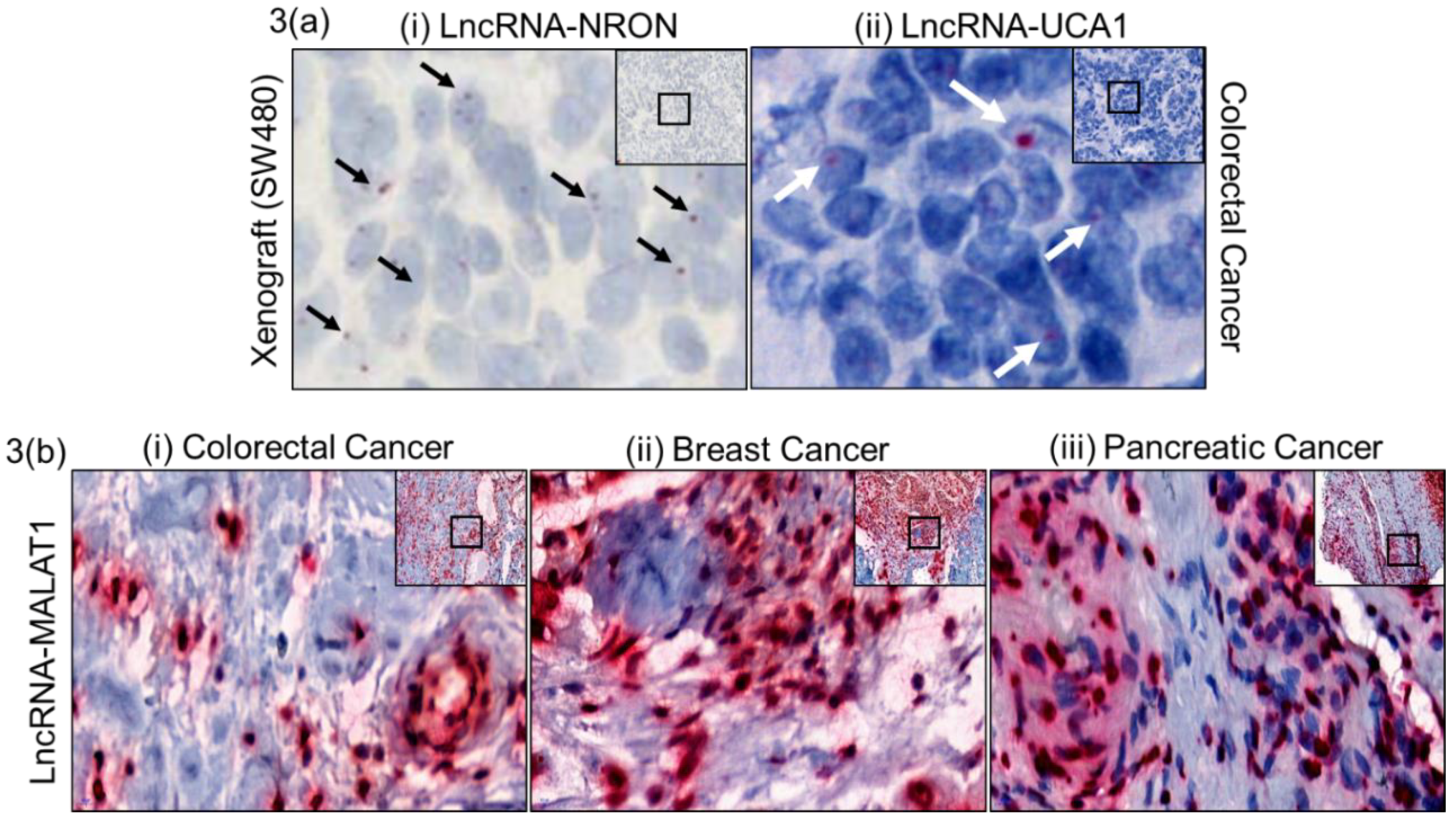
2.2. Assay Validation
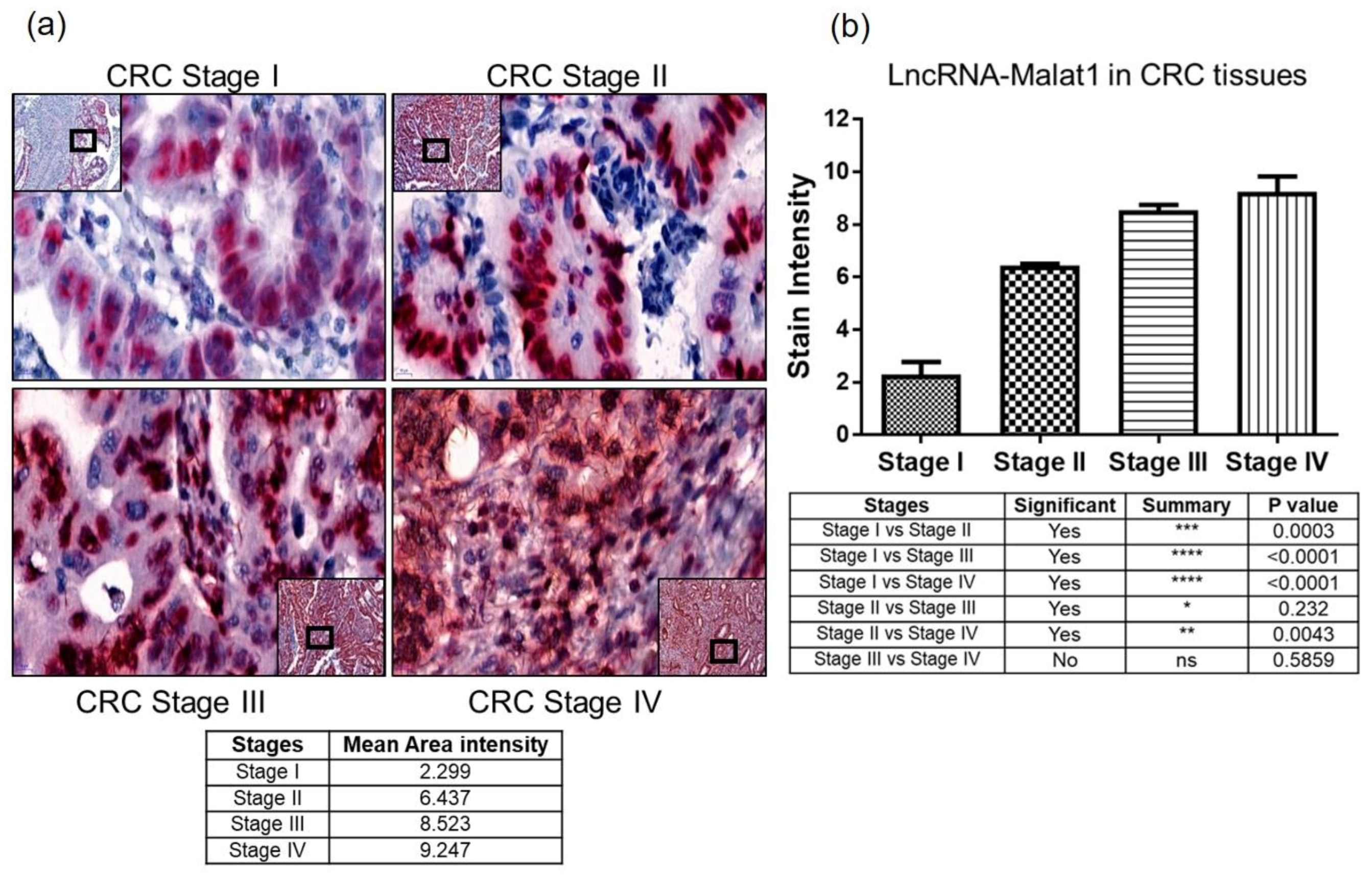
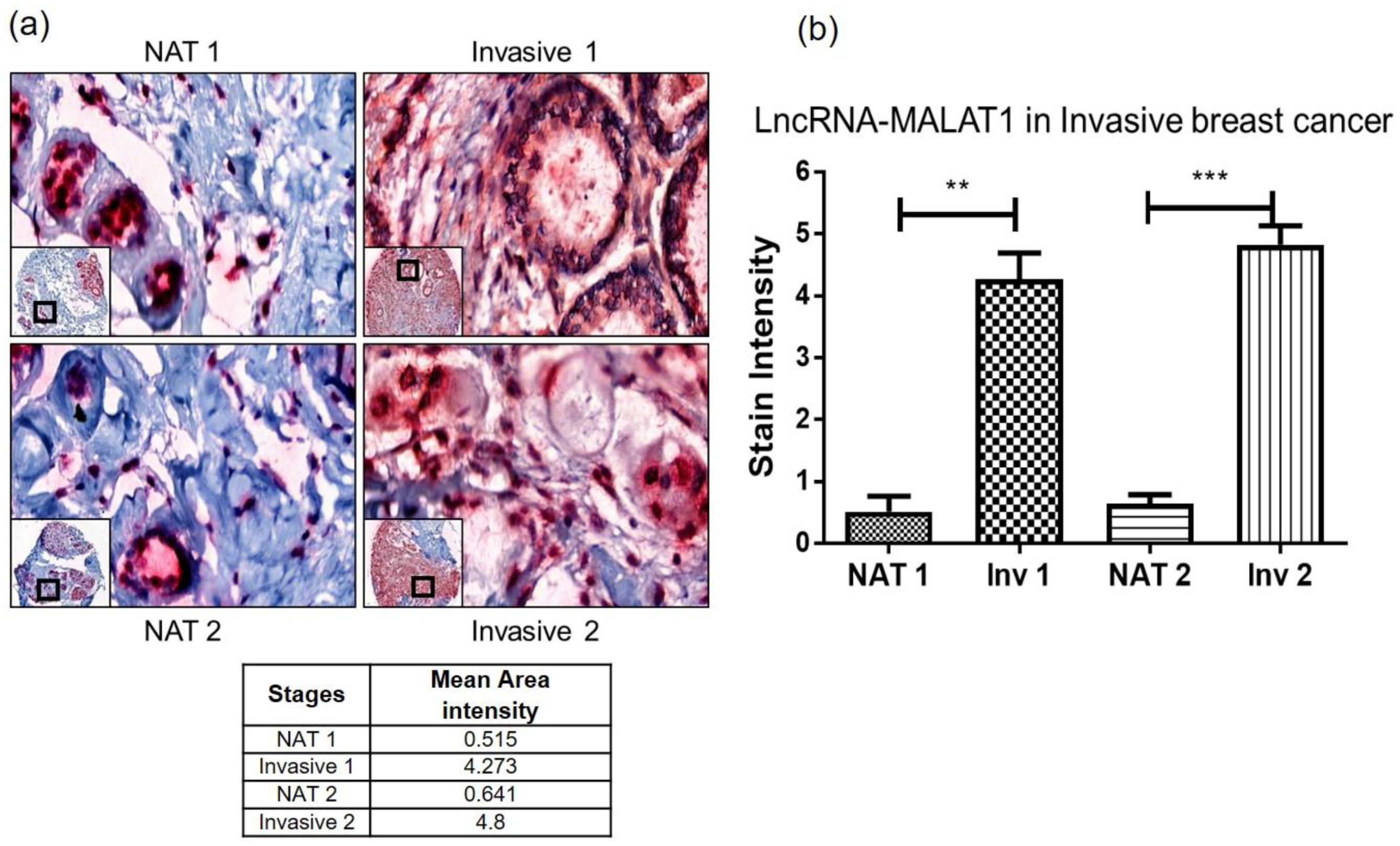
2.3. Quantitative Measure of Progression and Invasiveness
3. Discussion
4. Materials and Methods
4.1. Research Involving Human Tissues
4.2. Chromogenic Staining in FFPE Tissues
4.3. Quantitation (ImageJ) Analysis
Summary of ImageJ Analysis
4.4. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, W.; Zhang, R.; Li, J.; Li, S.; Ma, Y.; Jin, W.; Wang, K. Overexpressed long noncoding RNA CRNDE with distinct alternatively spliced isoforms in multiple cancers. Front. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Richtig, G.; Haemmerle, M.; Pichler, M. From biomarkers to therapeutic targets-the promises and perils of long non-coding RNAs in cancer. Cancer Metastasis Rev. 2018, 37, 83–105. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Hodges, T.R.; Song, R.; Gong, Y.; Calin, G.A.; Heimberger, A.B.; Zhao, H. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol. Carcinog. 2018, 57, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Wang, K.; Zhang, R.; Zou, S. Long noncoding RNA MALAT1: A potential novel prognostic biomarkers in cancers based on Meta-analysis. Zhong Nan Da Xue Xue Bao Yi Xue Ban/J. Cent. South Univ. Med. Sci. 2016, 41, 1163–1167. [Google Scholar]
- Kong, H.; Wu, Y.; Zhu, M.; Zhai, C.; Qian, J.; Gao, X.; Wang, S.; Hou, Y.; Lu, S.; Zhu, H. Long non-coding RNAs: Novel prognostic biomarkers for liver metastases in patients with early stage colorectal cancer. Oncotarget 2016, 7, 50428–50436. [Google Scholar] [CrossRef] [PubMed]
- Matos, L.L.; Trufelli, D.C.; de Matos, M.G.; da Silva Pinhal, M.A. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark. Insights 2010, 5, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, R.; Nielsen, P.S.; Jensen, T.H. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA 2005, 11, 1745–1748. [Google Scholar] [CrossRef] [PubMed]
- Weiszmann, R.; Hammonds, A.S.; Celniker, S.E. Determination of gene expression patterns using high-throughput RNA in situ hybridization to whole-mount Drosophila embryos. Nat. Protoc. 2009, 4, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Weaver, D.L.; Olsen, D.; Peng, Z.; Ashikaga, T.; Evans, M.F. Long non-coding RNA chromogenic in situ hybridisation signal pattern correlation with breast tumour pathology. J. Clin. Pathol. 2016, 69, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.T.; Ma, X.J.; Luo, Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Velcheti, V.; Schalper, K.A.; Carvajal, D.E.; Anagnostou, V.K.; Syrigos, K.N.; Sznol, M.; Herbst, R.S.; Gettinger, S.N.; Chen, L.; Rimm, D.L. Programmed death ligand-1 expression in non-small cell lung cancer. Lab. Investig. J. Tech. Methods Pathol. 2014, 94, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Guertler, R.; Naim, S.; Nixdorf, S.; Fedier, A.; Hacker, N.F.; Heinzelmann-Schwarz, V. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS ONE 2013, 8, e59180. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef] [PubMed]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Findlay, G.M.; Bandukwala, H.S.; Oberdoerffer, S.; Baust, B.; Li, Z.; Schmidt, V.; Hogan, P.G.; Sacks, D.B.; Rao, A. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc. Natl. Acad. Sci. USA 2011, 108, 11381–11386. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liang, G.; Yang, S.; Sui, J.; Yao, W.; Shen, X.; Zhang, Y.; Peng, H.; Hong, W.; Xu, S.; et al. Dysregulated lncRNA-UCA1 contributes to the progression of gastric cancer through regulation of the PI3K-Akt-mTOR signaling pathway. Oncotarget 2017, 8, 93476–93491. [Google Scholar] [PubMed]
- Wang, Z.Q.; Cai, Q.; Hu, L.; He, C.Y.; Li, J.F.; Quan, Z.W.; Liu, B.Y.; Li, C.; Zhu, Z.G. Long noncoding RNA UCA1 induced by SP1 promotes cell proliferation via recruiting EZH2 and activating AKT pathway in gastric cancer. Cell Death Dis. 2017, 8, e2839. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhou, N.; Watabe, K.; Lu, Z.; Wu, F.; Xu, M.; Mo, Y.Y. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014, 5, e1008. [Google Scholar] [CrossRef] [PubMed]
- Jadaliha, M.; Zong, X.; Malakar, P.; Ray, T.; Singh, D.K.; Freier, S.M.; Jensen, T.; Prasanth, S.G.; Karni, R.; Ray, P.S.; et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 2016, 7, 40418–40436. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Cui, Y.; Liu, L.F.; Ren, W.B.; Li, Q.Q.; Zhou, X.; Li, Y.L.; Li, Y.; Bai, X.Y.; Zu, X.B. High Expression of Long Noncoding RNA MALAT1 Indicates a Poor Prognosis and Promotes Clinical Progression and Metastasis in Bladder Cancer. Clin. Genitourin. Cancer 2017, 15, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.T.; Shi, D.B.; Wang, Y.W.; Li, X.X.; Xu, Y.; Tripathi, P.; Gu, W.L.; Cai, G.X.; Cai, S.J. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3174–3181. [Google Scholar] [PubMed]
- Handa, H.; Kuroda, Y.; Kimura, K.; Masuda, Y.; Hattori, H.; Alkebsi, L.; Matsumoto, M.; Kasamatsu, T.; Kobayashi, N.; Tahara, K.I.; et al. Long non-coding RNA MALAT1 is an inducible stress response gene associated with extramedullary spread and poor prognosis of multiple myeloma. Br. J. Haematol. 2017, 179, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Pan, X.; Wang, X.; Jiao, X.; Zheng, J.; Li, Z.; Huo, Y. Long noncoding RNA MALAT1 promotes cell proliferation through suppressing miR-205 and promoting SMAD4 expression in osteosarcoma. Oncotarget 2017, 8, 106648–106660. [Google Scholar] [CrossRef] [PubMed]
- Xiping, Z.; Bo, C.; Shifeng, Y.; Feijiang, Y.; Hongjian, Y.; Qihui, C.; Binbin, T. Roles of MALAT1 in development and migration of triple negative and Her-2 positive breast cancer. Oncotarget 2018, 9, 2255–2267. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.S.; Chi, Y.Y.; Xue, J.Y.; Liu, M.Y.; Huang, S.; Mo, M.; Zhou, S.L.; Wu, J. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget 2016, 7, 37957–37965. [Google Scholar] [CrossRef] [PubMed]
- Soudyab, M.; Iranpour, M.; Ghafouri-Fard, S. The Role of Long Non-Coding RNAs in Breast Cancer. Arch. Iran. Med. 2016, 19, 508–517. [Google Scholar] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripathi, M.K.; Zacheaus, C.; Doxtater, K.; Keramatnia, F.; Gao, C.; Yallapu, M.M.; Jaggi, M.; Chauhan, S.C. Z Probe, An Efficient Tool for Characterizing Long Non-Coding RNA in FFPE Tissues. Non-Coding RNA 2018, 4, 20. https://doi.org/10.3390/ncrna4030020
Tripathi MK, Zacheaus C, Doxtater K, Keramatnia F, Gao C, Yallapu MM, Jaggi M, Chauhan SC. Z Probe, An Efficient Tool for Characterizing Long Non-Coding RNA in FFPE Tissues. Non-Coding RNA. 2018; 4(3):20. https://doi.org/10.3390/ncrna4030020
Chicago/Turabian StyleTripathi, Manish K., Chidi Zacheaus, Kyle Doxtater, Fatemeh Keramatnia, Cuilan Gao, Murali M. Yallapu, Meena Jaggi, and Subhash C. Chauhan. 2018. "Z Probe, An Efficient Tool for Characterizing Long Non-Coding RNA in FFPE Tissues" Non-Coding RNA 4, no. 3: 20. https://doi.org/10.3390/ncrna4030020
APA StyleTripathi, M. K., Zacheaus, C., Doxtater, K., Keramatnia, F., Gao, C., Yallapu, M. M., Jaggi, M., & Chauhan, S. C. (2018). Z Probe, An Efficient Tool for Characterizing Long Non-Coding RNA in FFPE Tissues. Non-Coding RNA, 4(3), 20. https://doi.org/10.3390/ncrna4030020





