Multi-Gene Phylogenetic Approach for Identification and Diversity Analysis of Bipolaris maydis and Curvularia lunata Isolates Causing Foliar Blight of Zea mays
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Maydis-Leaf-Blight-Symptomatic Samples from Major Maize-Growing Regions of India
Sampling
2.2. Isolation and Morphological Characterization of Pathogens
2.3. Molecular Characterization
2.3.1. DNA Extraction, PCR, and Sequencing
2.3.2. Amplification and Purification of PCR Products
2.3.3. Sequence Alignment and Phylogenetic Analyses
2.3.4. Statistical Analysis
| Isolates Code | Host | Location | Identified Organism | ITS1 and ITS4 Accession Number | GAPDH Accession Number | LSU Accession Number | D1 and D2 Region of LSU Accession Number | NAIMCC Accession Number | Reference |
|---|---|---|---|---|---|---|---|---|---|
| E43 | Zea mays | Hariharpur, Sant Kabir Nagar, Uttar Pradesh, India | Curvularia lunata | MH104636 | LC543643 | MT416023 | LC546663 | NAIMCC-F-03913 | This study |
| E36 | Zea mays | Ludhiana, Punjab, India | Curvularia lunata | MH104637 | LC543644 | MT416024 | LC546664 | Yet to be submitted | This study |
| E50 | Zea mays | Pantnagar, Udham Singh Nagar, Uttarakhand, India | Bipolaris maydis | MH145411 | LC543968 | MT416015 | LC546657 | NAIMCC-F-03997 | This study |
| E7 | Zea mays | Nagla, Udham Singh Nagar, Uttarakhand, India | Bipolaris maydis | MH145412 | LC508971 | MT416016 | LC546658 | NAIMCC-F-03919 | This study |
| E10 | Zea mays | HardasChak, Khagaria, Bihar, India | Bipolaris maydis | MH145413 | LC543969 | MT416017 | LC546659 | NAIMCC-F-03990 | This study |
| E15 | Zea mays | Godargama, Begusarai, Bihar, India | Bipolaris maydis | MH145414 | LC538220 | MT416018 | LC546660 | NAIMCC-F-03991 | This study |
| E13 | Zea mays | Khargajepur, MauNath Bhanjan Uttar Pradesh, India | Curvularia lunata | MH183189 | LC524133 | MT416004 | LC546646 | NAIMCC-F-03917 | This study |
| E27 | Zea mays | KhiriyaMau Nath BhanjanUttar Pradesh, India | Bipolaris maydis | MH183190 | LC544122 | MT416005 | LC546647 | NAIMCC-F-03916 | This study |
| E25 | Zea mays | Kasuet, MaunathBhanjanUttar Pradesh, India | Bipolaris maydis | MH183191 | LC529408 | MT416006 | LC546648 | NAIMCC-F-03988 | This study |
| E37 | Zea mays | Ludhiana, Punjab, India | Curvularia lunata | MH183192 | LC544123 | MT416007 | LC546649 | NAIMCC-F-03915 | This study |
| E41 | Zea mays | Azamgarh, Uttar Pradesh, India | Curvularia lunata | MH183193 | LC529749 | MT416008 | LC546650 | NAIMCC-F-03914 | This study |
| E16 | Zea mays | Sari Tole SanicharaAsthan, Samastipur, Bihar, India | Curvularia lunata | MH183194 | LC538355 | MT416009 | LC546651 | Yet to be submitted | This study |
| E39 | Zea mays | Mau Nath BhanjanUttar Pradesh, India | Curvularia lunata | MH183195 | LC544124 | MT416010 | LC546652 | Yet to be submitted | This study |
| E12 | Zea mays | Singhaw, FatehpurUttarPradesh, India | Curvularia lunata | MH183196 | LC508972 | MT416011 | LC546653 | NAIMCC-F-03918 | This study |
| E19 | Zea mays | Saidpur Ama, Begusarai, Bihar | Bipolaris maydis | MH183197 | LC541581 | MT416012 | LC546654 | NAIMCC-F-03992 | This study |
| E34 | Zea mays | Ludhiana, Punjab, India | Curvularia lunata | MH183198 | LC545392 | MT416013 | LC546655 | Yet to be submitted | This study |
| E3 | Zea mays | Godargama, Begusarai, Bihar, India | Bipolaris maydis | MT799977 | LC538353 | MT799989 | LC546665 | NAIMCC-F-03989 | This study |
| E14 | Zea mays | Lakho, Begusarai, Bihar, India | Curvularia lunata | MT799978 | LC538354 | MT799990 | LC546666 | NAIMCC-F-04001 | This study |
| E24 | Zea mays | Jigni, Rohtas, Bihar, India | Bipolaris maydis | MT524321 | LC552295 | MT516300 | MT533838 | NAIMCC-F-03993 | This study |
| E30 | Zea mays | BhindKund, Ballia, Uttar Pradesh, India | Bipolaris maydis | MT524322 | LC552288 | MT516301 | MT533839 | NAIMCC-F-03998 | This study |
| E35 | Zea mays | Ludhiana, Punjab, India | Curvularia lunata | MT524323 | LC552292 | MT516302 | MT533840 | Yet to be submitted | This study |
| E28 | Zea mays | Bhar, MauNathBhanjan, Uttar Pradesh, India | Curvularia lunata | MT524324 | LC552287 | MT516303 | MT533841 | Yet to be submitted | This study |
| E33 | Zea mays | Khojpur, Jalandhar, Punjab, India | Curvularia lunata | MT524325 | LC552290 | MT516304 | MT533842 | NAIMCC-F-03986 | This study |
| E6 | Zea mays | Talwandi Bharo, Jalandhar, Punjab, India | Curvularia lunata | MT524326 | LC552294 | MT516305 | MT533843 | Yet to be submitted | This study |
| E38 | Zea mays | Jalandhar, Punjab, India | Bipolaris maydis | MT524327 | LC552293 | MT516306 | MT533844 | NAIMCC-F-03996 | This study |
| E32 | Zea mays | Allowal, Punjab, India | Curvularia lunata | MT524328 | LC552289 | MT516307 | MT533845 | Yet to be submitted | This study |
| E31 | Zea mays | BarharaChargaha, Maharajganj, Uttar Pradesh, India | Curvularia lunata | MT524329 | LC552683 | MT516308 | MT533846 | NAIMCC-F-04002 | This study |
| E26 | Zea mays | Mustafabad, FaizabadUttar Pradesh, India | Bipolaris maydis | MT524331 | LC552286 | MT516310 | MT533848 | NAIMCC-F-03994 | This study |
| AR 5183 | Zea mays | Japan | Bipolaris maydis | KM230390 | KM034848 | NA | NA | NA | [11] |
| CBS 136.29 | Zea mays | Japan | Bipolaris maydis | KJ909769 | KM034845 | NA | NA | NA | [11] |
| AR 5182 | Zea mays | Japan | Bipolaris maydis | KM230388 | KM034844 | NA | NA | NA | [11] |
| CBS 137271/C5 | Zea mays | USA | Bipolaris maydis | AF071325 | KM034846 | NA | NA | NA | [11] |
| CBS137271/C5 | Zea mays | USA | Bipolaris maydis | AF071325 | KM034846 | KM243280 | NA | NA | [11] |
| Zea mays | NA | ||||||||
| CBS157.34 | Indonesia | Bipolaris maydis | JX256430 | JX276442 | - | NA | NA | [42] | |
| CPC 28832 | Triticum aestivum | Thailand | Bipolaris sorokiniana | MF490812 | MF490834 | - | NA | NA | [59] |
| M 1122/C4 | Zea mays | USA | Bipolaris maydis | KM230389 | KM034847 | - | NA | NA | [11] |
| CBS 108941 | NA | = | Dreschlera erythrospila | AY004782 | AY004813 | - | NA | NA | [60] |
| MFLUCC 10-0706 | Oryza sativa, | Thailand | Curvularia lunata | JX256431 | JX276443 | JX256398 | NA | NA | [11] |
| CBS730.96 | Human lung biopsy | USA | Curvularia lunata | JX256429 | JX276441 | JX256396 | NA | NA | [11] |
| MFLUCC 10-0695 | Panicum sp. | Thailand | Curvularia lunata | JX256432 | JX276444 | JX256399 | NA | NA | [11] |
| CBS 136.29 | Zea mays | Japan | Bipolaris maydis | KJ909769 | KM034845 | - | NA | NA | [11] |
| CBS 307.64 | Zea mays | USA | Bipolaris maydis | HF934925 | HG779085 | HF934875 | NA | NA | [61] |
| CBS 130.26 | Zea mays | - | Bipolaris maydis | HF934923 | HG779084 | HF934873 | NA | NA | [61] |
| CBS573.73 | Zea mays | USA | Bipolaris maydis | HF934924 | NA | MH872501 | HF934881 | NA | [61] |
| CBS 307.84 | Avena sativa | Sweden | Pyrenophora avenicola | MK539972 | MK540180 | MK540042 | NA | NA | [62] |
| CBS330.53 | = | Japan | Rhizopus oryzae | MH857229 | NA | MH868766 | NA | NA | [63] |
| ICMP 6128 | Cynodon dactylon | New Zealand | Bipolaris cynodontis | JX256412 | JX276427 | JX256380 | NA | NA | [42] |
| BRIP 12898 | Melinis munitiflora | Australia | Bipolaris melinidis | JN601035 | JN600972 | JX256411 | NA | NA | [42] |
| CBS 280.91 | Microlaenae stipoidis | Australia | Bipolaris microlaenae | JN601032 | JN600974 | JN600995 | NA | NA | [42] |
| MFLUCC 10-0694 | Oryza sativa | Thailand | Bipolaris oryzae | JX256413 | JX276428 | JX256381 | NA | NA | [42] |
| BRIP 12790 | Cynodon dactylon | Australia | Bipolaris peregianensis | JN601034 | JN600977 | JN601000 | NA | NA | [42] |
| MFLUCC 10-0705 | Panicum sp. | Thailand | Curvularia alcornii | JX256421 | JX276434 | JX256388 | NA | NA | [42] |
| MFLUCC 10-0687 | Oryza sativa | Thailand | Curvularia asianensis | JX256422 | JX276435 | JX256389 | NA | NA | [42] |
| CBS 193.62 | Air | Pakistan | Curvularia ellisii | JN192375 | JN600963 | JN600985 | NA | NA | [42] |
| ICMP 6160 | Gladiolus sp. | New Zealand | Curvularia gladioli | JX256426 | JX276438 | JX256393 | NA | NA | [42] |
| BRIP 23186a | - | Australia | Curvularia graminicola | JN192376 | JN600964 | JN600986 | NA | NA | [42] |
| BRIP 15933 | Chloris gayana | Australia | Curvularia hawaiiensis | JN601028 | JN600965 | JN600987 | NA | NA | [42] |
| CBS 284.91 | Heteropogon contortus | Australia | Curvularia heteropogonis | JN192379 | JN600969 | JN600990 | NA | NA | [42] |
| ICMP 6172 | Ischaemum indicum | Solomon Islands | Curvularia ischaemi | JX256428 | JX276440 | JX256395 | NA | NA | [42] |
| BRIP 15882a | Eragrostis interrupta | Australia | Curvularia ovariicola | JN601031 | JN600971 | JN600992 | NA | NA | [42] |
| BRIP 13165a | Sporobolus fertilis | Australia | Curvularia ravenelii | JN192386 | JN600978 | JN601001 | NA | NA | [42] |
| CBS 274.52a | Soil | Spain | Curvularia spicifera | JN192387 | JN600979 | JX256400 | NA | NA | [42] |
| BRIP 12375 | Unknown | Australia | Curvularia tripogonis | JN192388 | JN600980 | JN601002 | NA | NA | [42] |
| CBS 146.63 | Unknown | India | Curvularia tuberculata | MH858243 | LT715830 | MH869845 | NA | NA | [42] |
| CBS 192.29 | Japan | Curvularia coicis | MH855040 | HG779130 | MH866505 | NA | NA | [63] | |
| BRIP 14845 | Coffea arabica | Kenya | B. coffeana | KJ415525 | KJ415421 | KJ415478 | NA | NA | [64] |
| BRIP 12530 | Dactyloctenium radulan | Australia | B. clavate | KJ415524 | KJ415422 | KJ415477 | NA | NA | [64] |
| CBS274.91 | Eleusine indica | USA | Bipolaris eleusines | KJ909768 | KM034820 | KM243289 | NA | NA | [11] |
| BRIP 14838 | Croton sp. | - | Bipolaris crotonis | KJ415526 | KJ415420 | KJ415479 | NA | NA | [64] |
| CBS 109894 | C. dactylon | Hungary | Bipolaris cynodontis | KJ909767 | KM034838 | KM243288 | NA | NA | [11] |
| CBS241.92 | Hevea sp. | Nigeria | B.heveae | KJ909763 | KM034843 | KM243294 | NA | NA | [11] |
| BRIP 14840 | Gossypium sp. | Kenya | B. gossypina | KJ415528 | KJ415418 | KJ415481 | NA | NA | [11] |
| BRIP 15613 | Microlaena stipoides | Australia | B. microlaenae | JN601032 | JN600974 | JN600995 | NA | NA | [18] |
| CBS 199.29 | P. miliaceum | Japan | B. panici-miliace | KJ909773 | KM042896 | KM243281 | NA | NA | [18] |
| BRIP 12790 | C. dactylon | Australia | B. peregianensis | JN601034 | JN600977 | JN601000 | NA | NA | [18] |
| BRIP 14839 | E. coracana | Zambia | B. pluriseptata | KJ41553 | KJ415414 | KJ41548 | NA | NA | [64] |
| IMI 228224 | Salvinia auriculata | Brazil | B. salviniae | KJ922390 | KM034829 | KM243283 | NA | NA | [11] |
| CBS 120.24 | Italy | B. sorokiniana | KJ909776 | KM034821 | KM243278 | NA | NA | [11] | |
| CBS 624.68 | Dichanthium annulatum | USA | C. robusta | KJ909783 | KM083613 | KM243297 | NA | NA | [11] |
| CBS 349.90 | S. creber | Australia | C. riley | KJ909766 | KM083612 | KM243267 | NA | NA | [11] |
| CBS 239.48 | - | USA | Curvularia portulacae | MH856324 | LT715903 | MH867878 | NA | NA | [63] |
| CBS 350.90a | Perotis rara | Australia | Curvularia perotidis | JN192385 | HG779138 | JN600999 | NA | NA | [18] |
| CBS 156.35 | Air | Jawa | C. pallescens | KJ922380 | KM083606 | KM243269 | NA | NA | [11] |
| CBS 160.58 | Eleusine Indica | USA | C. nodulosa | JN601033 | JN600975 | JN600997 | NA | NA | [11] |
| CBS 656.74 | Desert soil | Egypt | C. subpapendorfii | KJ909777 | KM061791 | KM243266 | NA | NA | [11] |
| CBS 327.64 | Avena sativa | USA | B. victoriae | KJ909778 | KM034811 | KM243271 | NA | NA | [11] |
| BRIP 17186 | Heliconia psittacorum | Australia | Bipolaris heliconiae | KJ415530 | KJ415417 | KJ415483 | NA | NA | [64] |
| BRIP 15900 | Sorghum bicolor | Australia | Curvularia sorghina | KJ415558 | KJ415388 | KJ415512 | NA | NA | [64] |
| BRIP 14845 | Coffea arabica | Kenya | Bipolaris coffeana | KJ415525 | KJ415421 | KJ415478 | NA | NA | [64] |
| E29 | Zea mays | India | Curvularia geniculata | MT524330 | LC552684 | MT533847 | NA | NA | [49] |
3. Results
3.1. Morphological Identification of Bipolaris Isolates (Maydis Leaf Blight)
3.2. Morphological Identification of Curvularia Isolates (Maize Leaf Spot)
3.3. Pathogenicity of the Isolate
3.4. Phylogenetic Study of Bipolaris Isolates (Causal Organism of Maydis Leaf Blight)
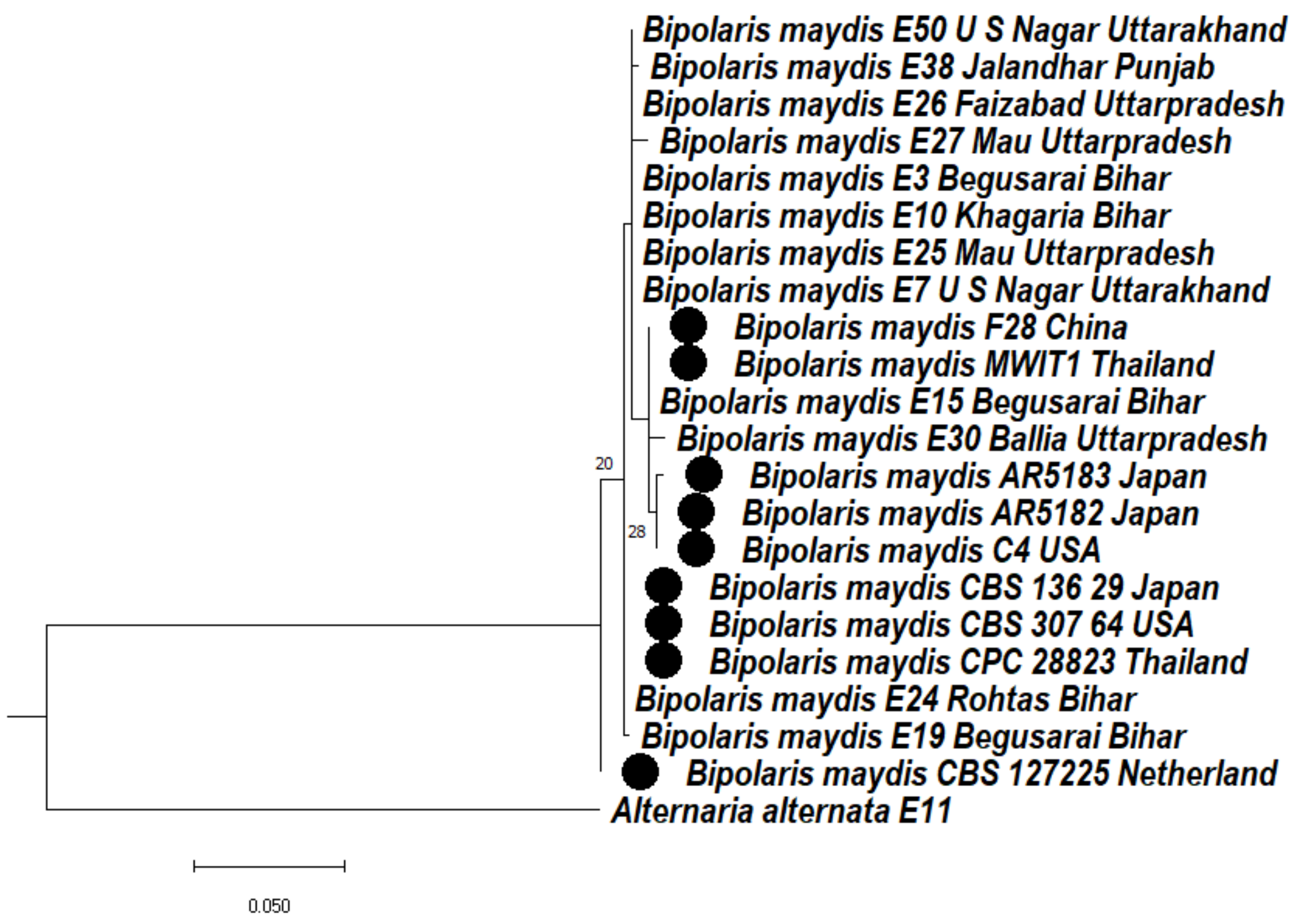
3.4.1. Phylogeny Based on the Ribosomal Marker ITS Regions of Bipolaris maydis
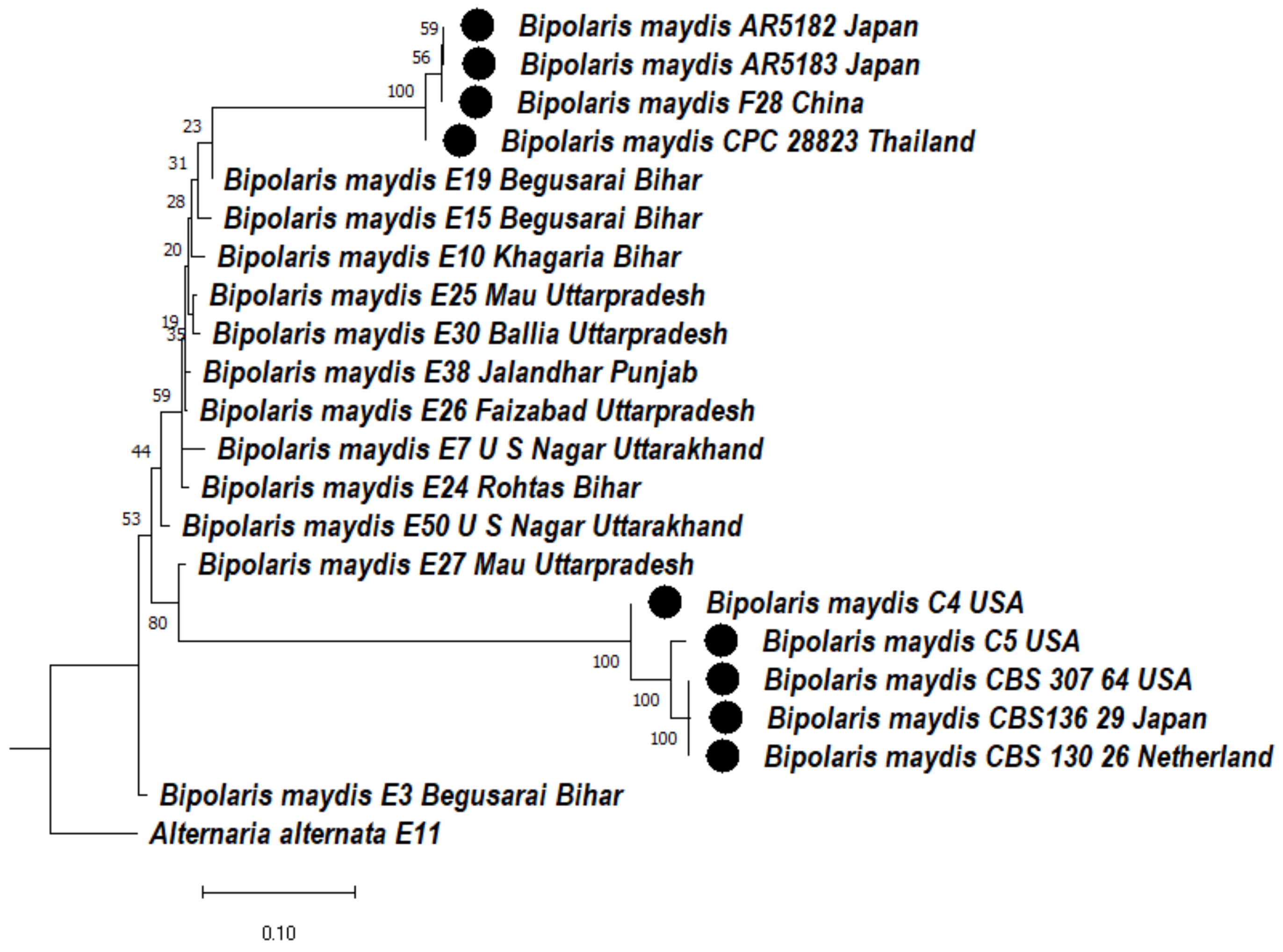
3.4.2. Phylogenetic Analysis of ITS + GAPDH Gene of Bipolaris maydis
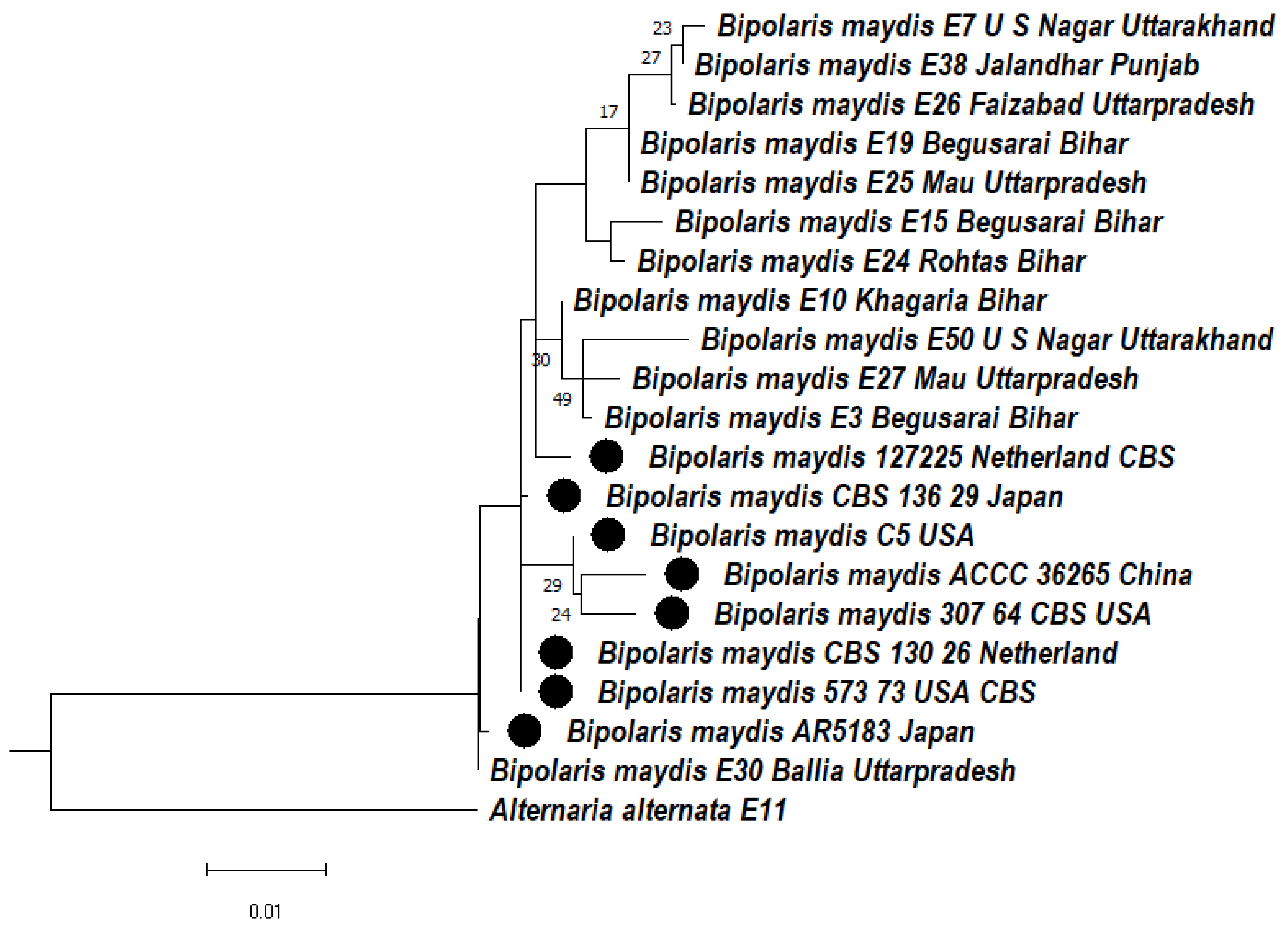
3.4.3. Phylogeny Based on the ITS + LSU Gene of Bipolaris maydis
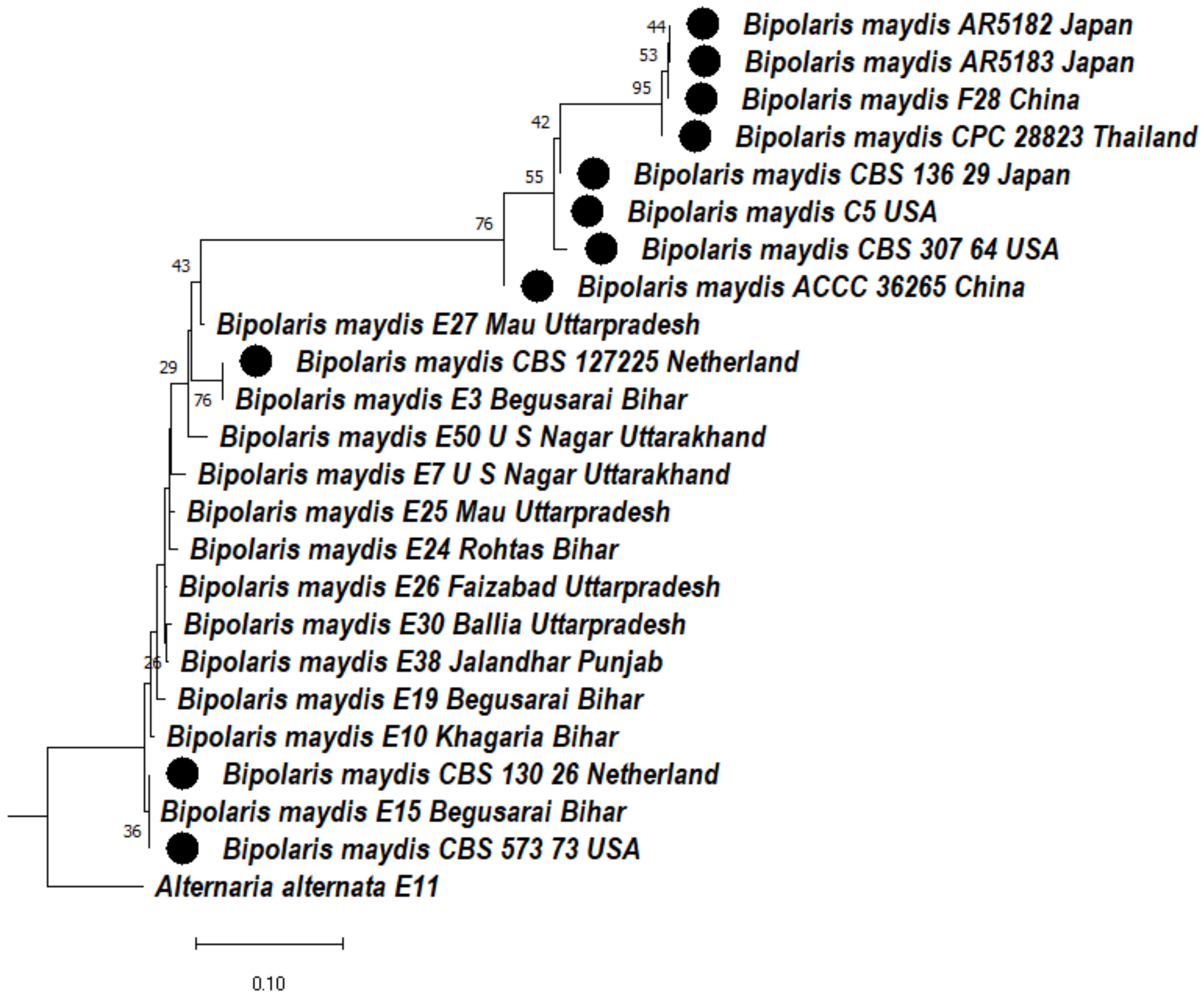
3.4.4. Phylogeny Based on the ITS + LSU + GAPDH Gene of Bipolaris maydis
3.5. Phylogenetic Study of Curvularia lunata Isolates (Causal Organism of Maize Leaf Spot)
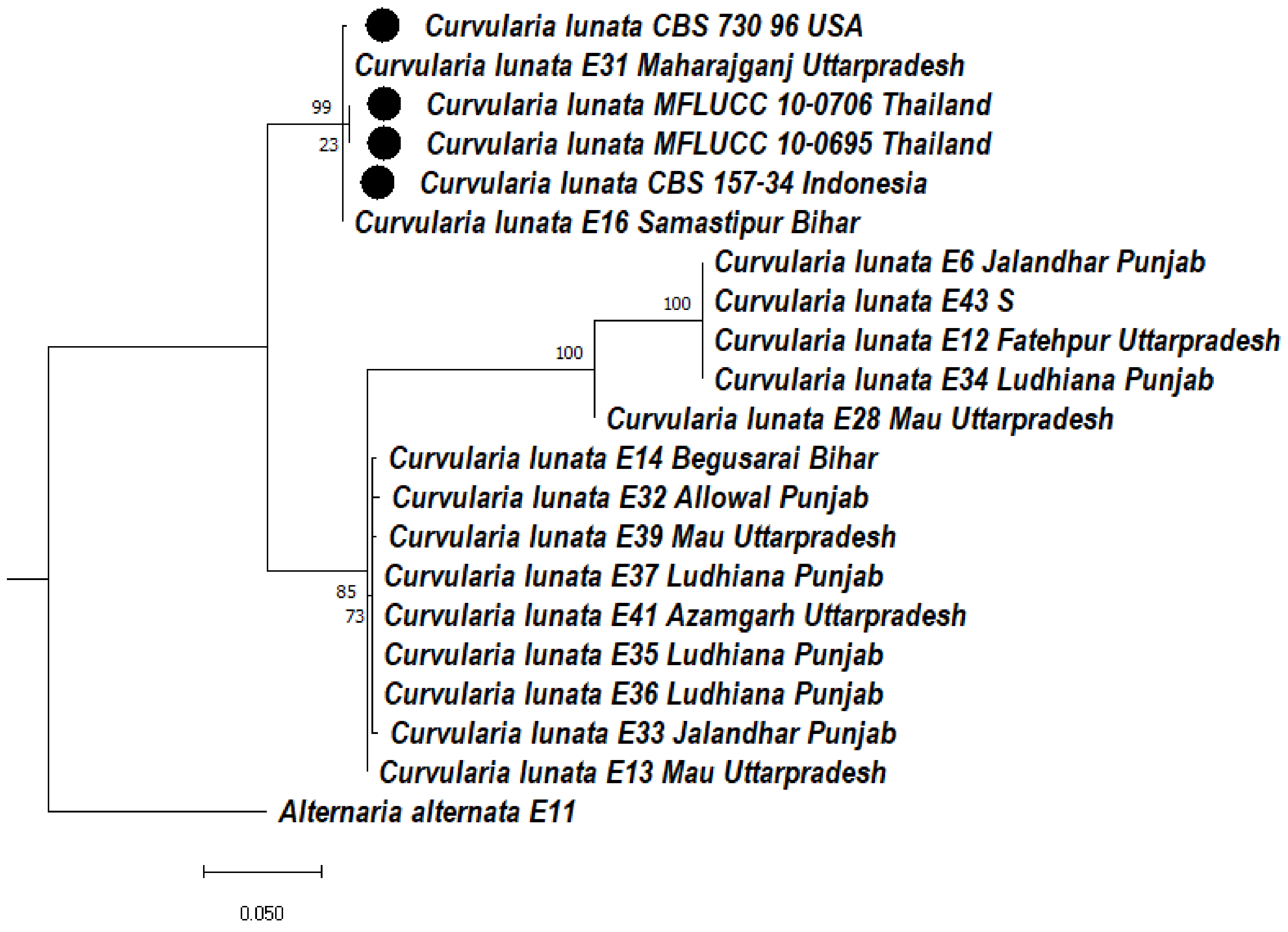
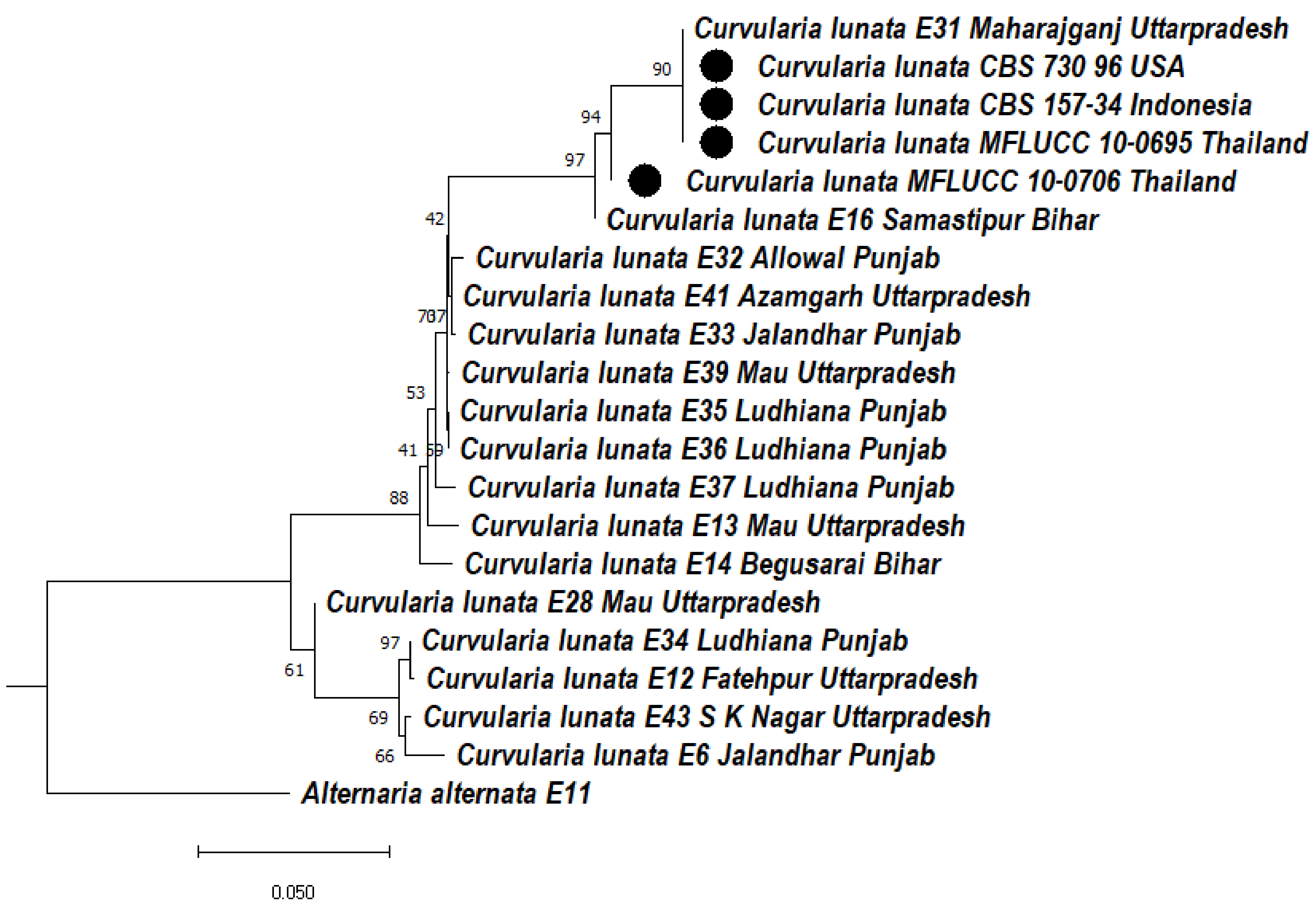
3.5.1. Phylogeny Based on the ITS of Curvularia lunata
3.5.2. Phylogeny Based on the ITS + GAPDH of Curvularia lunata
3.5.3. Phylogeny Based on the ITS + LSU of Curvularia lunata
3.5.4. Phylogeny Based on the Multigene of Curvularia lunata
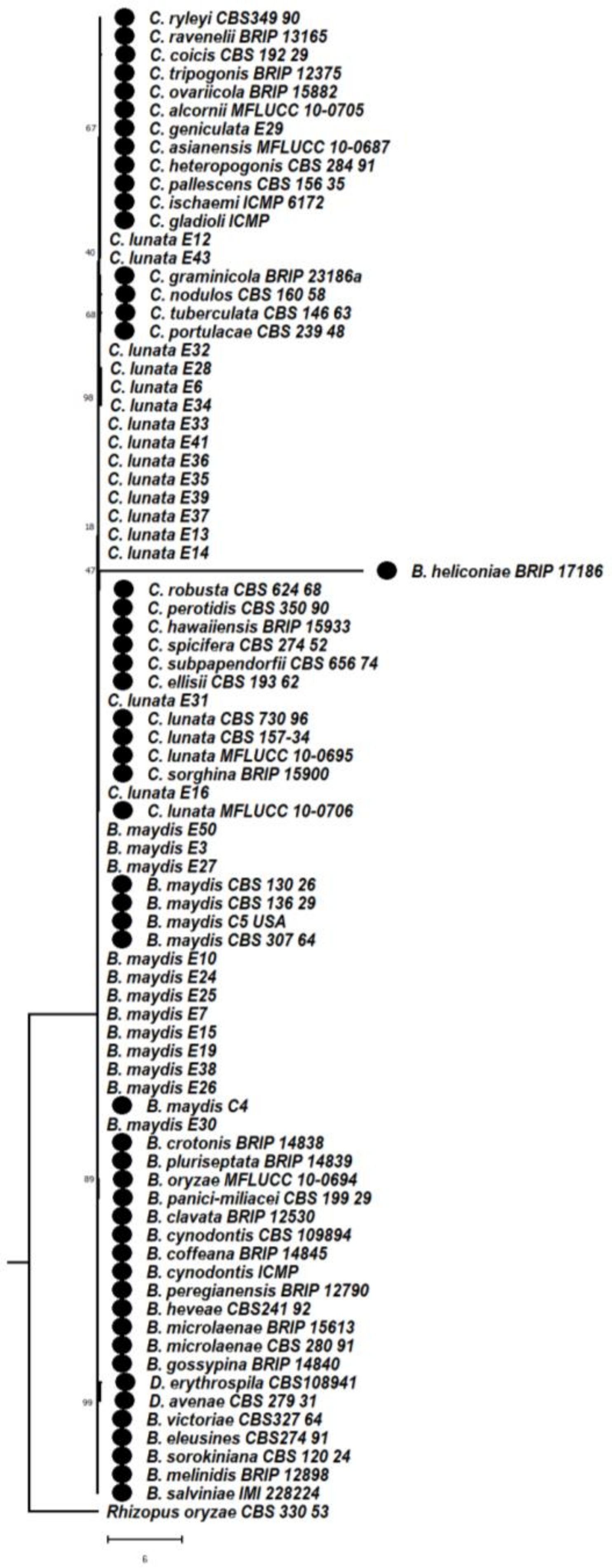
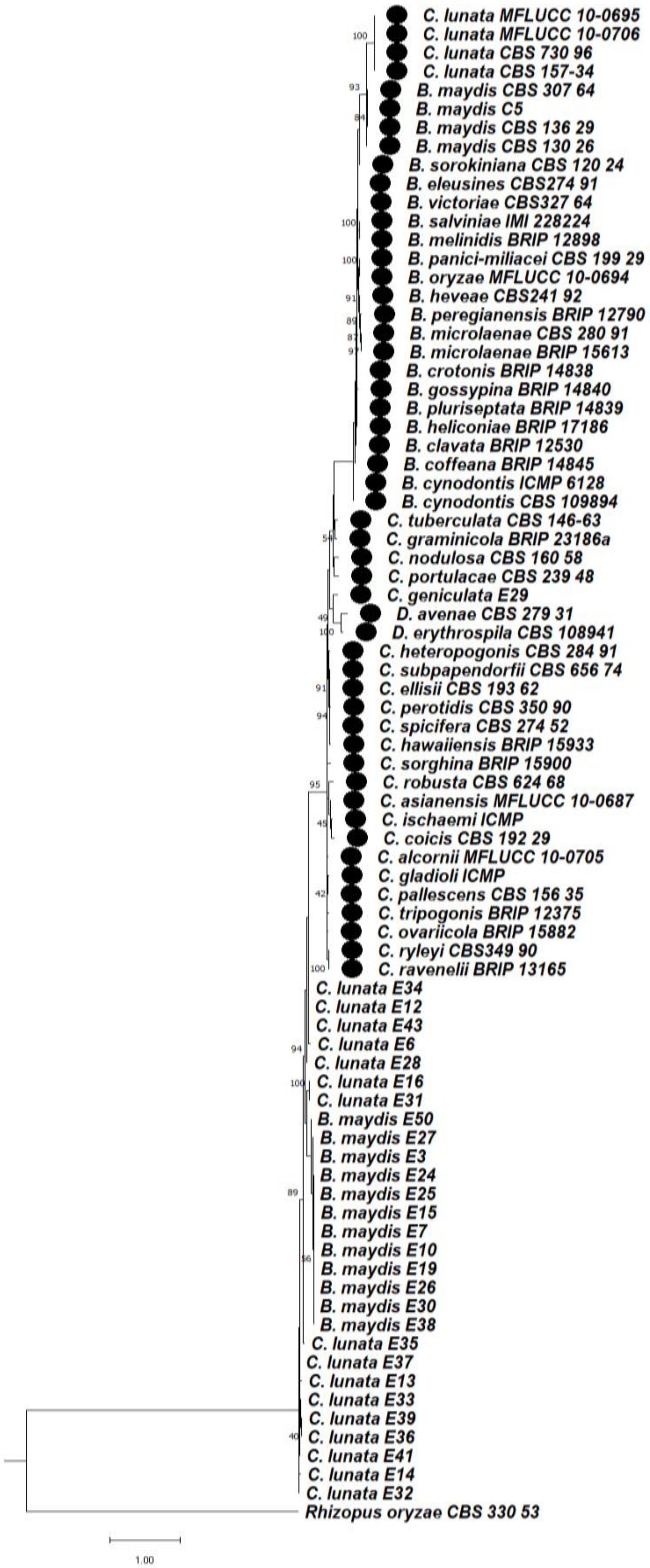
3.6. Phylogenetic Studies between Bipolaris and Curvularia Complex
3.6.1. Phylogeny Based on the Combined Sequences of ITS + GAPDH of Bipolaris maydis and Curvularia lunata
3.6.2. Phylogeny Based on the Combined Sequences of ITS + LSU of Bipolaris maydis and Curvularia lunata
3.6.3. Phylogeny Based on the Combined Regions Multigene for Bipolaris and Curvularia Complex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parihar, C.; Jat, S.; Singh, A.; Kumar, R.S.; Hooda, K.; GK, C.; Singh, D. Maize production technologies in India. DMR Tech. Bull. Dir. Maize Res. Pusa Campus New Delhi 2011, 30, 1–7. [Google Scholar]
- Ngoune Tandzi, L.; Mutengwa, C.S. Estimation of maize (Zea mays L.) yield per harvest area: Appropriate methods. Agronomy 2020, 10, 29. [Google Scholar] [CrossRef]
- Faostat, F. Food and Agriculture Organization of the United Nations-Statistic Division. 2019. Available online: https://www.fao.org/faostat/en/#data (accessed on 23 May 2021).
- Otieno, H.M. Assessment Of Key Kharif Season Maize Production Constraints in East Champaran District, India. Int. J. Sci. Technol. Res. 2019, 8, 920–924. [Google Scholar]
- Venkata Rao, P.; Subbaiah, G.; Veeraraghavaiah, R. Productivity and nutrient uptake of rice fallow maize (Zea mays L.) as influenced by plant density and fertilizer N under no-till conditions. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 826–836. [Google Scholar]
- Revilla, P.; Anibas, C.; Tracy, W. Sweet Corn Research around the World 2015–2020. Agronomy 2021, 11, 534. [Google Scholar] [CrossRef]
- Singh, M.; Mehra, R.; Malik, V.K. Evaluation of Maize Genotypes against Maydis Leaf Blight Caused by Bipolaris maydis (Nisikado and Miyake) Shoemaker under Artificial Epiphytotic Conditions. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 1006–1013. [Google Scholar] [CrossRef]
- Kumar, R.; Hooda, K.; Olakh, D.; Kaur, H.; Malik, V.; Kumar, S. Reaction of QPM inbred lines against Maydish Leaf Blight (MLB) and Charcol Rot. Electron. J. Plant Breed. 2013, 4, 1280–1283. [Google Scholar]
- Malik, V.K.; Singh, M.; Hooda, K.S.; Yadav, N.K.; Chauhan, P.K. Efficacy of newer molecules, bioagents and botanicals against maydis leaf blight and banded leaf and sheath blight of maize. Plant Pathol. J. 2018, 34, 121. [Google Scholar] [CrossRef]
- Ellis, M. Dematiaceous Hyphomycetes, Commonwealth Mycol. Inst. Kew Surrey Lond. Engl. 1971, 1, 1–608. [Google Scholar]
- Manamgoda, D.; Rossman, A.Y.; Castlebury, L.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Hyde, K.D. The genus bipolaris. Stud. Mycol. 2014, 79, 221–288. [Google Scholar] [CrossRef]
- Sivanesan, A. Graminicolous Species of Bipolaris, Curvularia, Drechslera, Exserohilum and Their Teleomorphs; CAB International: Wallingford, UK, 1987. [Google Scholar]
- Tan, Y.P.; Crous, P.W.; Shivas, R.G. Eight novel Bipolaris species identified from John L. Alcorn’s collections at the Queensland Plant Pathology Herbarium (BRIP). Mycol. Prog. 2016, 15, 1203–1214. [Google Scholar] [CrossRef]
- Sun, X.; Qi, X.; Wang, W.; Liu, X.; Zhao, H.; Wu, C.; Chang, X.; Zhang, M.; Chen, H.; Gong, G. Etiology and Symptoms of Maize Leaf Spot Caused by Bipolaris spp. in Sichuan, China. Pathogens 2020, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Chand, D.; Maurya, A.; Sharma, R.; Agarwal, P. Pathogenic fungi intercepted in introduced transgenics during 2006–2013. Plant Pathol. Quar. 2014, 4, 99–107. [Google Scholar] [CrossRef]
- Yago, J.I.; Roh, J.-H.; Bae, S.-D.; Yoon, Y.-N.; Kim, H.-J.; Nam, M.-H. The effect of seed-borne mycoflora from sorghum and foxtail millet seeds on germination and disease transmission. Mycobiology 2011, 39, 206–218. [Google Scholar] [CrossRef][Green Version]
- Bengyella, L.; Yekwa, E.L.; Nawaz, K.; Iftikhar, S.; Tambo, E.; Alisoltani, A.; Feto, N.A.; Roy, P. Global invasive Cochliobolus species: Cohort of destroyers with implications in food losses and insecurity in the twenty-first century. Arch. Microbiol. 2018, 200, 119–135. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Cai, L.; Bahkali, A.H.; Chukeatirote, E.; Hyde, K.D. Cochliobolus: An overview and current status of species. Fungal Divers. 2011, 51, 3–42. [Google Scholar] [CrossRef]
- Gogoi, R.; Singh, S.; Singh, P.K.; Kulanthaivel, S.; Rai, S. Genetic variability in the isolates of Bipolaris maydis causing maydis leaf blight of maize. Afr. J. Agric. Res. 2014, 9, 1906–1913. [Google Scholar]
- Sharma, R.; Rai, S. Assessment and quantitative analysis of losses due to Drechslera maydis in maize. In Proceedings of International Conference on Integrated Plant Disease Management for Sustainable Agriculture; Indian Phytopathological Society: Solan, India, 2000; pp. 246–248. [Google Scholar]
- Kirk, P.; Cannon, P.; Minter, D.; Stalpers, J. Ainsworth & Bisby’s Dictionary of the Fungi, 10th ed.; CAB International: Wallingford, UK, 2008. [Google Scholar]
- Gan, L.; Dai, Y.; Yang, X.; Du, Y.; Ruan, H.; Shi, N.; Chen, F. First report of leaf spot caused by Cochliobolus eragrostidis on corn (Zea mays L.) in Fujian Province, China. Plant. Dis. 2018, 102, 439. [Google Scholar] [CrossRef]
- Li, G.; Liu, K.; Xiao, S.; Lu, Y.; Xue, C.; Wang, G. First report of leaf spot of maize (Zea mays) caused by Bipolaris spicifera in China. Plant Dis. 2016, 100, 855. [Google Scholar] [CrossRef]
- Tsai, J.; Tsai, W.; Chen, J. Pathogenicity of Exserohilum rostratum on corn and weeds in the corn fields. Plant Pathol. Bull. 2001, 10, 181–186. [Google Scholar]
- Weems, J.D.; Bradley, C.A. Exserohilum turcicum race population distribution in the north central United States. Plant Dis. 2018, 102, 292–299. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Rossman, A.Y.; Castlebury, L.A.; Chukeatirote, E.; Hyde, K.D. A taxonomic and phylogenetic re-appraisal of the genus Curvularia (Pleosporaceae): Human and plant pathogens. Phytotaxa 2015, 212, 175–198. [Google Scholar] [CrossRef]
- Arabi, M.; Jawhar, M. Molecular and pathogenic variation identified among isolates of Cochliobolus sativus. Aust. Plant Pathol. 2007, 36, 17–21. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Groenewald, J.; Cai, L.; Chen, Q.; Marincowitz, S.; Barnes, I.; Bensch, K.; Braun, U.; Camporesi, E.; Damm, U. Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 2017, 86, 99–216. [Google Scholar] [CrossRef]
- Manamgoda, D.; Cai, L.; McKenzie, E.; Chukeatirote, E.; Hyde, K. Two new Curvularia species from northern Thailand. Sydowia 2012, 64, 255–266. [Google Scholar]
- Tekpinar, A.; Kalmer, A. Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwig. 2019, 109, 187–224. [Google Scholar] [CrossRef]
- Smith, M.E.; Henkel, T.W.; Uehling, J.K.; Fremier, A.K.; Clarke, H.D.; Vilgalys, R. The Ectomycorrhizal Fungal Community in a Neotropical Forest Dominated by the Endemic Dipterocarp Pakaraimaea dipterocarpacea. PLoS ONE 2013, 8, e55160. [Google Scholar] [CrossRef]
- Harrower, E.; Ammirati, J.F.; Cappuccino, A.A.; Ceska, O.; Kranabetter, J.M.; Kroeger, P.; Lim, S.; Taylor, T.; Berbee, M.L. Cortinarius species diversity in British Columbia and molecular phylogenetic comparison with European specimen sequences. Botany 2011, 89, 799–810. [Google Scholar] [CrossRef]
- Schoch, C.L.; Robbertse, B.; Robert, V.; Vu, D.; Cardinali, G.; Irinyi, L.; Meyer, W.; Nilsson, R.H.; Hughes, K.; Miller, A.N.; et al. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for Fungi. Database 2014, 2014, 21. [Google Scholar] [CrossRef]
- Stielow, J.B.; Lévesque, C.A.; Seifert, K.A.; Meyer, W.; Irinyi, L.; Smits, D.; Renfurm, R.; Verkley, G.J.M.; Groenewald, M.; Chaduli, D.; et al. One fungus, which genes? Development and Assessment of Universal Primers for Potential Secondary Fungal DNA Barcodes. Persoonia 2015, 35, 242–263. [Google Scholar] [CrossRef]
- Porter, T.M.; Shokralla, S.; Baird, D.; Golding, G.B.; Hajibabaei, M. Ribosomal DNA and Plastid Markers Used to Sample Fungal and Plant Communities from Wetland Soils Reveals Complementary Biotas. PLoS ONE 2016, 11, e0142759. [Google Scholar] [CrossRef][Green Version]
- Raja, H.A.; El-Elimat, T.; Oberlies, N.H. Minutisphaerales (Dothideomycetes, Ascomycota): A new order of freshwater ascomycetes including a new family, Minutisphaeraceae, and two new species from North Carolina, USA. Mycologia 2015, 107, 845–862. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among Ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Berbee, M.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Boonmee, S.; Zhang, Y.; Chomnunti, P.; Chukeatirote, E.; Tsui, C.K.; Bahkali, A.H.; Hyde, K.D. Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Divers. 2011, 51, 63–102. [Google Scholar] [CrossRef]
- Hunter, G.C.; Wingfield, B.D.; Crous, P.W.; Wingfield, M.J. A multi-gene phylogeny for species of Mycosphaerella occurring on Eucalyptus leaves. Stud. Mycol. 2006, 55, 147–161. [Google Scholar] [CrossRef]
- Liu, J.-K.; Phookamsak, R.; Jones, E.G.; Zhang, Y.; Ko-Ko, T.W.; Hu, H.-L.; Boonmee, S.; Doilom, M.; Chukeatirote, E.; Bahkali, A.H. Astrosphaeriella is polyphyletic, with species in Fissuroma gen. nov., and Neoastrosphaeriella gen. nov. Fungal Divers. 2011, 51, 135–154. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Cai, L.; McKenzie, E.H.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Shivas, R.G.; Tan, Y.P.; Hyde, K.D. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers. 2012, 56, 131–144. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Dong, Y.; Jayawardena, R.S.; Jeewon, R.; Phukhamsakda, C.; Bundhun, D.; Hyde, K.D.; Sheng, J. A polyphasic approach to delineate species in Bipolaris. Fungal Divers. 2020, 102, 225–256. [Google Scholar] [CrossRef]
- Dai, Y.; Yang, X.; Gan, L.; Chen, F.; Ruan, H.; Du, Y.; Shi, N.; Gao, Z. First report of southern leaf blight caused by Cochliobolus heterostrophus on corn (Zea mays) in Fujian Province, China. Plant. Dis. 2016, 100, 1781. [Google Scholar] [CrossRef]
- Alcorn, J. The taxonomy of “Helminthosporium” species. Ann. Rev. Phytopathol. 1988, 26, 37–56. [Google Scholar] [CrossRef]
- Subramanian, C.; Jain, B. A revision of some graminicolous Helminthosporia. Curr. Sci. 1966, 35, 352–355. [Google Scholar]
- Mullen, J.M. Plant Disease Diagnosis. In Plant Pathology Concepts and Laboratory Exercises, 2nd ed.; Trigiano, R.N., Windham, M.T., Windham, A.S., Eds.; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Hooda, K.S.; Bagaria, P.K.; Khokhar, M.; Kaur, H.; Rakshit, S. Mass Screening Techniques for Resistance to Maize Diseases; ICAR-Indian Institute of Maize Research, PAU Campus: Ludhiana, India, 2018. [Google Scholar]
- Manzar, N.; Kashyap, A.S.; Sharma, P.K.; Saxena, A.K. First report of leaf spot of maize caused by Curvularia geniculata in India. Plant Dis. 2021, 105, 4155. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- O’donnell, K. The fungal holomorph: Mitotic, meiotic and pleomorphic speciation in fungal systematics. Fusarium Its Near Relat. 1993, 1, 225–233. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; Jeewon, R.; Ghobad-Nejhad, M.; Wanasinghe, D.N.; Liu, N.; Phillips, A.J.; Oliveira-Filho, J.R.C.; da Silva, G.A.; Gibertoni, T.B. One stop shop II: Taxonomic update with molecular phylogeny for important phytopathogenic genera: 26–50 (2019). Fungal Divers. 2019, 94, 41–129. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Raza, M.; Zhang, Z.-F.; Hyde, K.D.; Diao, Y.-Z.; Cai, L. Culturable plant pathogenic fungi associated with sugarcane in southern China. Fungal Divers. 2019, 99, 1–104. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Trillas, M.I.; Casanova, E.; Cotxarrera, L.; Ordov’as, J.; Borrero, C.; Avil’es, M. Composts from agricultural waste and the Trichoderma asperellum strain T-34 suppress Rhizoctonia solani in cucumber seedlings. Biol. Control 2006, 39, 32–38. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Senwanna, C.; Cheewangkoon, R.; Crous, P.W. New species and records of Bipolaris and Curvularia from Thailand. Mycosphere 2017, 8, 1555–1573. [Google Scholar] [CrossRef]
- Zhang, G.; Berbee, M.L. TITLE Pyrenophora phylogenetics inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. J. Mycol. 2001, 93, 1048–1063. [Google Scholar] [CrossRef]
- Madrid, H.; Da Cunha, K.; Gené, J.; Dijksterhuis, J.; Cano, J.; Sutton, D.; Guarro, J.; Crous, P. Novel Curvularia species from clinical specimens. Pers. Mol. Phyl. Evol. Fungi 2014, 33, 48. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Iturrieta-González, I.; García, D.; Gené, J.; Groenewald, J.; Cai, L.; Chen, Q.; Quaedvlieg, W.; Schumacher, R. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019, 94, 1–124. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.; Cardinali, G.; Houbraken, J. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Tan, Y.P.; Madrid, H.; Crous, P.W.; Shivas, R.G. Johnalcornia gen. et. comb. nov., and nine new combinations in Curvularia based on molecular phylogenetic analysis. Aust. Plant. Pathol. 2014, 43, 589–603. [Google Scholar] [CrossRef]
- Payak, M.; Sharma, R. Maize diseases and approaches to their management in India. Int. J. Pest. Manag. 1985, 31, 302–310. [Google Scholar] [CrossRef]
- Drechsleb, C. A species of Hebninthosporium distinct from Helminthosporium sacohari, causing brown stripe of Sugar Cane. Phytopathology 1928, 18, 135–136. [Google Scholar]
- Luttrell, E. A taxonomic revision of Helminthosporium sativum and related species. Am. J. Bot. 1955, 42, 57–68. [Google Scholar] [CrossRef]
- Potter, P.E.; Maynard, J.B.; Pryor, W.A. Sedimentology of Shale: Study Guide and Reference Source; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Leonard, K.; Suggs, E.G. Setosphaeria prolata, the ascigerous state of Exserohilum prolatum. Mycologia 1974, 66, 281–297. [Google Scholar] [CrossRef]
- Pal, I.; Singh, V.; Gogoi, R.; Hooda, K.; Bedi, N. Characterization of Bipolaris maydis isolates of different maize cropping zones of India. Ind. Phytopathol. 2015, 68, 63–66. [Google Scholar]
- Morejon, K.R.; Moraes, M.H.D.; Bach, E.E. Identification of Bipolaris bicolor and Bipolaris sorokiniana on wheat seeds (Triticum aestivum L.) in Brazil. Braz. J. Microbiol. 2006, 37, 247–250. [Google Scholar] [CrossRef][Green Version]
- Chun, S.-C.; Loo, H.-M.; Lee, S.-H.; Jung, I.-M. A seedborne fungus Bipolaris spicifera detected from imported grass seeds. Plant Pathol. J. 2003, 19, 133–137. [Google Scholar] [CrossRef][Green Version]
- Lakshmi, P. Differential interactions between Helminthosporium maydis Nisikado & Miyake incitant of maydis leaf blight and cultivars of Zea mays LM Sc.(Ag.) Thesis. IARI 1984, 1, 26–78. [Google Scholar]
- Harlapur, S.; Kulkarni, M.; Yeshoda, H.; Srikant, K. Variability in Exserohilum turcicum (Pass.) Leonard and Suggs., causal agent of turcicum leaf blight of maize. Karn. J. Agric. Sci. 2007, 20, 665–666. [Google Scholar]
- Addangadi, K.C.; Harlapur, S.I. Studies on Curvularia Leaf Spot of Maize. Ph.D. Thesis, University of Agricultural Sciences Dharwad, Dharwad, India, 2015. [Google Scholar]



| S. No. | Sampling Sites | District Coordinates (Latitude and Longitude) | Maydis-Leaf-Blight-Symptomatic Samples (186) | Maize Leaf Spot Samples (129) |
|---|---|---|---|---|
| 1. | Jaunpur, Uttar Pradesh | 25.62 N; 82.98 E | 09 | 07 |
| 2. | Sant Kabir Nagar, Uttar Pradesh | 25.59 N; 82.98 E | 06 | 04 |
| 3. | Faizabad, Uttar Pradesh | 26.73 N; 82.15 E | 04 | 03 |
| 4. | Maharajganj, Uttar Pradesh | 27.22 N; 83.82 E | 09 | 05 |
| 5. | MauNathBhanjan, Uttar Pradesh | 25.87 N; 83.47 E | 15 | 04 |
| 6. | Ballia, Uttar Pradesh | 25.77 N; 84.16 E | 13 | 07 |
| 7. | Azamgarh, Uttar Pradesh | 25.73 N; 82.99 E | 09 | 05 |
| 8. | Udham Singh Nagar, Uttarakhand | 28.99 N; 79.51 E | 13 | 05 |
| 9. | Almora, Uttarakhand | 29.62 N; 79.67 E | 07 | 08 |
| 10. | Haldwani, Uttarakhand | 29.16 N; 79.51 E | 06 | 03 |
| 11. | Samastipur, Bihar | 25.87 N; 85.81 E | 08 | 11 |
| 12. | Khagaria, Bihar | 25.51 N; 86.50 E | 12 | 07 |
| 13. | Begusarai, Bihar | 25.38 N; 86.18 E | 18 | 12 |
| 14. | Rohtas, Bihar | 24.63 N; 83.92 E | 06 | 06 |
| 15. | Baruni, Bihar | 25.45 N; 86.00 E | 09 | 07 |
| 16. | Bhagalpur, Bihar | 25.25 N; 86.95 E | 06 | 04 |
| 17. | Katihar, Bihar | 25.39 N; 87.63 E | 08 | 02 |
| 18. | Ludhiana, Punjab | 30.90 N; 75.79 E | 09 | 11 |
| 19. | Jalandhar, Punjab | 30.99 N; 75.74 E | 11 | 06 |
| 20. | Allowal, Punjab | 31.15 N; 75.67 E | 08 | 12 |
| Group | Colony Characteristics | Conidia | ||
|---|---|---|---|---|
| Average Length (µm) | Average Width (µm) | Shape | ||
| A (n = 20) | Grey with white spots, rough raised mycelia, irregular margin | 80.44 ± 0.71 a (64–89) | 14.64 ± 0.28 ab (13–16) | Fusiform, slightly curved, dark brown, 4–9 distoseptate |
| B (n = 9) | Grey, smooth raised mycelia, irregular margin | 65.38 ± 0.64 e (53–71) | 12.43 ± 0.59 d (11–13) | Fusiform, slightly curved, light brown, 4 distoseptate |
| C (n = 25) | Grey, smooth appressed mycelia, irregular margin | 74.05 ± 0.14 c (63–97) | 15.14 ± 0.35 a (14–16) | Brown, 6–8 distoseptate |
| D (n = 11) | Blackish grey with white spot, smooth appressed mycelia, regular margin | 76.68 ± 0.54 b (68–84) | 13.44 ± 0.44 c (12–14) | Brown, 4–5 distoseptate |
| E (n = 12) | Whitish grey, rough raised mycelia, irregular margin | 71.29 ± 0.80 d (51–83) | 13.89 ± 0.38 bc (12–15) | Light brown, 5–9 distoseptate |
| Group | Colony Characteristics | Conidia | ||
|---|---|---|---|---|
| Average Length | Average Width | Shape | ||
| F (n = 15) | Black, smooth velvety mycelia, regular margin | 20.86 ± 0.85 bc (19–22) | 10.23 ± 0.35 b (8–11) | Light brown, 3–4 distoseptate |
| G (n = 8) | Grey, smooth floccose mycelia, irregular margin | 27.29 ± 0.79 a (25–29) | 12.71 ± 0.72 a (12–13) | Light yellow, 3–4 distoseptate |
| H (n = 21) | Grey, smooth appressed mycelia, irregular margin | 29.15 ± 0.27 a (25–35) | 11.96 ± 0.78 a (10–12) | Brown, 4–5 distoseptate |
| I (n = 14) | Blackish grey, smooth appressed mycelia, regular margin | 19.69 ± 0.46 c (16–24) | 9.58 ± 0.44 b (9–10) | Light brown, 3–4 distoseptate |
| J (n = 16) | Whitish grey, smooth velvety mycelia, regular margin | 22.82 ± 0.24 b (22–23) | 8.77 ± 0.67 b (8–9) | Light brown, 3–4 distoseptate |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manzar, N.; Kashyap, A.S.; Maurya, A.; Rajawat, M.V.S.; Sharma, P.K.; Srivastava, A.K.; Roy, M.; Saxena, A.K.; Singh, H.V. Multi-Gene Phylogenetic Approach for Identification and Diversity Analysis of Bipolaris maydis and Curvularia lunata Isolates Causing Foliar Blight of Zea mays. J. Fungi 2022, 8, 802. https://doi.org/10.3390/jof8080802
Manzar N, Kashyap AS, Maurya A, Rajawat MVS, Sharma PK, Srivastava AK, Roy M, Saxena AK, Singh HV. Multi-Gene Phylogenetic Approach for Identification and Diversity Analysis of Bipolaris maydis and Curvularia lunata Isolates Causing Foliar Blight of Zea mays. Journal of Fungi. 2022; 8(8):802. https://doi.org/10.3390/jof8080802
Chicago/Turabian StyleManzar, Nazia, Abhijeet Shankar Kashyap, Avantika Maurya, Mahendra Vikram Singh Rajawat, Pawan Kumar Sharma, Alok Kumar Srivastava, Manish Roy, Anil Kumar Saxena, and Harsh Vardhan Singh. 2022. "Multi-Gene Phylogenetic Approach for Identification and Diversity Analysis of Bipolaris maydis and Curvularia lunata Isolates Causing Foliar Blight of Zea mays" Journal of Fungi 8, no. 8: 802. https://doi.org/10.3390/jof8080802
APA StyleManzar, N., Kashyap, A. S., Maurya, A., Rajawat, M. V. S., Sharma, P. K., Srivastava, A. K., Roy, M., Saxena, A. K., & Singh, H. V. (2022). Multi-Gene Phylogenetic Approach for Identification and Diversity Analysis of Bipolaris maydis and Curvularia lunata Isolates Causing Foliar Blight of Zea mays. Journal of Fungi, 8(8), 802. https://doi.org/10.3390/jof8080802






