Functions of RNAi Pathways in Ribosomal RNA Regulation
Abstract
1. Introduction
1.1. Ribosomal DNA Loci and Ribosome Biogenesis
1.2. Small RNA Pathways
2. The Role of siRNAs in the Nucleolar Dominance
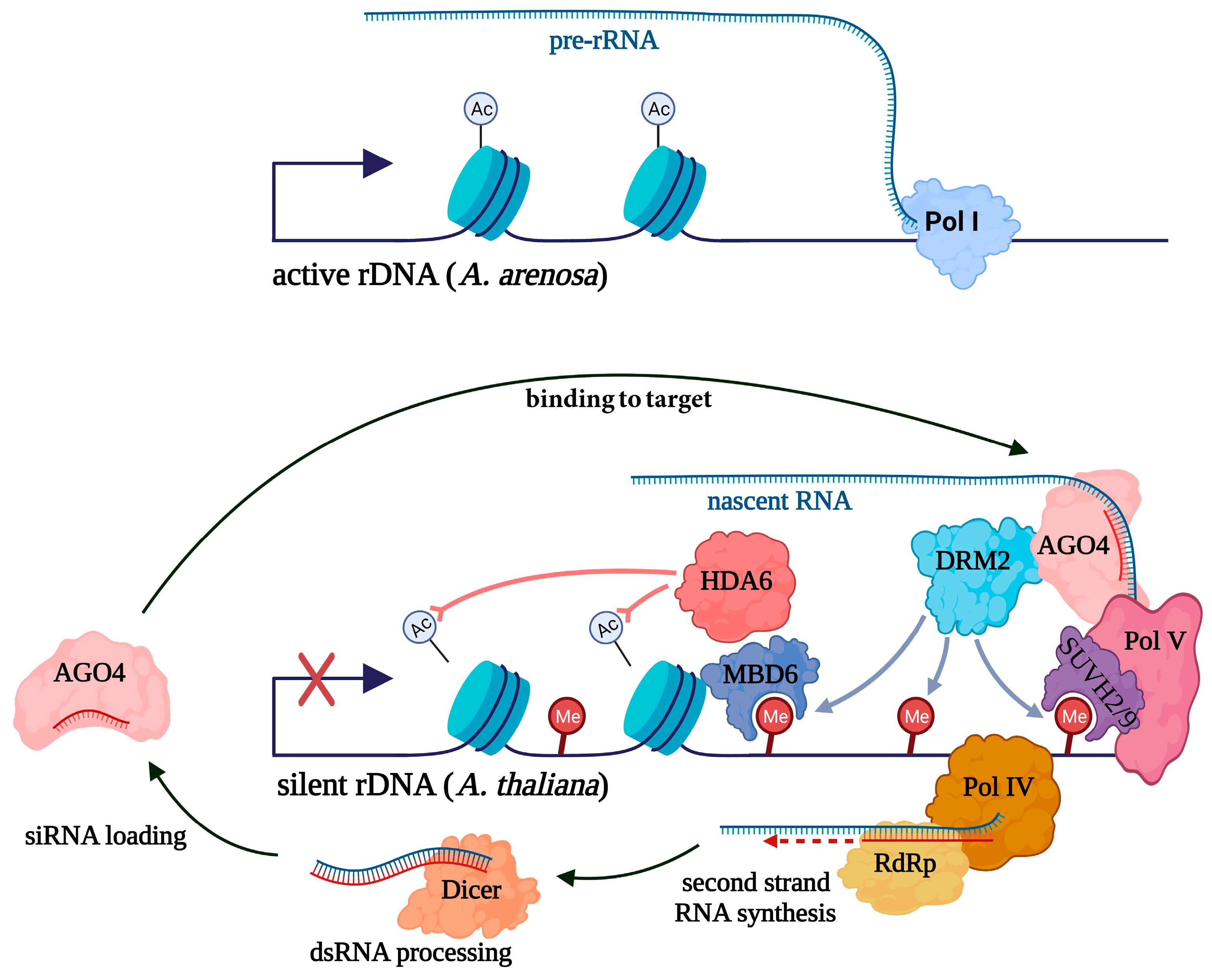
3. siRNAs Induce Selective Repression of 5S rRNA Gene Variants in Plants
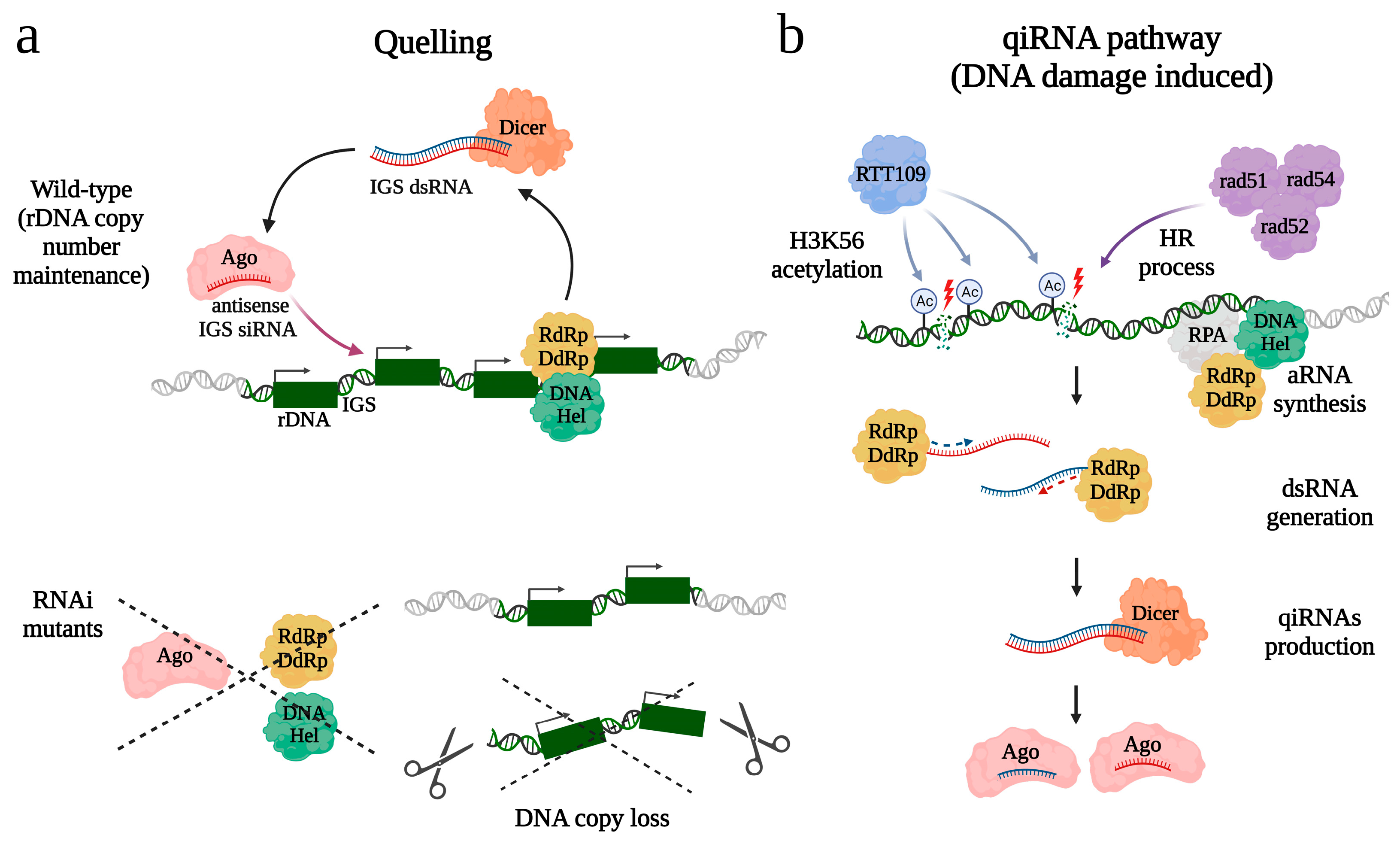
4. RNAi Pathways and rDNA Integrity
5. Aberrant rRNAs Serve as Sources of Antisense Small RNAs That Can Induce rDNA Silencing
6. Implications of the piRNA Pathway in rRNA Regulation
7. Potential Mechanisms of rDNA Silencing by RNAi in Animals
8. Conclusions and Perspective
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef] [PubMed]
- McStay, B. Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination. Genes. Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef]
- Ciganda, M.; Williams, N. Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip. Rev. RNA 2011, 2, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.D.; Karbstein, K. Quality control ensures fidelity in ribosome assembly and cellular health. J. Cell Biol. 2023, 222, e202209115. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar] [CrossRef] [PubMed]
- Grummt, I. The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma 2013, 122, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Verheyden, Y.; Goedert, L.; Leucci, E. Control of nucleolar stress and translational reprogramming by lncRNAs. Cell Stress. 2018, 3, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Pirogov, S.A.; Gvozdev, V.A.; Klenov, M.S. Long Noncoding RNAs and Stress Response in the Nucleolus. Cells 2019, 8, 668. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Kuroda, T.; Kishimoto, H.; Wang, C.; Iwama, A.; Kimura, K. Downregulation of rRNA transcription triggers cell differentiation. PLoS ONE 2014, 9, e98586. [Google Scholar] [CrossRef]
- Woolnough, J.L.; Atwood, B.L.; Liu, Z.; Zhao, R.; Giles, K.E. The Regulation of rRNA Gene Transcription during Directed Differentiation of Human Embryonic Stem Cells. PLoS ONE 2016, 11, e0157276. [Google Scholar] [CrossRef]
- Le Goff, S.; Boussaid, I.; Floquet, C.; Raimbault, A.; Hatin, I.; Andrieu-Soler, C.; Salma, M.; Leduc, M.; Gautier, E.F.; Guyot, B.; et al. p53 activation during ribosome biogenesis regulates normal erythroid differentiation. Blood 2021, 137, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hu, X.; Qiu, D.; Zhang, Z.; Lei, L. rDNA Transcription in Developmental Diseases and Stem Cells. Stem Cell Rev. Rep. 2023, 19, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Buszczak, M. Ribosome biogenesis and function in development and disease. Development 2023, 150, dev201187. [Google Scholar] [CrossRef] [PubMed]
- Breznak, S.M.; Kotb, N.M.; Rangan, P. Dynamic regulation of ribosome levels and translation during development. Semin. Cell Dev. Biol. 2023, 136, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Thomas, G.; Volarevic, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Schofer, C.; Weipoltshammer, K. Nucleolus and chromatin. Histochem. Cell Biol. 2018, 150, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Srivastava, R.; Ahn, S.H. The Epigenetic Pathways to Ribosomal DNA Silencing. Microbiol. Mol. Biol. Rev. 2016, 80, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Herdman, C.; Mars, J.C.; Stefanovsky, V.Y.; Tremblay, M.G.; Sabourin-Felix, M.; Lindsay, H.; Robinson, M.D.; Moss, T. A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription. PLoS Genet. 2017, 13, e1006899. [Google Scholar] [CrossRef]
- Zentner, G.E.; Saiakhova, A.; Manaenkov, P.; Adams, M.D.; Scacheri, P.C. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011, 39, 4949–4960. [Google Scholar] [CrossRef]
- Li, J.; Langst, G.; Grummt, I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006, 25, 5735–5741. [Google Scholar] [CrossRef]
- Hori, Y.; Engel, C.; Kobayashi, T. Regulation of ribosomal RNA gene copy number, transcription and nucleolus organization in eukaryotes. Nat. Rev. Mol. Cell Biol. 2023, 24, 414–429. [Google Scholar] [CrossRef] [PubMed]
- Kindelay, S.M.; Maggert, K.A. Under the magnifying glass: The ups and downs of rDNA copy number. Semin. Cell Dev. Biol. 2023, 136, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Mohan, J.; Ritossa, F.M. Regulation of ribosomal RNA synthesis and its bearing on the bobbed phenotype in Drosophila melanogaster. Dev. Biol. 1970, 22, 495–512. [Google Scholar] [CrossRef] [PubMed]
- Thompson, O.; Edgley, M.; Strasbourger, P.; Flibotte, S.; Ewing, B.; Adair, R.; Au, V.; Chaudhry, I.; Fernando, L.; Hutter, H.; et al. The million mutation project: A new approach to genetics in Caenorhabditis elegans. Genome Res. 2013, 23, 1749–1762. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.G.; Branco, A.T.; Godinho, S.A.; Yu, S.; Lemos, B. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 2485–2490. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.M.; Kurylo, C.M.; Dass, R.A.; Bojmar, L.; Lyden, D.; Vincent, C.T.; Blanchard, S.C. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 2018, 4, eaao0665. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.L.; Nelson, J.O.; Watase, G.J.; Warsinger-Pepe, N.; Yamashita, Y.M. Transgenerational dynamics of rDNA copy number in Drosophila male germline stem cells. Elife 2018, 7, e32421. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Li, H.; Perry, J.M.; Singh, V.P.; Unruh, J.; Yu, Z.; Zakari, M.; McDowell, W.; Li, L.; Gerton, J.L. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017, 13, e1006771. [Google Scholar] [CrossRef] [PubMed]
- Stults, D.M.; Killen, M.W.; Pierce, H.H.; Pierce, A.J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008, 18, 13–18. [Google Scholar] [CrossRef]
- McKnight, S.L.; Miller, O.L., Jr. Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell 1976, 8, 305–319. [Google Scholar] [CrossRef]
- Conconi, A.; Widmer, R.M.; Koller, T.; Sogo, J.M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 1989, 57, 753–761. [Google Scholar] [CrossRef]
- Stancheva, I.; Lucchini, R.; Koller, T.; Sogo, J.M. Chromatin structure and methylation of rat rRNA genes studied by formaldehyde fixation and psoralen cross-linking. Nucleic Acids Res. 1997, 25, 1727–1735. [Google Scholar] [CrossRef]
- Roussel, P.; Andre, C.; Comai, L.; Hernandez-Verdun, D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol. 1996, 133, 235–246. [Google Scholar] [CrossRef]
- Zillner, K.; Komatsu, J.; Filarsky, K.; Kalepu, R.; Bensimon, A.; Nemeth, A. Active human nucleolar organizer regions are interspersed with inactive rDNA repeats in normal and tumor cells. Epigenomics 2015, 7, 363–378. [Google Scholar] [CrossRef]
- Fefelova, E.A.; Pleshakova, I.M.; Mikhaleva, E.A.; Pirogov, S.A.; Poltorachenko, V.A.; Abramov, Y.A.; Romashin, D.D.; Shatskikh, A.S.; Blokh, R.S.; Gvozdev, V.A.; et al. Impaired function of rDNA transcription initiation machinery leads to derepression of ribosomal genes with insertions of R2 retrotransposon. Nucleic Acids Res. 2022, 50, 867–884. [Google Scholar] [CrossRef]
- Yao, R.W.; Xu, G.; Wang, Y.; Shan, L.; Luan, P.F.; Wang, Y.; Wu, M.; Yang, L.Z.; Xing, Y.H.; Yang, L.; et al. Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus. Mol. Cell 2019, 76, 767–783.e11. [Google Scholar] [CrossRef]
- Hori, Y.; Shimamoto, A.; Kobayashi, T. The human ribosomal DNA array is composed of highly homogenized tandem clusters. Genome Res. 2021, 31, 1971–1982. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef]
- Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009, 457, 413–420. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V.; et al. Molecular Mechanisms and Biological Functions of siRNA. Int. J. Biomed. Sci. 2017, 13, 48–57. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Author Correction: Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2018, 19, 808. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Rosace, D.; Lopez, J.; Blanco, S. Emerging roles of novel small non-coding regulatory RNAs in immunity and cancer. RNA Biol. 2020, 17, 1196–1213. [Google Scholar] [CrossRef]
- Pontes, O.; Li, C.F.; Costa Nunes, P.; Haag, J.; Ream, T.; Vitins, A.; Jacobsen, S.E.; Pikaard, C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 2006, 126, 79–92. [Google Scholar] [CrossRef]
- Preuss, S.B.; Costa-Nunes, P.; Tucker, S.; Pontes, O.; Lawrence, R.J.; Mosher, R.; Kasschau, K.D.; Carrington, J.C.; Baulcombe, D.C.; Viegas, W.; et al. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol. Cell 2008, 32, 673–684. [Google Scholar] [CrossRef]
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004, 2, E104. [Google Scholar] [CrossRef]
- Blevins, T.; Pontes, O.; Pikaard, C.S.; Meins, F., Jr. Heterochromatic siRNAs and DDM1 independently silence aberrant 5S rDNA transcripts in Arabidopsis. PLoS ONE 2009, 4, e5932. [Google Scholar] [CrossRef]
- Lee, H.C.; Chang, S.S.; Choudhary, S.; Aalto, A.P.; Maiti, M.; Bamford, D.H.; Liu, Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 2009, 459, 274–277. [Google Scholar] [CrossRef]
- Cecere, G.; Cogoni, C. Quelling targets the rDNA locus and functions in rDNA copy number control. BMC Microbiol. 2009, 9, 44. [Google Scholar] [CrossRef]
- Zhang, Z.; Chang, S.S.; Zhang, Z.; Xue, Z.; Zhang, H.; Li, S.; Liu, Y. Homologous recombination as a mechanism to recognize repetitive DNA sequences in an RNAi pathway. Genes. Dev. 2013, 27, 145–150. [Google Scholar] [CrossRef]
- Buhler, M.; Spies, N.; Bartel, D.P.; Moazed, D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol. 2008, 15, 1015–1023. [Google Scholar] [CrossRef]
- Cam, H.P.; Sugiyama, T.; Chen, E.S.; Chen, X.; FitzGerald, P.C.; Grewal, S.I. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005, 37, 809–819. [Google Scholar] [CrossRef]
- Chak, L.L.; Mohammed, J.; Lai, E.C.; Tucker-Kellogg, G.; Okamura, K. A deeply conserved, noncanonical miRNA hosted by ribosomal DNA. RNA 2015, 21, 375–384. [Google Scholar] [CrossRef]
- Zhou, X.; Feng, X.; Mao, H.; Li, M.; Xu, F.; Hu, K.; Guang, S. RdRP-synthesized antisense ribosomal siRNAs silence pre-rRNA via the nuclear RNAi pathway. Nat. Struct. Mol. Biol. 2017, 24, 258–269. [Google Scholar] [CrossRef]
- Zhu, C.; Yan, Q.; Weng, C.; Hou, X.; Mao, H.; Liu, D.; Feng, X.; Guang, S. Erroneous ribosomal RNAs promote the generation of antisense ribosomal siRNA. Proc. Natl. Acad. Sci. USA 2018, 115, 10082–10087. [Google Scholar] [CrossRef]
- Cherlin, T.; Magee, R.; Jing, Y.; Pliatsika, V.; Loher, P.; Rigoutsos, I. Ribosomal RNA fragmentation into short RNAs (rRFs) is modulated in a sex- and population of origin-specific manner. BMC Biol. 2020, 18, 38. [Google Scholar] [CrossRef]
- Wei, H.; Zhou, B.; Zhang, F.; Tu, Y.; Hu, Y.; Zhang, B.; Zhai, Q. Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PLoS ONE 2013, 8, e56842. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Klushevskaya, E.S.; Alembekov, I.R.; Bukreeva, A.S.; Kretova, A.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Kravatsky, Y.V. Fragments of rDNA Genes Scattered over the Human Genome Are Targets of Small RNAs. Int. J. Mol. Sci. 2022, 23, 3014. [Google Scholar] [CrossRef]
- Son, D.J.; Kumar, S.; Takabe, W.; Kim, C.W.; Ni, C.W.; Alberts-Grill, N.; Jang, I.H.; Kim, S.; Kim, W.; Won Kang, S.; et al. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat. Commun. 2013, 4, 3000. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Lopez, J.; Alonso, L.; Cardenas, D.B.; Artaza-Alvarez, H.; Hourcade Jde, D.; Martinez, S.; Brieno-Enriquez, M.A.; Del Mazo, J. Diversity and functional convergence of small noncoding RNAs in male germ cell differentiation and fertilization. RNA 2015, 21, 946–962. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, S.; Sunkar, R. Genome-wide discovery and analysis of phased small interfering RNAs in Chinese sacred lotus. PLoS ONE 2014, 9, e113790. [Google Scholar] [CrossRef] [PubMed]
- Tucker, S.; Vitins, A.; Pikaard, C.S. Nucleolar dominance and ribosomal RNA gene silencing. Curr. Opin. Cell Biol. 2010, 22, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhara, C.; Mohannath, G.; Blevins, T.; Pontvianne, F.; Pikaard, C.S. Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes. Dev. 2016, 30, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Greil, F.; Ahmad, K. Nucleolar dominance of the Y chromosome in Drosophila melanogaster. Genetics 2012, 191, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Pikaard, C.S. Nucleolar dominance: Uniparental gene silencing on a multi-megabase scale in genetic hybrids. Plant Mol. Biol. 2000, 43, 163–177. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Chandrasekhara, C.; McKinlay, A.; Enganti, R.; Fultz, D. Reaching for the off switch in nucleolar dominance. Plant J. 2023, 115, 1185–1192. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Robaszkiewicz, E.; Mykhailyk, S.; Wartini, J.; Pinski, A.; Kovarik, A.; Hasterok, R. To Be or Not to Be Expressed: The First Evidence of a Nucleolar Dominance Tissue-Specificity in Brachypodium hybridum. Front. Plant Sci. 2021, 12, 768347. [Google Scholar] [CrossRef]
- Pontes, O.; Lawrence, R.J.; Silva, M.; Preuss, S.; Costa-Nunes, P.; Earley, K.; Neves, N.; Viegas, W.; Pikaard, C.S. Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS ONE 2007, 2, e1157. [Google Scholar] [CrossRef]
- Chen, Z.J.; Pikaard, C.S. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes. Dev. 1997, 11, 2124–2136. [Google Scholar] [CrossRef]
- Earley, K.; Lawrence, R.J.; Pontes, O.; Reuther, R.; Enciso, A.J.; Silva, M.; Neves, N.; Gross, M.; Viegas, W.; Pikaard, C.S. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes. Dev. 2006, 20, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Earley, K.W.; Pontvianne, F.; Wierzbicki, A.T.; Blevins, T.; Tucker, S.; Costa-Nunes, P.; Pontes, O.; Pikaard, C.S. Mechanisms of HDA6-mediated rRNA gene silencing: Suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes. Dev. 2010, 24, 1119–1132. [Google Scholar] [CrossRef] [PubMed]
- Costa-Nunes, P.; Pontes, O.; Preuss, S.B.; Pikaard, C.S. Extra views on RNA-dependent DNA methylation and MBD6-dependent heterochromatin formation in nucleolar dominance. Nucleus 2010, 1, 254–259. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wierzbicki, A.T.; Ream, T.S.; Haag, J.R.; Pikaard, C.S. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009, 41, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, R.J.; Earley, K.; Pontes, O.; Silva, M.; Chen, Z.J.; Neves, N.; Viegas, W.; Pikaard, C.S. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 2004, 13, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Law, J.A. RNA Pol IV and V in gene silencing: Rebel polymerases evolving away from Pol II’s rules. Curr. Opin. Plant Biol. 2015, 27, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.M.; Du, J.; Hale, C.J.; Bischof, S.; Feng, S.; Chodavarapu, R.K.; Zhong, X.; Marson, G.; Pellegrini, M.; Segal, D.J.; et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 2014, 507, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Sackton, T.B.; Martinsen, L.; Lemos, B.; Eickbush, T.H.; Hartl, D.L. Y chromosome mediates ribosomal DNA silencing and modulates the chromatin state in Drosophila. Proc. Natl. Acad. Sci. USA 2012, 109, 9941–9946. [Google Scholar] [CrossRef]
- Warsinger-Pepe, N.; Li, D.; Yamashita, Y.M. Regulation of Nucleolar Dominance in Drosophila melanogaster. Genetics 2020, 214, 991–1004. [Google Scholar] [CrossRef]
- Schlesinger, S.; Selig, S.; Bergman, Y.; Cedar, H. Allelic inactivation of rDNA loci. Genes. Dev. 2009, 23, 2437–2447. [Google Scholar] [CrossRef]
- Mathieu, O.; Jasencakova, Z.; Vaillant, I.; Gendrel, A.V.; Colot, V.; Schubert, I.; Tourmente, S. Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell 2003, 15, 2929–2939. [Google Scholar] [CrossRef]
- Douet, J.; Tourmente, S. Transcription of the 5S rRNA heterochromatic genes is epigenetically controlled in Arabidopsis thaliana and Xenopus laevis. Heredity 2007, 99, 5–13. [Google Scholar] [CrossRef]
- Vaillant, I.; Tutois, S.; Cuvillier, C.; Schubert, I.; Tourmente, S. Regulation of Arabidopsis thaliana 5S rRNA Genes. Plant Cell Physiol. 2007, 48, 745–752. [Google Scholar] [CrossRef]
- Guetg, C.; Lienemann, P.; Sirri, V.; Grummt, I.; Hernandez-Verdun, D.; Hottiger, M.O.; Fussenegger, M.; Santoro, R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010, 29, 2135–2146. [Google Scholar] [CrossRef]
- Kim, J.A.; Kruhlak, M.; Dotiwala, F.; Nussenzweig, A.; Haber, J.E. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J. Cell Biol. 2007, 178, 209–218. [Google Scholar] [CrossRef]
- Peng, J.C.; Karpen, G.H. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 2007, 9, 25–35. [Google Scholar] [CrossRef]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef]
- Houseley, J.; Kotovic, K.; El Hage, A.; Tollervey, D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007, 26, 4996–5006. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, Q.; Sun, G.; Chen, S.; He, Q.; Li, S.; Liu, Y. Histone H3K56 acetylation is required for quelling-induced small RNA production through its role in homologous recombination. J. Biol. Chem. 2014, 289, 9365–9371. [Google Scholar] [CrossRef]
- Lee, H.C.; Aalto, A.P.; Yang, Q.; Chang, S.S.; Huang, G.; Fisher, D.; Cha, J.; Poranen, M.M.; Bamford, D.H.; Liu, Y. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 2010, 8, e1000496. [Google Scholar] [CrossRef]
- Nolan, T.; Cecere, G.; Mancone, C.; Alonzi, T.; Tripodi, M.; Catalanotto, C.; Cogoni, C. The RNA-dependent RNA polymerase essential for post-transcriptional gene silencing in Neurospora crassa interacts with replication protein A. Nucleic Acids Res. 2008, 36, 532–538. [Google Scholar] [CrossRef]
- Gao, M.; Wei, W.; Li, M.M.; Wu, Y.S.; Ba, Z.; Jin, K.X.; Li, M.M.; Liao, Y.Q.; Adhikari, S.; Chong, Z.; et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014, 24, 532–541. [Google Scholar] [CrossRef]
- d’Adda di Fagagna, F. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol. 2014, 24, 171–178. [Google Scholar] [CrossRef]
- Michalik, K.M.; Bottcher, R.; Forstemann, K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012, 40, 9596–9603. [Google Scholar] [CrossRef]
- Chen, H.; Kobayashi, K.; Miyao, A.; Hirochika, H.; Yamaoka, N.; Nishiguchi, M. Both OsRecQ1 and OsRDR1 are required for the production of small RNA in response to DNA-damage in rice. PLoS ONE 2013, 8, e55252. [Google Scholar] [CrossRef]
- Castel, S.E.; Ren, J.; Bhattacharjee, S.; Chang, A.Y.; Sanchez, M.; Valbuena, A.; Antequera, F.; Martienssen, R.A. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 2014, 159, 572–583. [Google Scholar] [CrossRef]
- Roche, B.; Arcangioli, B.; Martienssen, R.A. RNA interference is essential for cellular quiescence. Science 2016, 354, aah5651. [Google Scholar] [CrossRef]
- Mochida, S.; Yanagida, M. Distinct modes of DNA damage response in S. pombe G0 and vegetative cells. Genes. Cells 2006, 11, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Du, P.; Wang, X.; Yu, Y.Q.; Qiu, Y.H.; Li, W.; Gal-On, A.; Zhou, C.; Li, Y.; Ding, S.W. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14613–14618. [Google Scholar] [CrossRef] [PubMed]
- You, C.; He, W.; Hang, R.; Zhang, C.; Cao, X.; Guo, H.; Chen, X.; Cui, J.; Mo, B. FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis. Nat. Commun. 2019, 10, 4424. [Google Scholar] [CrossRef]
- Lange, H.; Sement, F.M.; Gagliardi, D. MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J. 2011, 68, 51–63. [Google Scholar] [CrossRef]
- Allmang, C.; Mitchell, P.; Petfalski, E.; Tollervey, D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000, 28, 1684–1691. [Google Scholar] [CrossRef]
- Okuda, E.K.; Gonzales-Zubiate, F.A.; Gadal, O.; Oliveira, C.C. Nucleolar localization of the yeast RNA exosome subunit Rrp44 hints at early pre-rRNA processing as its main function. J. Biol. Chem. 2020, 295, 11195–11213. [Google Scholar] [CrossRef]
- Hang, R.; Xu, Y.; Wang, X.; Hu, H.; Flynn, N.; You, C.; Chen, X. Arabidopsis HOT3/eIF5B1 constrains rRNA RNAi by facilitating 18S rRNA maturation. Proc. Natl. Acad. Sci. USA 2023, 120, e2301081120. [Google Scholar] [CrossRef]
- Schmidt, K.; Butler, J.S. Nuclear RNA surveillance: Role of TRAMP in controlling exosome specificity. Wiley Interdiscip. Rev. RNA 2013, 4, 217–231. [Google Scholar] [CrossRef]
- Joh, R.I.; Khanduja, J.S.; Calvo, I.A.; Mistry, M.; Palmieri, C.M.; Savol, A.J.; Ho Sui, S.J.; Sadreyev, R.I.; Aryee, M.J.; Motamedi, M. Survival in Quiescence Requires the Euchromatic Deployment of Clr4/SUV39H by Argonaute-Associated Small RNAs. Mol. Cell 2016, 64, 1088–1101. [Google Scholar] [CrossRef]
- Almeida, M.V.; Andrade-Navarro, M.A.; Ketting, R.F. Function and Evolution of Nematode RNAi Pathways. Noncoding RNA 2019, 5, 8. [Google Scholar] [CrossRef]
- Liao, S.; Chen, X.; Xu, T.; Jin, Q.; Xu, Z.; Xu, D.; Zhou, X.; Zhu, C.; Guang, S.; Feng, X. Antisense ribosomal siRNAs inhibit RNA polymerase I-directed transcription in C. elegans. Nucleic Acids Res. 2021, 49, 9194–9210. [Google Scholar] [CrossRef]
- Hirai, H.; Takemata, N.; Tamura, M.; Ohta, K. Facultative heterochromatin formation in rDNA is essential for cell survival during nutritional starvation. Nucleic Acids Res. 2022, 50, 3727–3744. [Google Scholar] [CrossRef]
- Wahba, L.; Hansen, L.; Fire, A.Z. An essential role for the piRNA pathway in regulating the ribosomal RNA pool in C. elegans. Dev. Cell 2021, 56, 2295–2312.e6. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, B.E.; Vijayasarathy, T.; Marks, T.N.; Cialek, C.A.; Reed, K.J.; Montgomery, T.A. Dual roles for piRNAs in promoting and preventing gene silencing in C. elegans. Cell Rep. 2021, 37, 110101. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.Z.; Chen, H.; Ozturk, A.R.; Tu, S.; Shirayama, M.; Tang, W.; Ding, Y.H.; Dai, S.Y.; Weng, Z.; Mello, C.C. Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 2018, 172, 937–951.e18. [Google Scholar] [CrossRef] [PubMed]
- Stolyarenko, A.D. Nuclear Argonaute Piwi Gene Mutation Affects rRNA by Inducing rRNA Fragment Accumulation, Antisense Expression, and Defective Processing in Drosophila ovaries. Int. J. Mol. Sci. 2020, 21, 1119. [Google Scholar] [CrossRef] [PubMed]
- Eickbush, T.H.; Eickbush, D.G. Integration, Regulation, and Long-Term Stability of R2 Retrotransposons. Microbiol. Spectr. 2015, 3, MDNA3-0011-2014. [Google Scholar] [CrossRef] [PubMed]
- Long, E.O.; Dawid, I.B. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell 1979, 18, 1185–1196. [Google Scholar] [CrossRef]
- Luo, Y.; Fefelova, E.; Ninova, M.; Chen, Y.A.; Aravin, A.A. Repression of interrupted and intact rDNA by the SUMO pathway in Drosophila melanogaster. Elife 2020, 9, e52416. [Google Scholar] [CrossRef] [PubMed]
- Raje, H.S.; Lieux, M.E.; DiMario, P.J. R1 retrotransposons in the nucleolar organizers of Drosophila melanogaster are transcribed by RNA polymerase I upon heat shock. Transcription 2018, 9, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Mikhaleva, E.A.; Leinsoo, T.A.; Ishizu, H.; Gvozdev, V.A.; Klenov, M.S. The nucleolar transcriptome regulates Piwi shuttling between the nucleolus and the nucleoplasm. Chromosome Res. 2019, 27, 141–152. [Google Scholar] [CrossRef]
- Sienski, G.; Donertas, D.; Brennecke, J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 2012, 151, 964–980. [Google Scholar] [CrossRef]
- Klenov, M.S.; Sokolova, O.A.; Yakushev, E.Y.; Stolyarenko, A.D.; Mikhaleva, E.A.; Lavrov, S.A.; Gvozdev, V.A. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl. Acad. Sci. USA 2011, 108, 18760–18765. [Google Scholar] [CrossRef]
- Wang, S.H.; Elgin, S.C. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. USA 2011, 108, 21164–21169. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Oe, A.; Nishida, K.M.; Yamashita, K.; Kajiya, A.; Hirano, S.; Matsumoto, N.; Dohmae, N.; Ishitani, R.; Saito, K.; et al. Crystal structure of Drosophila Piwi. Nat. Commun. 2020, 11, 858. [Google Scholar] [CrossRef]
- Buhler, M.; Verdel, A.; Moazed, D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 2006, 125, 873–886. [Google Scholar] [CrossRef]
- Guang, S.; Bochner, A.F.; Burkhart, K.B.; Burton, N.; Pavelec, D.M.; Kennedy, S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 2010, 465, 1097–1101. [Google Scholar] [CrossRef]
- Carmell, M.A.; Girard, A.; van de Kant, H.J.; Bourc’his, D.; Bestor, T.H.; de Rooij, D.G.; Hannon, G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 2007, 12, 503–514. [Google Scholar] [CrossRef]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Totoki, Y.; Toyoda, A.; Ikawa, M.; Asada, N.; Kojima, K.; Yamaguchi, Y.; Ijiri, T.W.; et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes. Dev. 2008, 22, 908–917. [Google Scholar] [CrossRef]
- Pezic, D.; Manakov, S.A.; Sachidanandam, R.; Aravin, A.A. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes. Dev. 2014, 28, 1410–1428. [Google Scholar] [CrossRef]
- Schopp, T.; Zoch, A.; Berrens, R.V.; Auchynnikava, T.; Kabayama, Y.; Vasiliauskaite, L.; Rappsilber, J.; Allshire, R.C.; O’Carroll, D. TEX15 is an essential executor of MIWI2-directed transposon DNA methylation and silencing. Nat. Commun. 2020, 11, 3739. [Google Scholar] [CrossRef]
- Kalantari, R.; Chiang, C.M.; Corey, D.R. Regulation of mammalian transcription and splicing by Nuclear RNAi. Nucleic Acids Res. 2016, 44, 524–537. [Google Scholar] [CrossRef]
- Sala, L.; Kumar, M.; Prajapat, M.; Chandrasekhar, S.; Cosby, R.L.; La Rocca, G.; Macfarlan, T.S.; Awasthi, P.; Chari, R.; Kruhlak, M.; et al. AGO2 silences mobile transposons in the nucleus of quiescent cells. Nat. Struct. Mol. Biol. 2023, 30, 1985–1995. [Google Scholar] [CrossRef]
- Sinkkonen, L.; Hugenschmidt, T.; Filipowicz, W.; Svoboda, P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS ONE 2010, 5, e12175. [Google Scholar] [CrossRef]
- Liang, X.H.; Crooke, S.T. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011, 39, 4875–4889. [Google Scholar] [CrossRef]
- Atwood, B.L.; Woolnough, J.L.; Lefevre, G.M.; Saint Just Ribeiro, M.; Felsenfeld, G.; Giles, K.E. Human Argonaute 2 Is Tethered to Ribosomal RNA through MicroRNA Interactions. J. Biol. Chem. 2016, 291, 17919–17928. [Google Scholar] [CrossRef]
- Stults, D.M.; Killen, M.W.; Williamson, E.P.; Hourigan, J.S.; Vargas, H.D.; Arnold, S.M.; Moscow, J.A.; Pierce, A.J. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 2009, 69, 9096–9104. [Google Scholar] [CrossRef]
- Killen, M.W.; Stults, D.M.; Adachi, N.; Hanakahi, L.; Pierce, A.J. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum. Mol. Genet. 2009, 18, 3417–3428. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shatskikh, A.S.; Fefelova, E.A.; Klenov, M.S. Functions of RNAi Pathways in Ribosomal RNA Regulation. Non-Coding RNA 2024, 10, 19. https://doi.org/10.3390/ncrna10020019
Shatskikh AS, Fefelova EA, Klenov MS. Functions of RNAi Pathways in Ribosomal RNA Regulation. Non-Coding RNA. 2024; 10(2):19. https://doi.org/10.3390/ncrna10020019
Chicago/Turabian StyleShatskikh, Aleksei S., Elena A. Fefelova, and Mikhail S. Klenov. 2024. "Functions of RNAi Pathways in Ribosomal RNA Regulation" Non-Coding RNA 10, no. 2: 19. https://doi.org/10.3390/ncrna10020019
APA StyleShatskikh, A. S., Fefelova, E. A., & Klenov, M. S. (2024). Functions of RNAi Pathways in Ribosomal RNA Regulation. Non-Coding RNA, 10(2), 19. https://doi.org/10.3390/ncrna10020019







