Co-Culture of White Rot Fungi Pleurotus ostreatus P5 and Bacillus amyloliquefaciens B2: A Strategy to Enhance Lipopeptide Production and Suppress of Fusarium Wilt of Cucumber
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Culture and Chemicals
2.2. PCR Amplification of B. amyloliquefaciens B2 Lipopeptide Genes
2.3. Extraction and Purification of Lipopeptides
2.4. Identification of Lipopeptides by LC-MS/MS
2.5. Co-Cultivation of B. amyloliquefaciens B2 and P. ostreatus P5
2.6. Gene Expression of CLP Biosynthetic Genes
2.7. The Effect of Co-Culture Filtrate on FOC Growth
2.8. Greenhouse Experiment
2.9. FOC Abundance Detection
2.10. Biolog EcoPlate Analysis
2.11. Statistical Analysis
3. Results
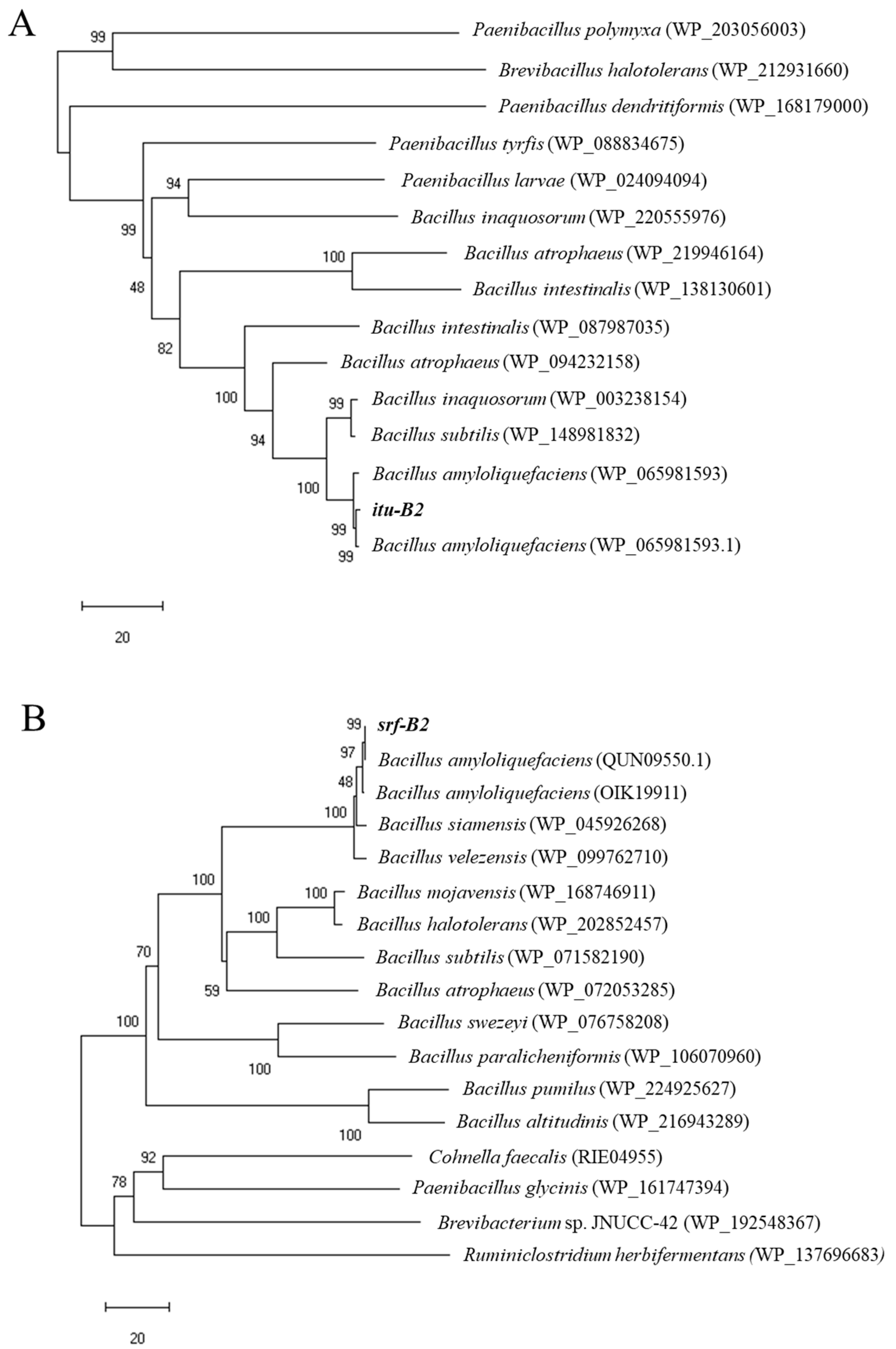
3.1. Identification of Lipopeptide Biosynthetic Genes from B. amyloliquefaciens B2
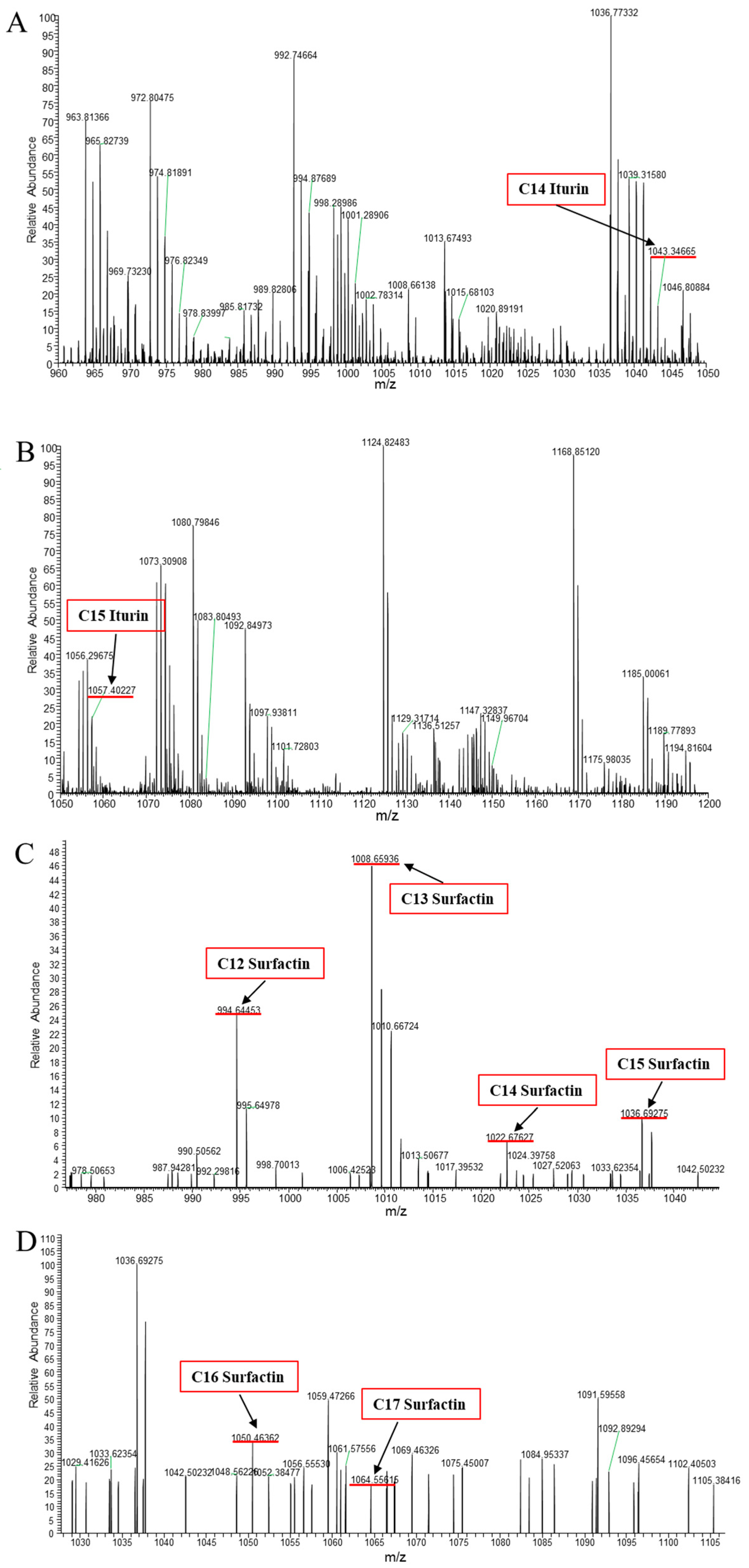
3.2. Identification of CLPs Produced by B. amyloliquefaciens B2
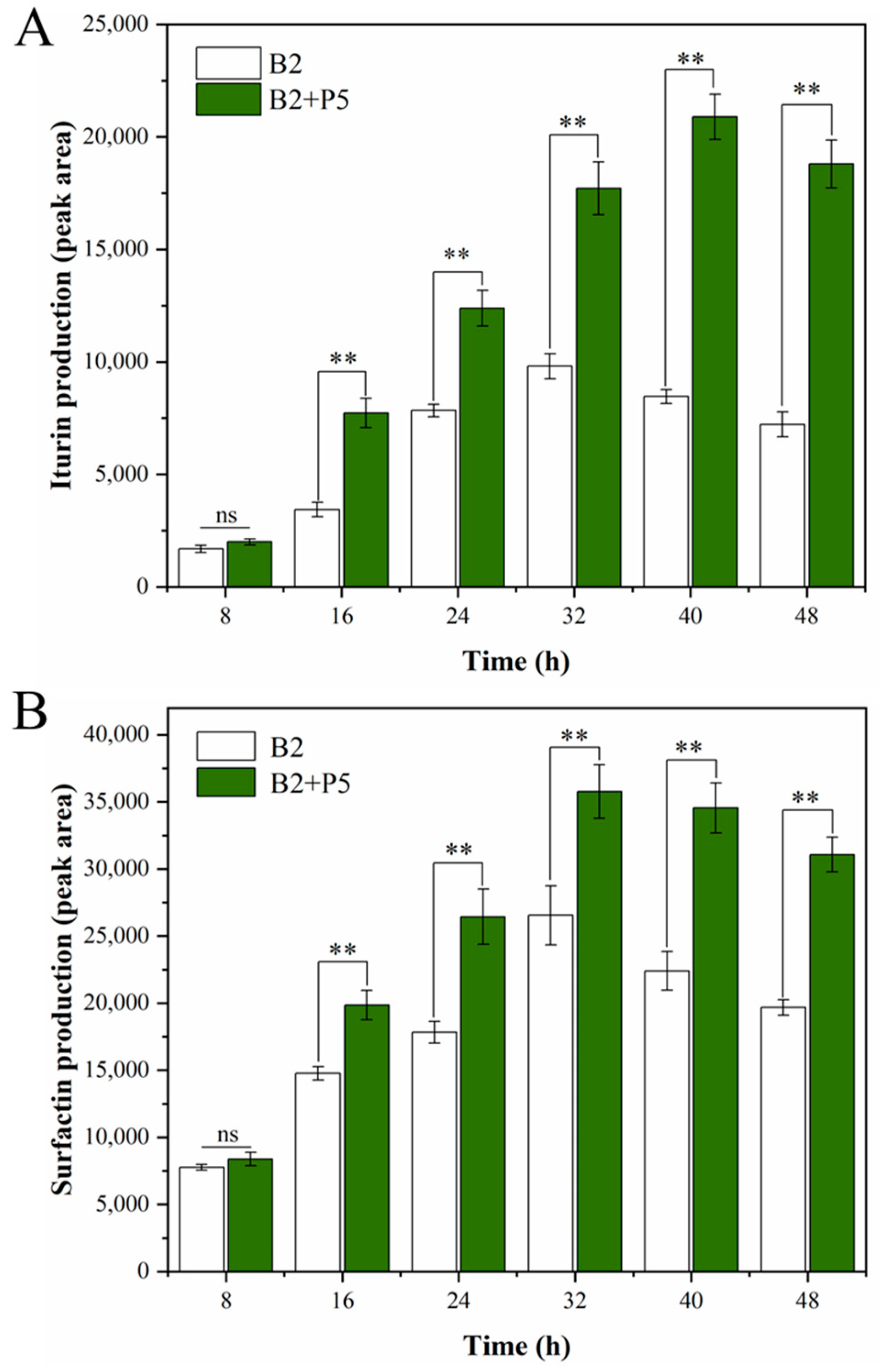
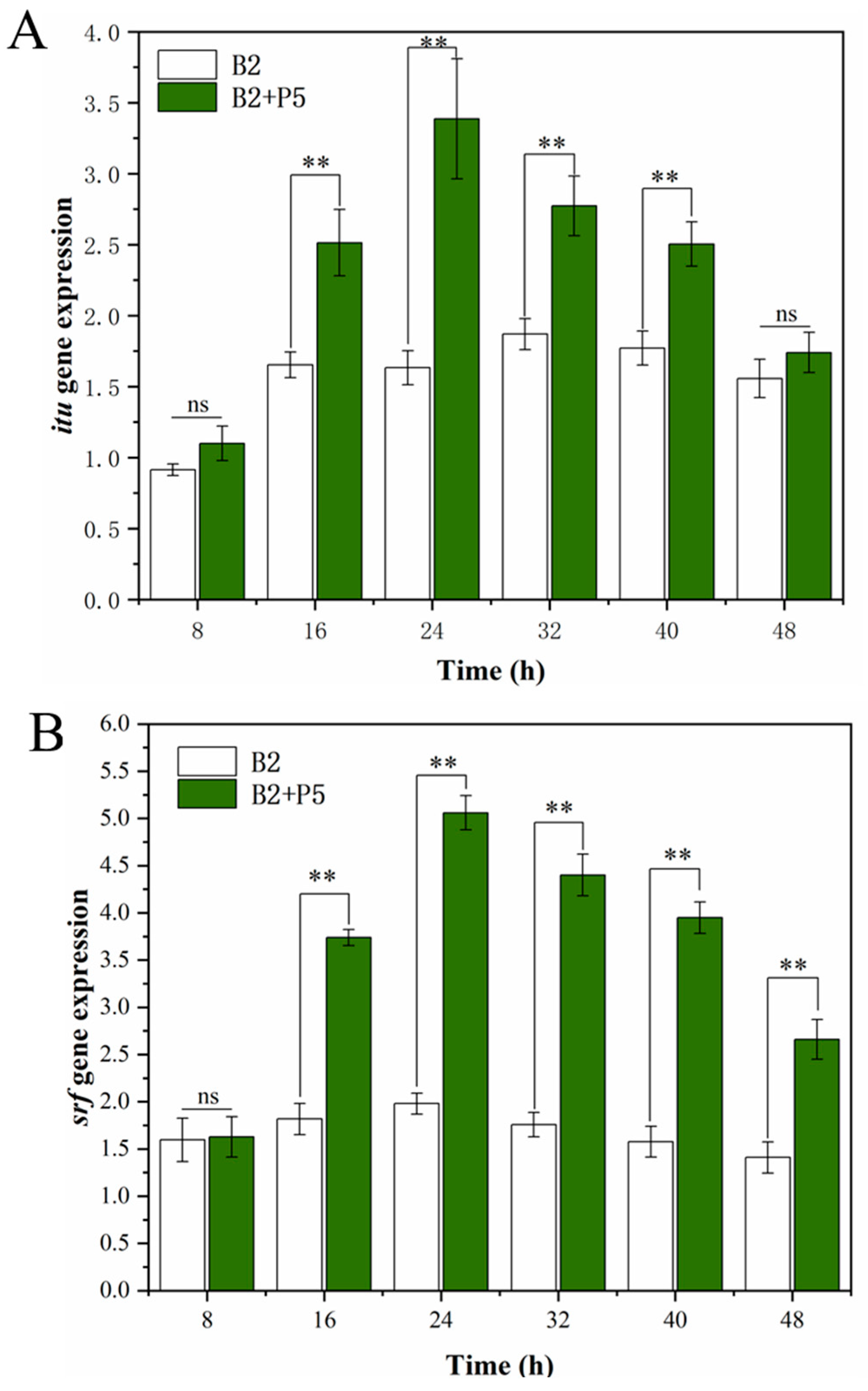
3.3. Effect of P. ostreatus P5 on the Production of CLPs and Transcription of Their Encoding Genes by B. amyloliquefaciens B2
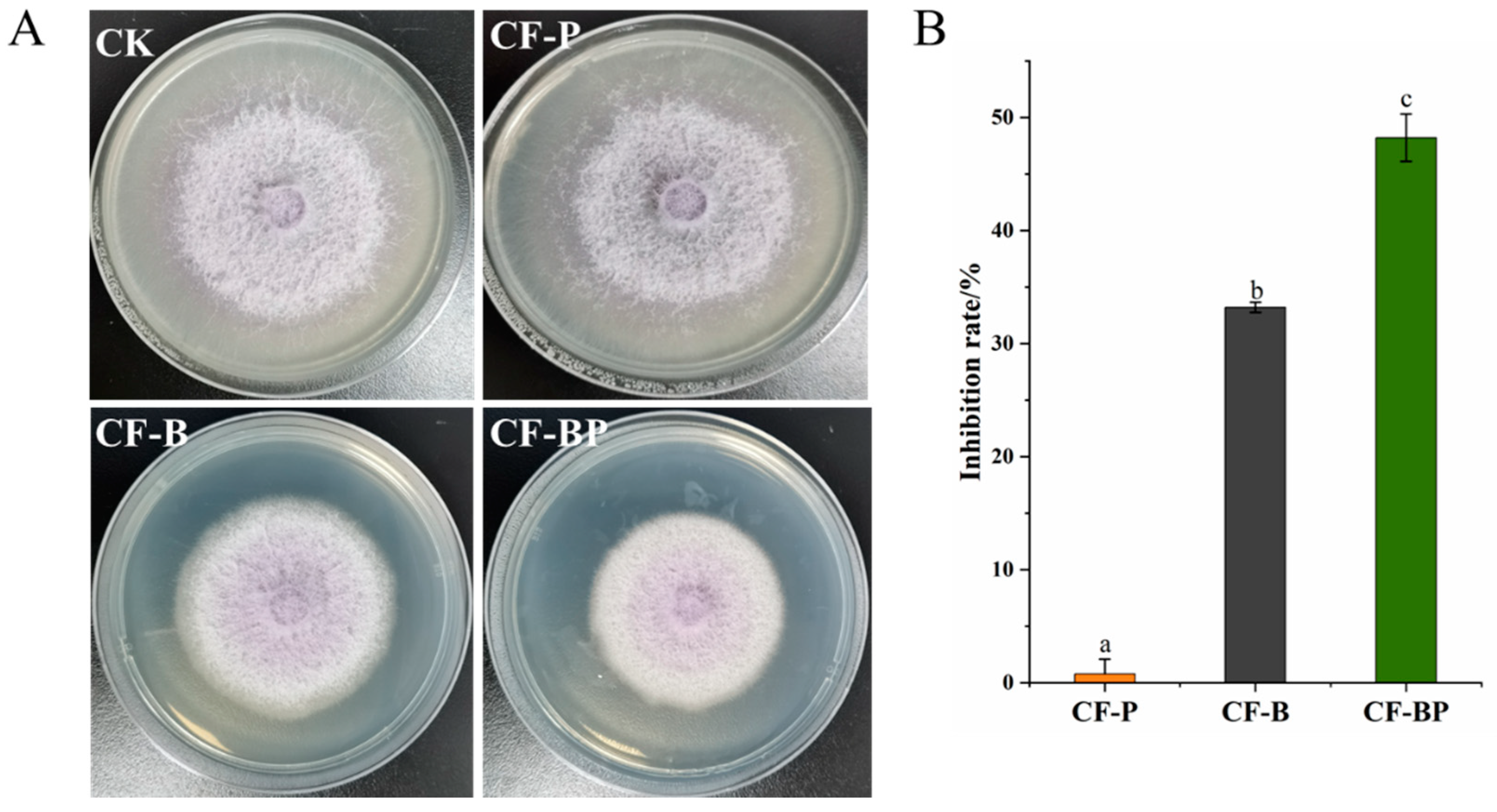
3.4. Inhibition of FOC Growth by Co-Culture Filtrate
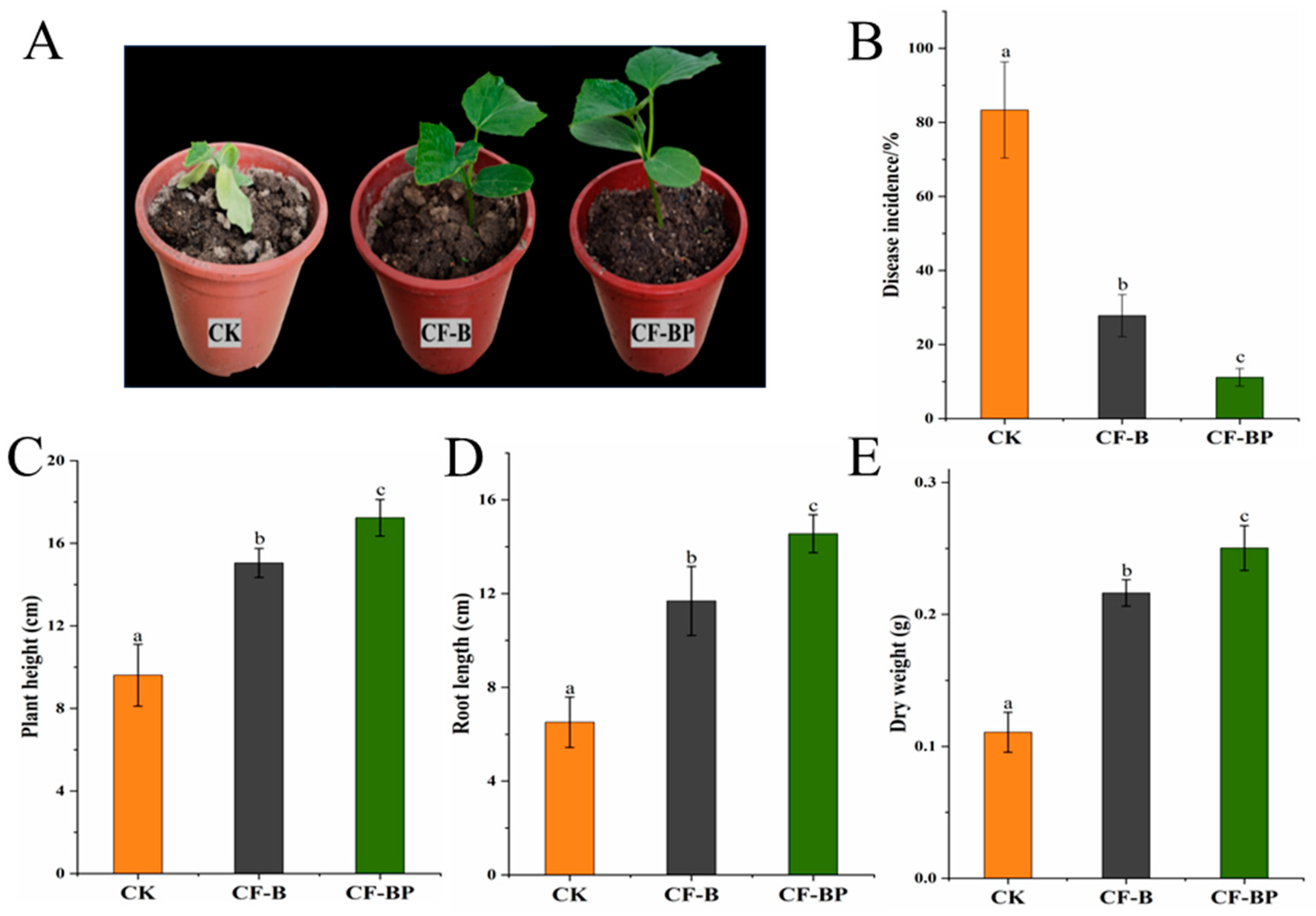
3.5. Effect of Co-Culture Filtrate against Cucumber Fusarium Wilt
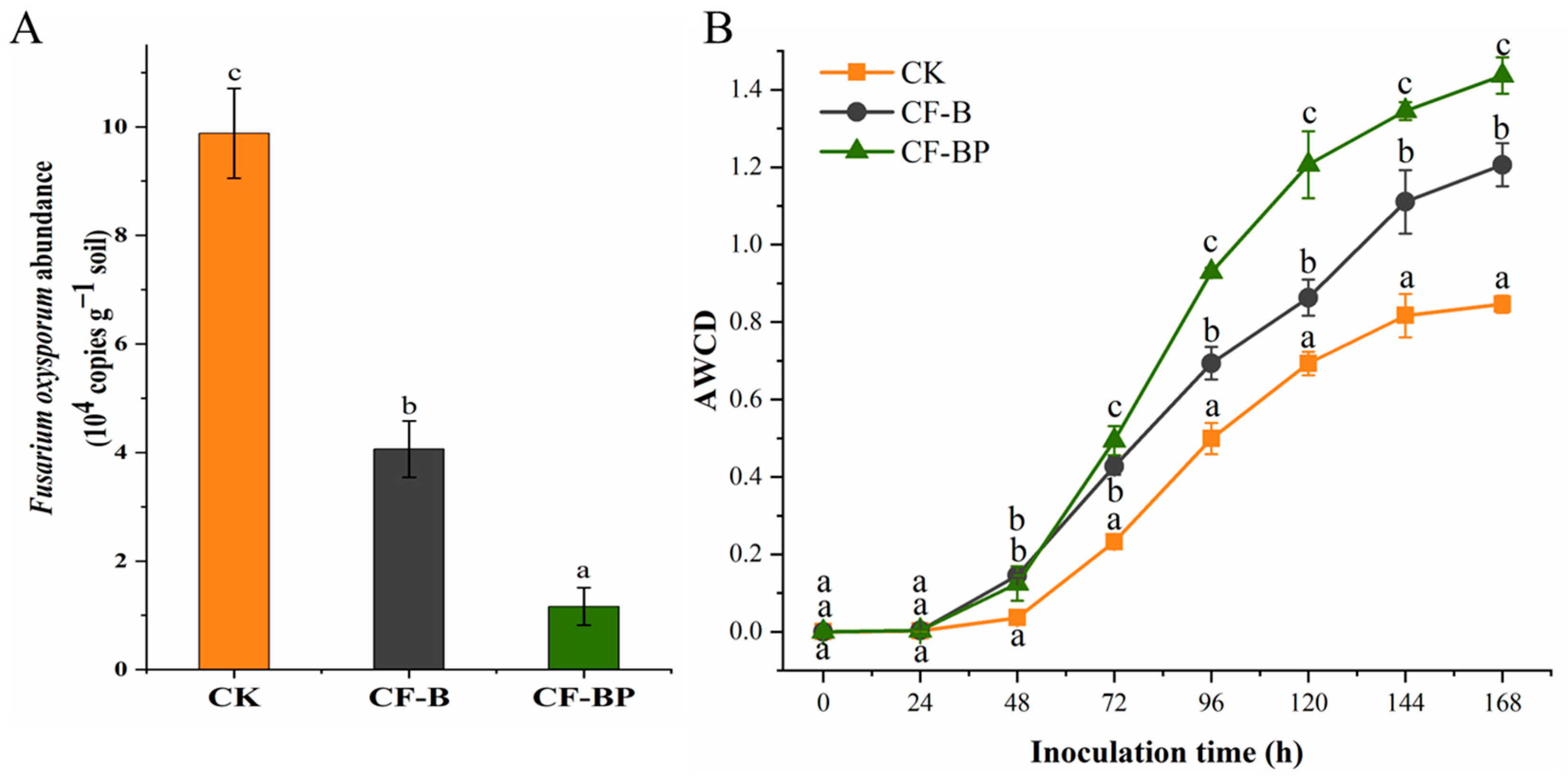
3.6. Effect of Culture Filtrate on the Abundances of Rhizosphere FOC and Microbial Metabolic Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, A.; Elrys, A.S.; Liu, L.; Xia, Q.; Wang, B.; Li, Y.; Dan, X.; Iqbal, M.; Zhao, J.; Huang, X. Deciphering the Synergies of Reductive Soil Disinfestation Combined with Biochar and Antagonistic Microbial Inoculation in Cucumber Fusarium Wilt Suppression Through Rhizosphere Microbiota Structure. Microb. Ecol. 2022, 85, 980–997. [Google Scholar] [CrossRef] [PubMed]
- Lievens, B.; Claes, L.; Vakalounakis, D.J.; Vanachter, A.C.; Thomma, B.P. A robust identification and detection assay to discriminate the cucumber pathogens Fusarium oxysporum f. sp. cucumerinum and f. sp. radicis-cucumerinum. Environ. Microbiol. 2007, 9, 2145–2161. [Google Scholar]
- Cao, Y.; Zhang, Z.; Ling, N.; Yuan, Y.; Zheng, X.; Shen, B.; Shen, Q. Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol. Fertil. Soils 2011, 47, 495–506. [Google Scholar] [CrossRef]
- Huang, N.; Wang, W.; Yao, Y.; Zhu, F.; Wang, W.; Chang, X. The influence of different concentrations of bio-organic fertilizer on cucumber Fusarium wilt and soil microflora alterations. PLoS ONE 2017, 12, e0171490. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cai, X.Y.; Xu, M.; Tian, F. Enhanced Biocontrol of Cucumber Fusarium Wilt by Combined Application of New Antagonistic Bacteria Bacillus amyloliquefaciens B2 and Phenolic Acid-Degrading Fungus Pleurotus ostreatus P5. Front. Microbiol. 2021, 12, 700142. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, F.; Yang, J.; Fan, Y.; Yu, L.; Li, Z.; Jiang, N.; An, J.; Jiao, Z.; Wang, C. Isolation and identification of Bacillus mojavensis YL-RY0310 and its biocontrol potential against Penicillium expansum and patulin in apples. Biol. Control 2023, 182, 105239. [Google Scholar] [CrossRef]
- Regassa, A.B.; Taegyu, C.; Lee, Y.S. Supplementing biocontrol efficacy of Bacillus velezensis against Glomerella cingulata. Physiol. Mol. Plant Pathol. 2018, 102, 173–179. [Google Scholar] [CrossRef]
- Almoneafy, A.A.; Kakar, K.U.; Nawaz, Z.; Li, B.; Saand, M.A.; Chun-lan, Y.; Xie, G.-L. Tomato plant growth promotion and antibacterial related-mechanisms of four rhizobacterial Bacillus strains against Ralstonia solanacearum. Symbiosis 2014, 63, 59–70. [Google Scholar] [CrossRef]
- Sarwar, A.; Brader, G.; Corretto, E.; Aleti, G.; Abaidullah, M.; Sessitsch, A.; Hafeez, F.Y. Qualitative analysis of biosurfactants from Bacillus species exhibiting antifungal activity. PLoS ONE 2018, 13, e0198107. [Google Scholar]
- Ma, Z.; Zhang, S.; Zhang, S.; Wu, G.; Shao, Y.; Mi, Q.; Liang, J.; Sun, K.; Hu, J. Isolation and characterization of a new cyclic lipopeptide surfactin from a marine-derived Bacillus velezensis SH-B74. J. Antibiot. 2020, 73, 863–867. [Google Scholar] [CrossRef]
- Solanki, M.K.; Singh, R.K.; Srivastava, S.; Kumar, S.; Kashyap, P.L.; Srivastava, A.K. Characterization of antagonistic-potential of two Bacillus strains and their biocontrol activity against Rhizoctonia solani in tomato. J. Basic Microbiol. 2015, 55, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Souto, G.; Correa, O.; Montecchia, M.; Kerber, N.; Pucheu, N.; Bachur, M.; Garcia, A. Genetic and functional characterization of a Bacillus sp. strain excreting surfactin and antifungal metabolites partially identified as iturin-like compounds. J. Appl. Microbiol. 2004, 97, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Villegas-Escobar, V.; González-Jaramillo, L.M.; Ramirez, M.; Moncada, R.N.; Sierra-Zapata, L.; Orduz, S.; Romero-Tabarez, M. Lipopeptides from Bacillus sp. EA-CB0959: Active metabolites responsible for in vitro and in vivo control of Ralstonia solanacearum. Biol. Control 2018, 125, 20–28. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, S.A.; Mou, H.; Ma, Y.; Li, M.; Hu, X. Characterization of lipopeptide biosurfactants produced by Bacillus licheniformis MB01 from marine sediments. Front. Microbiol. 2017, 8, 871. [Google Scholar] [CrossRef]
- Zihalirwa Kulimushi, P.; Argüelles Arias, A.; Franzil, L.; Steels, S.; Ongena, M. Stimulation of fengycin-type antifungal lipopeptides in Bacillus amyloliquefaciens in the presence of the maize fungal pathogen Rhizomucor variabilis. Front. Microbiol. 2017, 8, 850. [Google Scholar] [CrossRef]
- Cossus, L.; Roux-Dalvai, F.; Kelly, I.; Nguyen, T.T.A.; Antoun, H.; Droit, A.; Tweddell, R.J. Interactions with plant pathogens influence lipopeptides production and antimicrobial activity of Bacillus subtilis strain PTB185. Biol. Control 2021, 154, 104497. [Google Scholar] [CrossRef]
- Sarwar, A.; Hassan, M.N.; Imran, M.; Iqbal, M.; Majeed, S.; Brader, G.; Sessitsch, A.; Hafeez, F.Y. Biocontrol activity of surfactin A purified from Bacillus NH-100 and NH-217 against rice bakanae disease. Microbiol. Res. 2018, 209, 1–13. [Google Scholar] [CrossRef]
- Bartolini, M.; Cogliati, S.; Vileta, D.; Bauman, C.; Ramirez, W.; Grau, R. Stress-responsive alternative sigma factor SigB plays a positive role in the antifungal proficiency of Bacillus subtilis. Appl. Environ. Microbiol. 2019, 85, e00178-19. [Google Scholar] [CrossRef]
- Akone, S.H.; Mándi, A.; Kurtán, T.; Hartmann, R.; Lin, W.; Daletos, G.; Proksch, P. Inducing secondary metabolite production by the endophytic fungus Chaetomium sp. through fungal–bacterial co-culture and epigenetic modification. Tetrahedron 2016, 72, 6340–6347. [Google Scholar] [CrossRef]
- Li, T.; Tang, J.; Karuppiah, V.; Li, Y.; Xu, N.; Chen, J. Co-culture of Trichoderma atroviride SG3403 and Bacillus subtilis 22 improves the production of antifungal secondary metabolites. Biol. Control 2020, 140, 104122. [Google Scholar] [CrossRef]
- Ma, Q.; Cong, Y.; Feng, L.; Liu, C.; Yang, W.; Xin, Y.; Chen, K. Effects of mixed culture fermentation of Bacillus amyloliquefaciens and Trichoderma longibrachiatum on its constituent strains and the biocontrol of tomato Fusarium wilt. J. Appl. Microbiol. 2022, 132, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, X.; Zhang, M.; Zhu, Y.; Zhuo, R. Removal of heavy-metal pollutants by white rot fungi: A mini review. J. Clean. Prod. 2022, 353, 131681. [Google Scholar] [CrossRef]
- Yin, J.; Sui, Z.; Li, Y.; Yang, H.; Yuan, L.; Huang, J. A new function of white-rot fungi Ceriporia lacerata HG2011: Improvement of biological nitrogen fixation of broad bean (Vicia faba). Microbiol. Res. 2022, 256, 126939. [Google Scholar] [CrossRef] [PubMed]
- Sui, Z.; Yuan, L. White-rot fungus Ceriporia lacerata HG2011 improved the biological nitrogen fixation of soybean and the underlying mechanisms. Plant Soil 2023, 486, 425–439. [Google Scholar] [CrossRef]
- Sui, Z.; Yin, J.; Huang, J.; Yuan, L. Phosphorus mobilization and improvement of crop agronomic performances by a new white-rot fungus Ceriporia lacerata HG2011. J. Sci. Food Agric. 2022, 102, 1640–1650. [Google Scholar] [CrossRef]
- Borges, J.; Cardoso, P.; Lopes, I.; Figueira, E.; Venâncio, C. Exploring the Potential of White-Rot Fungi Exudates on the Amelioration of Salinized Soils. Agriculture 2023, 13, 382. [Google Scholar] [CrossRef]
- Wang, H.W.; Xu, M.; Cai, X.Y.; Feng, T.; Xu, W.L. Application of spent mushroom substrate suppresses Fusarium wilt in cucumber and alters the composition of the microbial community of the cucumber rhizosphere. Eur. J. Soil Biol. 2020, 101, 103245. [Google Scholar] [CrossRef]
- Farzand, A.; Moosa, A.; Zubair, M.; Khan, A.R.; Ayaz, M.; Massawe, V.C.; Gao, X. Transcriptional profiling of diffusible lipopeptides and fungal virulence genes during Bacillus amyloliquefaciens EZ1509-mediated suppression of Sclerotinia sclerotiorum. Phytopathology 2020, 110, 317–326. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Xin, J.; Chen, X.; Xu, T.; He, J.; Pan, Z.; Zhang, C. Metabolomic profiles of the liquid state fermentation in co-culture of Eurotium amstelodami and Bacillus licheniformis. Front. Microbiol. 2023, 14, 1080743. [Google Scholar] [CrossRef]
- Jordan, S.; Junker, A.; Helmann, J.D.; Mascher, T. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: Identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 2006, 188, 5153–5166. [Google Scholar] [CrossRef]
- Liu, H.; Li, T.; Li, Y.; Wang, X.; Chen, J. Effects of Trichoderma atroviride SG3403 and Bacillus subtilis 22 on the biocontrol of wheat head blight. J. Fungi 2022, 8, 1250. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Zhu, Y.X.; Xu, M.; Cai, X.Y.; Tian, F. Co-application of spent mushroom substrate and PGPR alleviates tomato continuous cropping obstacle by regulating soil microbial properties. Rhizosphere 2022, 23, 100563. [Google Scholar] [CrossRef]
- Farzand, A.; Moosa, A.; Zubair, M.; Khan, A.R.; Massawe, V.C.; Tahir, H.A.S.; Sheikh, T.M.M.; Ayaz, M.; Gao, X. Suppression of Sclerotinia sclerotiorum by the induction of systemic resistance and regulation of antioxidant pathways in tomato using fengycin produced by Bacillus amyloliquefaciens FZB42. Biomolecules 2019, 9, 613. [Google Scholar] [CrossRef] [PubMed]
- Nanjundan, J.; Ramasamy, R.; Uthandi, S.; Ponnusamy, M. Antimicrobial activity and spectroscopic characterization of surfactin class of lipopeptides from Bacillus amyloliquefaciens SR1. Microb. Pathog. 2019, 128, 374–380. [Google Scholar] [CrossRef]
- Farhaoui, A.; El Alami, N.; Khadiri, M.; Ezrari, S.; Radouane, N.; Baala, M.; Tahiri, A.; Lahlali, R. Biological control of diseases caused by Rhizoctonia solani AG-2-2 in sugar beet (Beta vulgaris L.) using plant growth-promoting rhizobacteria (PGPR). Physiol. Mol. Plant Pathol. 2023, 124, 101966. [Google Scholar] [CrossRef]
- Malakar, C.; Barman, D.; Kalita, M.C.; Deka, S. A biosurfactant-producing novel bacterial strain isolated from vermicompost having multiple plant growth-promoting traits. J. Basic Microbiol. 2023, 63, 746–758. [Google Scholar] [CrossRef]
- Zhang, X.; Li, B.; Wang, Y.; Guo, Q.; Lu, X.; Li, S.; Ma, P. Lipopeptides, a novel protein, and volatile compounds contribute to the antifungal activity of the biocontrol agent Bacillus atrophaeus CAB-1. Appl. Microbiol. Biotechnol. 2013, 97, 9525–9534. [Google Scholar] [CrossRef]
- Li, B.; Li, Q.; Xu, Z.; Zhang, N.; Shen, Q.; Zhang, R. Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production. Front. Microbiol. 2014, 5, 636. [Google Scholar] [CrossRef]
- Paraszkiewicz, K.; Bernat, P.; Siewiera, P.; Moryl, M.; Paszt, L.S.; Trzciński, P.; Jałowiecki, Ł.; Płaza, G. Agricultural potential of rhizospheric Bacillus subtilis strains exhibiting varied efficiency of surfactin production. Sci. Hortic. 2017, 225, 802–809. [Google Scholar] [CrossRef]
- Cawoy, H.; Debois, D.; Franzil, L.; De Pauw, E.; Thonart, P.; Ongena, M. Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Microb. Biotechnol. 2015, 8, 281–295. [Google Scholar] [CrossRef]
- Benoit, I.; van den Esker, M.H.; Patyshakuliyeva, A.; Mattern, D.J.; Blei, F.; Zhou, M.; Dijksterhuis, J.; Brakhage, A.A.; Kuipers, O.P.; de Vries, R.P. Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ. Microbiol. 2015, 17, 2099–2113. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Khan, A.A. Determining the antifungal activity and characterization of Bacillus siamensis AMU03 against Macrophomina phaseolina (Tassi) Goid. Indian Phytopathol. 2020, 73, 507–516. [Google Scholar] [CrossRef]
- Gu, Q.; Yang, Y.; Yuan, Q.; Shi, G.; Wu, L.; Lou, Z.; Huo, R.; Wu, H.; Borriss, R.; Gao, X. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus Fusarium graminearum. Appl. Environ. Microbiol. 2017, 83, e01075-17. [Google Scholar] [CrossRef]
- Farzand, A.; Moosa, A.; Zubair, M.; Khan, A.R.; Hanif, A.; Tahir, H.A.S.; Gao, X. Marker assisted detection and LC-MS analysis of antimicrobial compounds in different Bacillus strains and their antifungal effect on Sclerotinia sclerotiorum. Biol. Control 2019, 133, 91–102. [Google Scholar] [CrossRef]
- Liu, L.; Jin, X.; Lu, X.; Guo, L.; Lu, P.; Yu, H.; Lv, B. Mechanisms of Surfactin from Bacillus subtilis SF1 against Fusarium foetens: A Novel Pathogen Inducing Potato Wilt. J. Fungi 2023, 9, 367. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, C.; Teruel, J.A.; Aranda, F.J.; Ortiz, A. Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim. Biophys. Acta (BBA)-Biomembr. 2003, 1611, 91–97. [Google Scholar] [CrossRef]
- Joshi, S.; Bharucha, C.; Desai, A.J. Production of biosurfactant and antifungal compound by fermented food isolate Bacillus subtilis 20B. Bioresour. Technol. 2008, 99, 4603–4608. [Google Scholar] [CrossRef]
- Chen, M.; Wang, J.; Liu, B.; Zhu, Y.; Xiao, R.; Yang, W.; Ge, C.; Chen, Z. Biocontrol of tomato bacterial wilt by the new strain Bacillus velezensis FJAT-46737 and its lipopeptides. BMC Microbiol. 2020, 20, 160. [Google Scholar] [CrossRef]
- Pal, G.; Saxena, S.; Kumar, K.; Verma, A.; Kumar, D.; Shukla, P.; Pandey, A.; Verma, S.K. Seed endophytic bacterium Bacillus velezensis and its lipopeptides acts as elicitors of defense responses against Fusarium verticillioides in maize seedlings. Plant Soil 2023. [Google Scholar] [CrossRef]
- Yamamoto, S.; Shiraishi, S.; Suzuki, S. Are cyclic lipopeptides produced by Bacillus amyloliquefaciens S13-3 responsible for the plant defence response in strawberry against Colletotrichum gloeosporioides? Lett. Appl. Microbiol. 2015, 60, 379–386. [Google Scholar] [CrossRef]
- Lam, V.B.; Meyer, T.; Arias, A.A.; Ongena, M.; Oni, F.E.; Höfte, M. Bacillus cyclic lipopeptides iturin and fengycin control rice blast caused by Pyricularia oryzae in potting and acid sulfate soils by direct antagonism and induced systemic resistance. Microorganisms 2021, 9, 1441. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.G.; Zhao, Y.Y.; Yang, Y.; Lu, F.; Dai, C.C. Endophytic fungus alleviates soil sickness in peanut crops by improving the carbon metabolism and rhizosphere bacterial diversity. Microb. Ecol. 2021, 82, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Másmela-Mendoza, J.E.; Moreno-Velandia, C.A. Bacillus velezensis supernatant mitigates tomato Fusarium wilt and affects the functional microbial structure in the rhizosphere in a concentration-dependent manner. Rhizosphere 2022, 21, 100475. [Google Scholar] [CrossRef]
| Lipopeptides | Gene | Primer Sequence (5′-3′) | Reference |
|---|---|---|---|
| Iturin A | ituA | ACCTCACCTTGATCGGCTATAC | [28] |
| TGGTGGGCGAAAGAAGTTTATG | |||
| Surfactin | srf | CGGTGATCTTGCGAAGCTTTAT | [28] |
| CGCTTTCGTTCTGCCATTCT | |||
| Fengycin | fen | CGGCCATTCGCTCATCTTTAT | [28] |
| GTTTCCGCTTCATCAGTCTCTTC |
| Material | Gene | Strain B2 |
|---|---|---|
| Iturin A | ituA | + |
| Surfactin | srf | + |
| Fengycin | fen | − |
| Lipopeptides | Fatty Acid Chain | Molecular Formula | Calculated (m/z) | Reference |
|---|---|---|---|---|
| [M + H]+ | ||||
| Iturin A | C14 | C48H74N12O14 | 1043.34665 | [13] |
| C15 | C49H76N12O14 | 1057.40227 | [13] | |
| Surfactin | C12 | C50H87N7O13 | 994.64453 | [14] |
| C13 | C51H89N7O13 | 1008.65936 | [14] | |
| C14 | C52H91N7O13 | 1022.67627 | [14] | |
| C15 | C53H93N7O13 | 1036.69275 | [14] | |
| C16 | C54H95N7O13 | 1050.46362 | [9] | |
| C17 | C55H97N7O13 | 1064.55615 | [9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Shi, Y.; Fan, D.-L.; Kang, Y.-J.; Yan, X.-L.; Wang, H.-W. Co-Culture of White Rot Fungi Pleurotus ostreatus P5 and Bacillus amyloliquefaciens B2: A Strategy to Enhance Lipopeptide Production and Suppress of Fusarium Wilt of Cucumber. J. Fungi 2023, 9, 1049. https://doi.org/10.3390/jof9111049
Xu M, Shi Y, Fan D-L, Kang Y-J, Yan X-L, Wang H-W. Co-Culture of White Rot Fungi Pleurotus ostreatus P5 and Bacillus amyloliquefaciens B2: A Strategy to Enhance Lipopeptide Production and Suppress of Fusarium Wilt of Cucumber. Journal of Fungi. 2023; 9(11):1049. https://doi.org/10.3390/jof9111049
Chicago/Turabian StyleXu, Man, Ying Shi, De-Ling Fan, Yi-Jin Kang, Xin-Li Yan, and Hong-Wei Wang. 2023. "Co-Culture of White Rot Fungi Pleurotus ostreatus P5 and Bacillus amyloliquefaciens B2: A Strategy to Enhance Lipopeptide Production and Suppress of Fusarium Wilt of Cucumber" Journal of Fungi 9, no. 11: 1049. https://doi.org/10.3390/jof9111049
APA StyleXu, M., Shi, Y., Fan, D.-L., Kang, Y.-J., Yan, X.-L., & Wang, H.-W. (2023). Co-Culture of White Rot Fungi Pleurotus ostreatus P5 and Bacillus amyloliquefaciens B2: A Strategy to Enhance Lipopeptide Production and Suppress of Fusarium Wilt of Cucumber. Journal of Fungi, 9(11), 1049. https://doi.org/10.3390/jof9111049





