Characterization of the NRAMP Gene Family in the Arbuscular Mycorrhizal Fungus Rhizophagus irregularis
Abstract
:1. Introduction
2. Materials and Methods
2.1. RiSMFs Sequence Analyses
2.2. RNA Sequencing Analyses
2.3. Biological Materials and Growth Conditions
2.4. RNA Extraction and Gene Expression Analyses
2.5. Heterologous Expression
2.6. RiSMFs Subcellular Localization
2.7. Histochemical Staining of Fe
2.8. Mycorrhizal Colonization and ERM Development
2.9. Statistical Analyses
3. Results
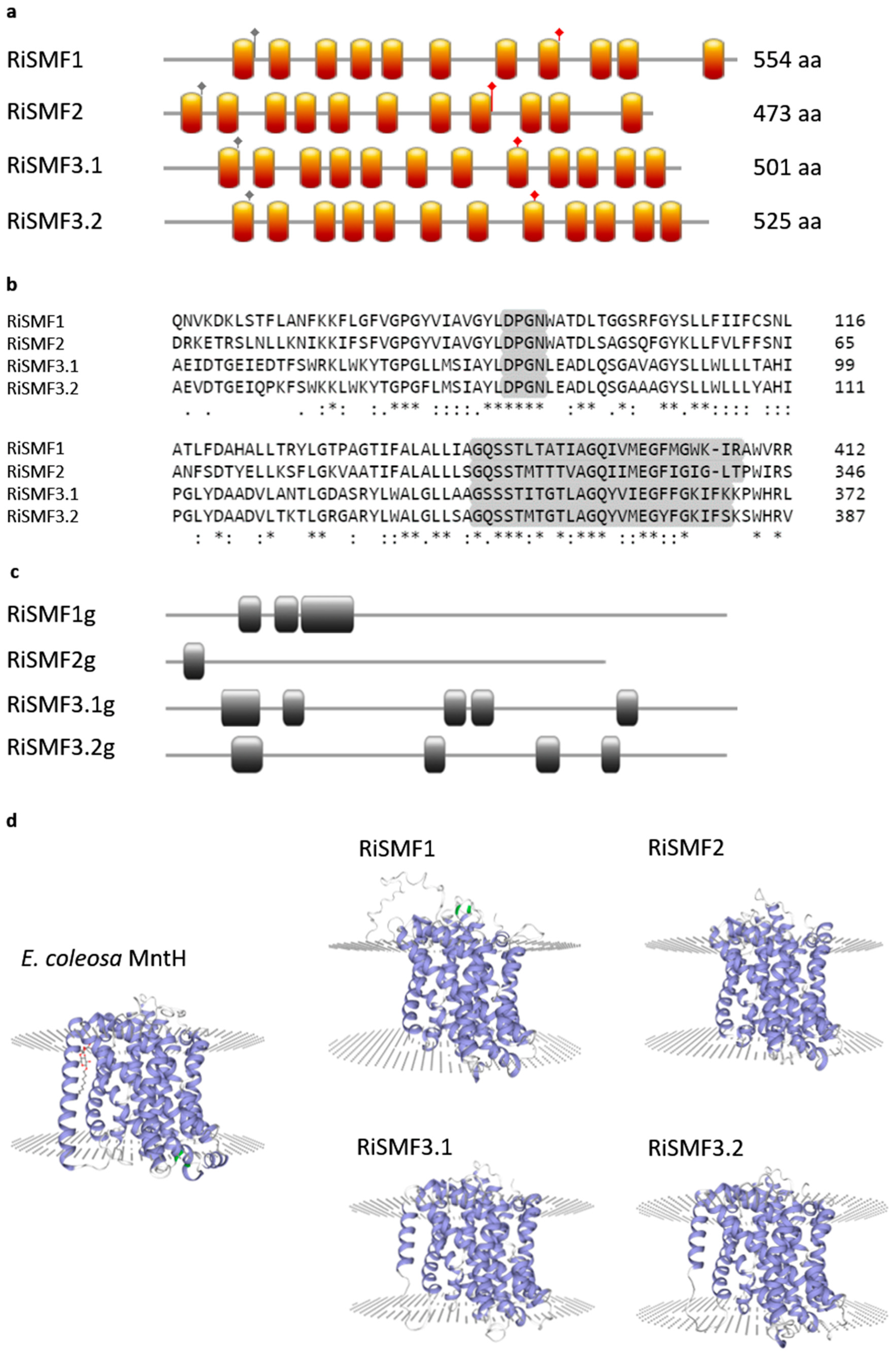
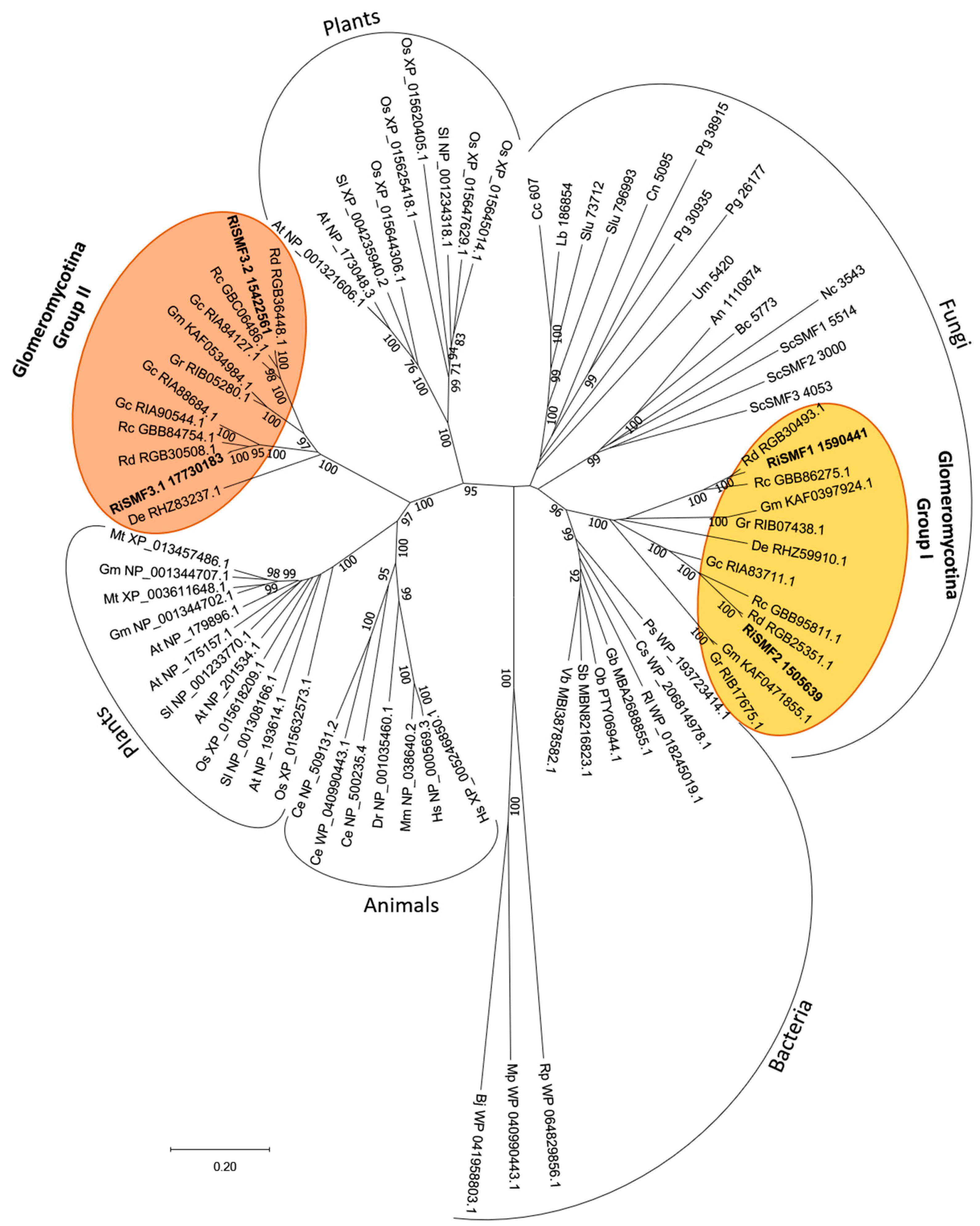
3.1. The NRAMP Family of R. irregularis
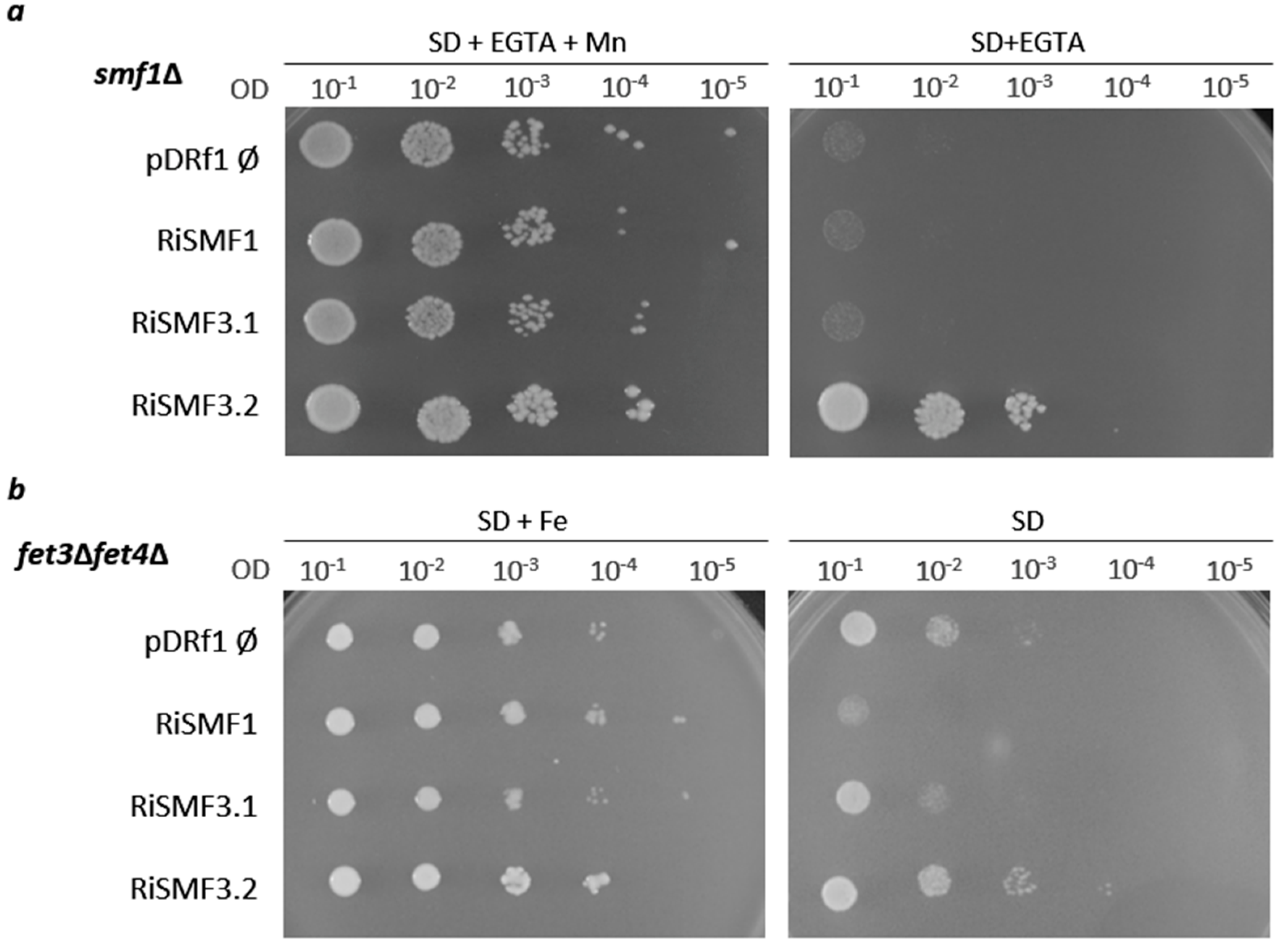
3.2. Functional Characterization of RiSMFs in Yeast
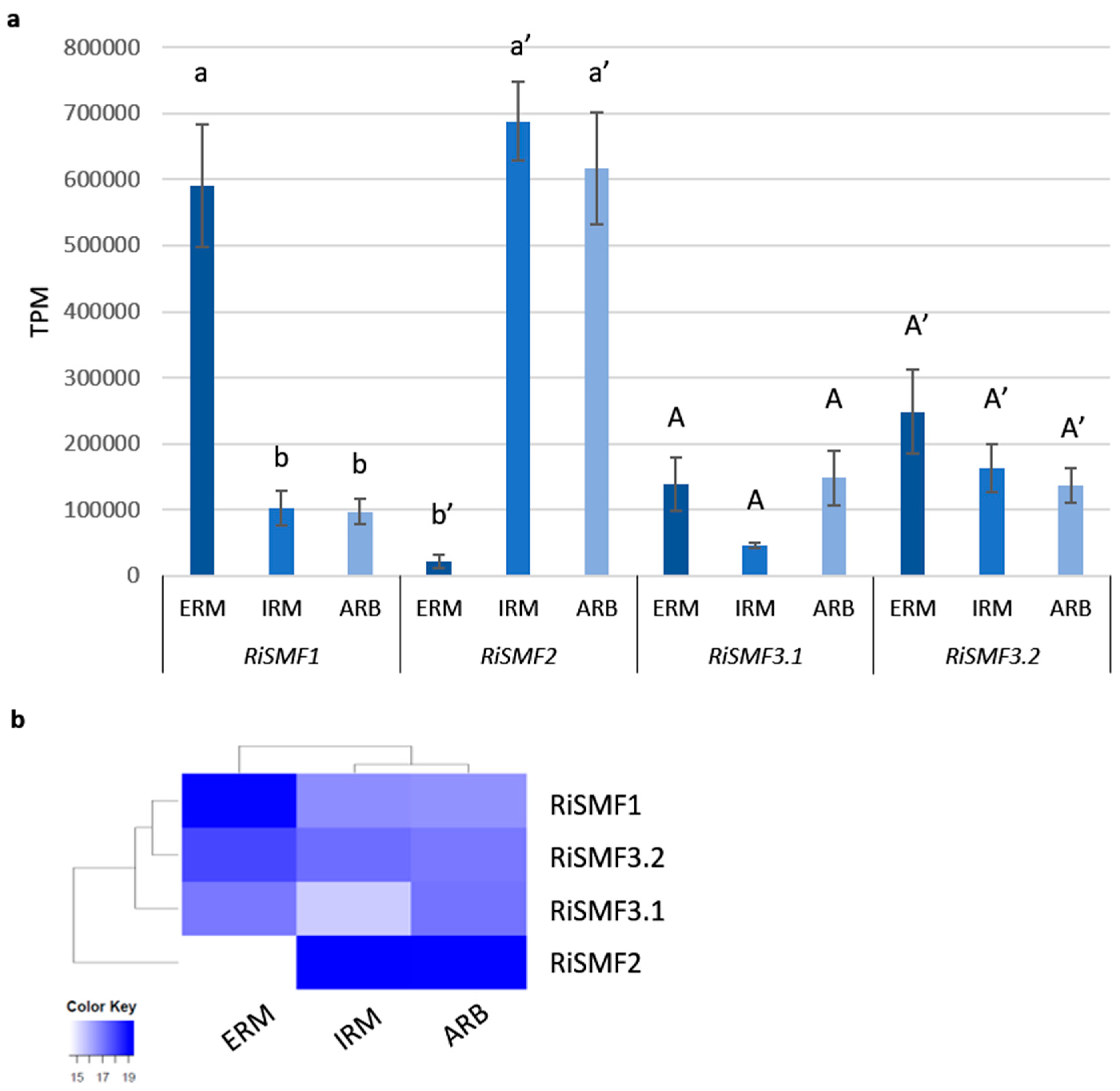
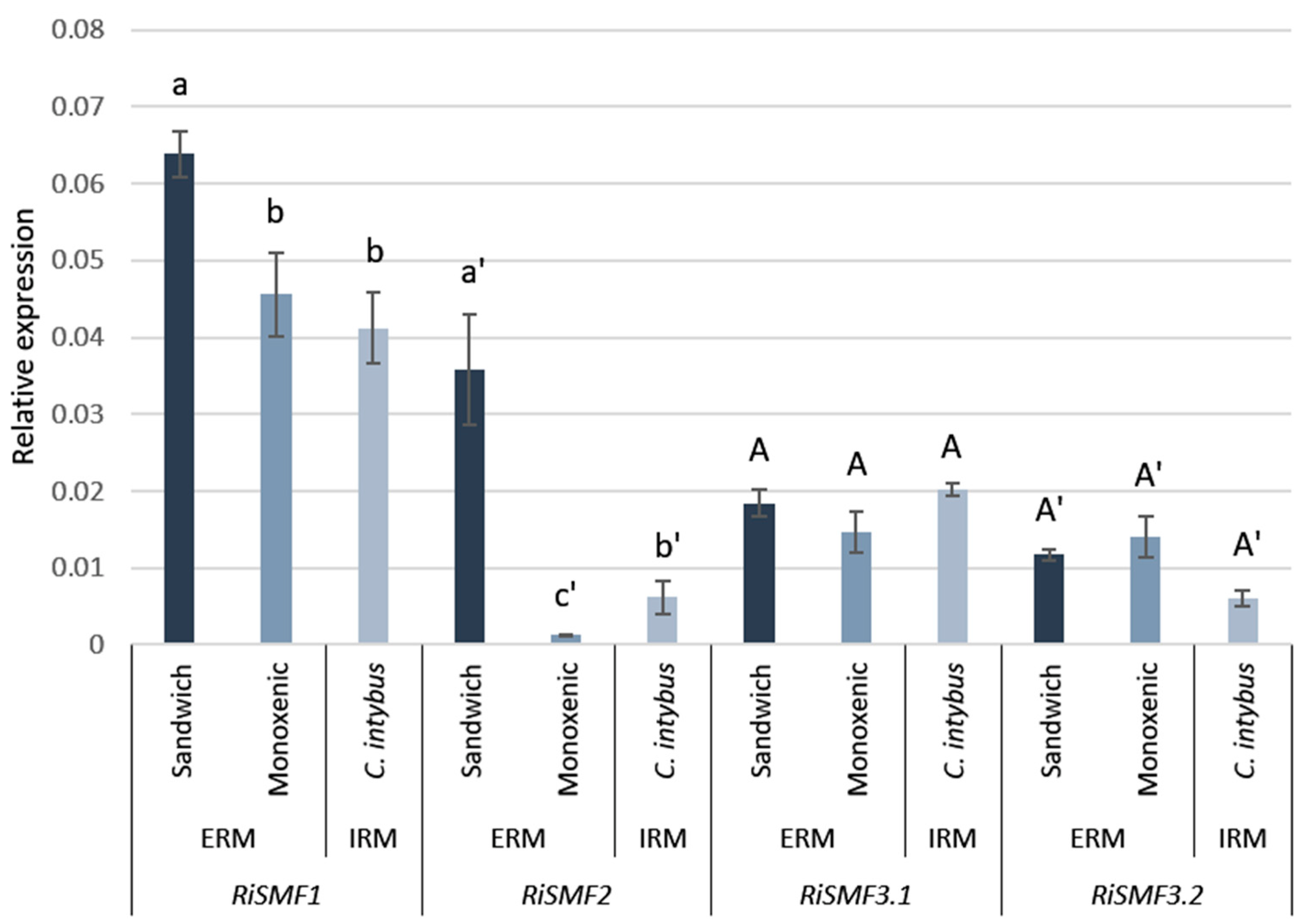
3.3. RiSMF Genes Are Differentially Expressed in Various Fungal Structures
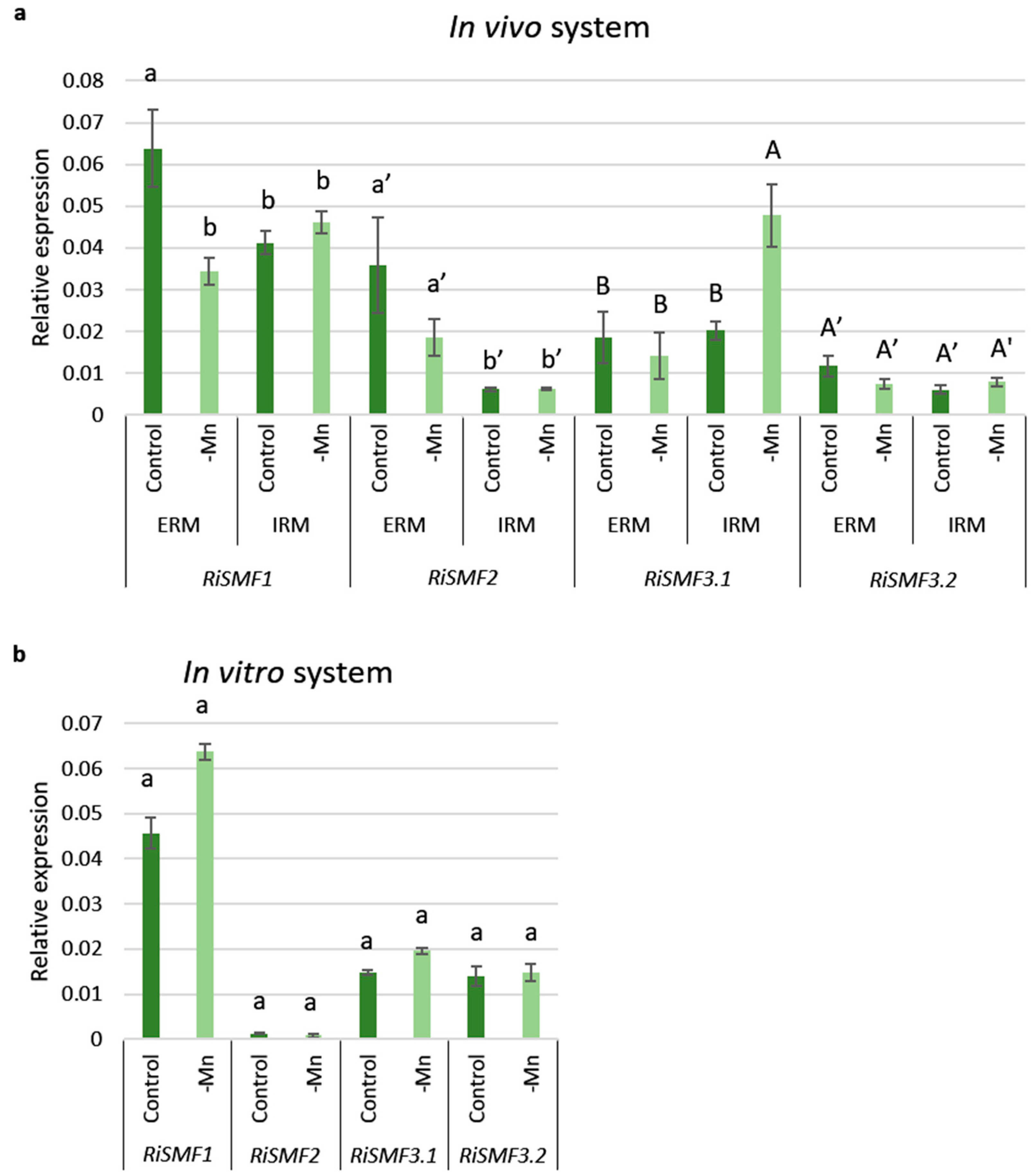
3.4. Mn Starvation Regulates RiSMF1 and RiSMF3.1 Expression
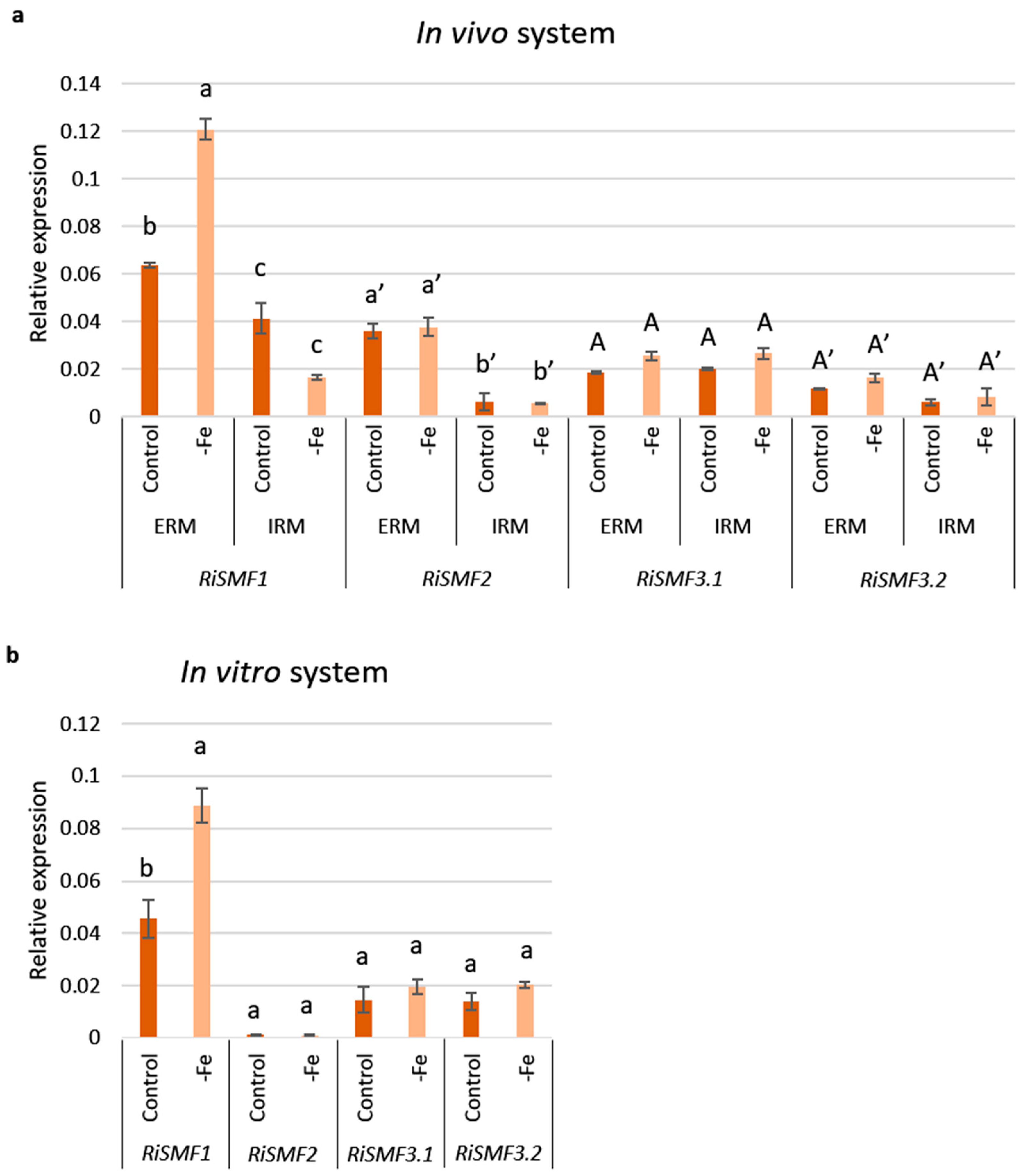
3.5. Impact of Fe Availability on RiSMFs Expression
3.6. Impact of Mn and Fe Deficiency on R. irregularis Development
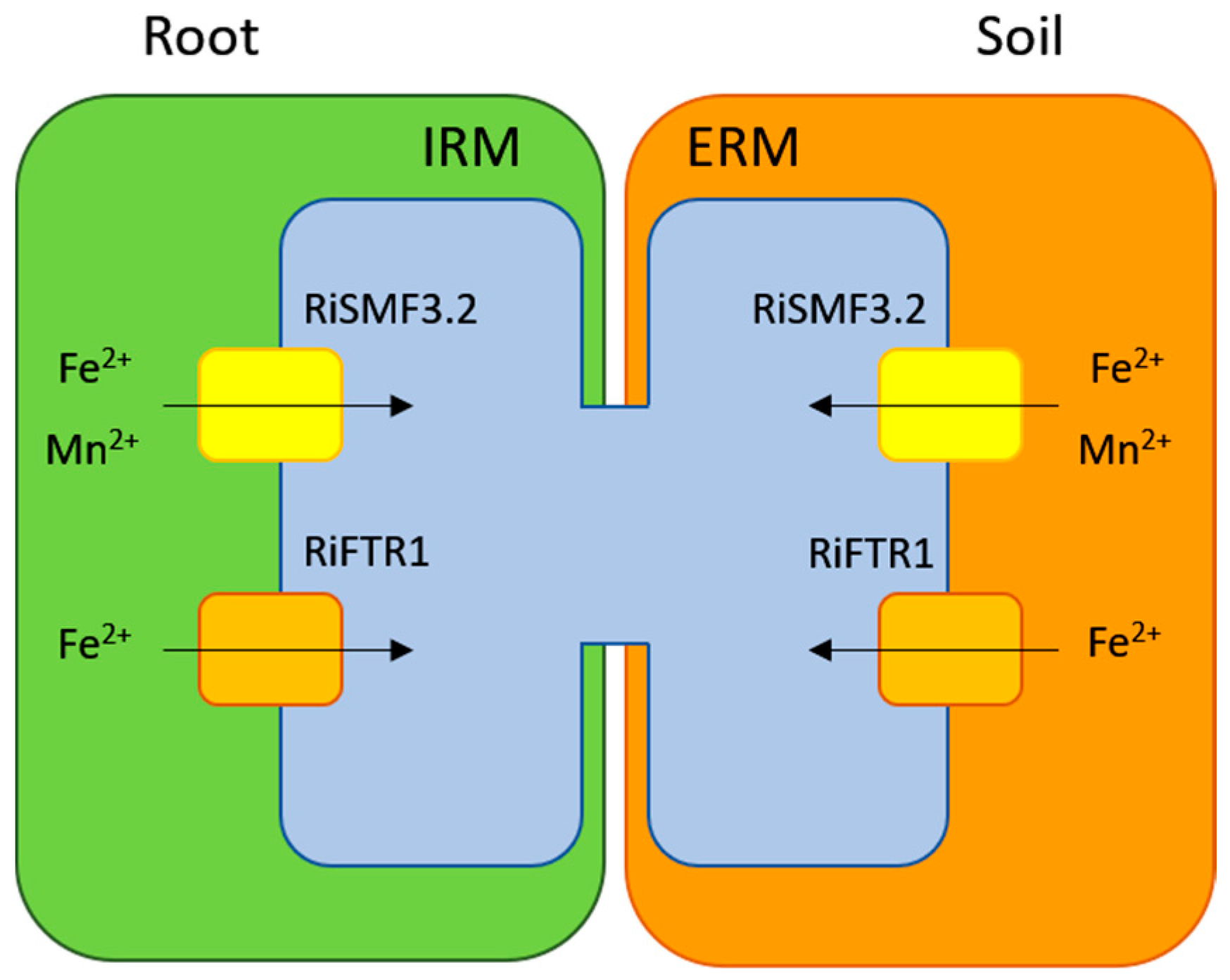
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marschner, H. Function of mineral nutrients: Micronutrients. In Mineral Nutrition of Higher Plants, 2nd ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 1995; pp. 313–404. [Google Scholar] [CrossRef]
- Schutzendubel, A.; Polle, A. Plant responses to abiotic stresses: Heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002, 53, 1351–1365. [Google Scholar] [CrossRef] [PubMed]
- Festa, R.A.; Thiele, D.J. Copper: An essential metal in biology. Curr. Biol. 2011, 21, R877–R883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, J.L.; Williams, L.E. Transition metal transporters in plants. J. Exp. Bot. 2003, 54, 2601–2613. [Google Scholar] [CrossRef]
- Nevo, Y.; Nelson, N. The NRAMP family of metal-ion transporters. Biochim. Biophys. Acta 2006, 1763, 609–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cellier, M.; Privé, G.; Belouchi, A.; Kwan, T.; Rodrigues, V.; Chia, W.; Gros, P. Nramp defines a family of membrane proteins. Proc. Natl. Acad. Sci. USA 1995, 92, 10089–10093. [Google Scholar] [CrossRef] [Green Version]
- West, A.H.; Clark, D.J.; Martin, J.; Neupert, W.; Hartl, F.U.; Horwich, A.L. Two related genes encoding extremely hydrophobic proteins suppress a lethal mutation in the yeast mitochondrial processing enhancing protein. J. Biol. Chem. 1992, 267, 24625–24633. [Google Scholar] [CrossRef]
- Chen, X.-Z.; Peng, J.-B.; Cohen, A.; Nelson, H.; Nelson, N.; Hediger, M.A. Yeast SMF1 Mediates H+-coupled Iron Uptake with Concomitant Uncoupled Cation Currents. J. Biol. Chem. 1999, 274, 35089–35094. [Google Scholar] [CrossRef] [Green Version]
- Portnoy, M.E.; Jensen, L.T.; Culotta, V.C. The distinct methods by which manganese and iron regulate the Nramp transporters in yeast. Biochem. J. 2002, 362, 119–124. [Google Scholar] [CrossRef]
- Portnoy, M.E.; Liu, X.F.; Culotta, V.C. Saccharomyces cerevisiae Expresses Three Functionally Distinct Homologues of the Nramp Family of Metal Transporters. Mol. Cell. Biol. 2000, 20, 7893–7902. [Google Scholar] [CrossRef]
- Agranoff, D.; Collins, L.; Kehres, D.; Harrison, T.; Maguire, M.; Krishna, S. The Nramp orthologue of Cryptococcus neoformans is a pH-dependent transporter of manganese, iron, cobalt and nickel. Biochem. J. 2005, 385, 225–232. [Google Scholar] [CrossRef] [Green Version]
- Tabuchi, M.; Yoshida, T.; Takegawa, K.; Kishi, F. Functional analysis of the human NRAMP family expressed in fission yeast. Biochem. J. 1999, 344, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Sugiura, R.; Kita, A.; Saito, M.; Deng, L.; He, Y.; Yabin, L.; Fujita, Y.; Takegawa, K.; Shuntoh, H.; et al. Pmr1, a P-type ATPase, and Pdt1, an Nramp homologue, cooperatively regulate cell morphogenesis in fission yeast: The importance of Mn2+ homeostasis. Genes Cells 2004, 9, 71–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fejes, B.; Ouedraogo, J.-P.; Fekete, E.; Sándor, E.; Flipphi, M.; Soós, Á.; Molnár, P.; Kovács, B.; Kubicek, C.P.; Tsang, A.; et al. The effects of external Mn2+ concentration on hyphal morphology and citric acid production are mediated primarily by the NRAMP-family transporter DmtA in Aspergillus niger. Microb. Cell Factories 2020, 19, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bensen, E.S.; Martin, S.J.; Li, M.; Berman, J.; Davis, D.A. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 2004, 54, 1335–1351. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Nagai, Y.; Kawagishi, H.; Hirai, H. Functional characterization of the manganese transporter smf2 homologue gene, PsMnt, of Phanerochaete sordida YK-624 via homologous overexpression. FEMS Microbiol. Lett. 2018, 365, 50. [Google Scholar] [CrossRef]
- Wei, Y.F.; Li, T.; Li, L.F.; Wang, J.L.; Cao, G.H.; Zhao, Z.W. Functional and transcript analysis of a novel metal transporter gene EpNramp from a dark septate endophyte (Exophiala pisciphila). Ecotoxicol. Environ. Saf. 2016, 124, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Toh-e, A.; Ohkusu, M.; Ishiwada, N.; Watanabe, A.; Kamei, K. Genetic system underlying responses of Cryptococcus neoformans to cadmium. Curr. Genet. 2022, 68, 125–141. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef] [Green Version]
- Azcón-Aguilar, C.; Barea, J.M. Nutrient cycling in the mycorrhizosphere. J. Soil Sci. Plant Nutr. 2015, 15, 372–396. [Google Scholar] [CrossRef] [Green Version]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef] [Green Version]
- Casieri, L.; Ait Lahmidi, N.; Doidy, J.; Veneault-Fourrey, C.; Migeon, A.; Bonneau, L.; Courty, P.-E.; Garcia, K.; Charbonnier, M.; Delteil, A.; et al. Biotrophic transportome in mutualistic plant–fungal interactions. Mycorrhiza 2013, 23, 597–625. [Google Scholar] [CrossRef]
- Ferrol, N.; Tamayo, E.; Vargas, P. The heavy metal paradox in arbuscular mycorrhizas: From mechanisms to biotechnological applications. J. Exp. Bot. 2016, 67, 6253–6265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivero, J.; Lidoy, J.; Llopis-Giménez, Á.; Herrero, S.; Flors, V.; Pozo, M.J. Mycorrhizal symbiosis primes the accumulation of antiherbivore compounds and enhances herbivore mortality in tomato. J. Exp. Bot. 2021, 72, 5038–5050. [Google Scholar] [CrossRef] [PubMed]
- Quiroga, G.; Erice, G.; Aroca, R.; Chaumont, F.; Ruiz-Lozano, J.M. Enhanced Drought Stress Tolerance by the Arbuscular Mycorrhizal Symbiosis in a Drought-Sensitive Maize Cultivar Is Related to a Broader and Differential Regulation of Host Plant Aquaporins than in a Drought-Tolerant Cultivar. Front. Plant Sci. 2017, 8, 1056. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.; Kamran, M.; Fang, Y.; Wang, Q.; Cao, H.; Yang, G.; Deng, L.; Wang, Y.; Zhou, Y.; Anastopoulos, I.; et al. Arbuscular mycorrhizal fungi-induced mitigation of heavy metal phytotoxicity in metal contaminated soils: A critical review. J. Hazard. Mater. 2021, 402, 123919. [Google Scholar] [CrossRef]
- Tamayo, E.; Knight, S.A.B.; Valderas, A.; Dancis, A.; Ferrol, N. The arbuscular mycorrhizal fungus Rhizophagus irregularis uses a reductive iron assimilation pathway for high-affinity iron uptake. Environ. Microbiol. 2018, 20, 1857–1872. [Google Scholar] [CrossRef]
- Gómez-Gallego, T.; Benabdellah, K.; Merlos, M.A.; Jiménez-Jiménez, A.M.; Alcon, C.; Berthomieu, P.; Ferrol, N. The Rhizophagus irregularis Genome Encodes Two CTR Copper Transporters That Mediate Cu Import Into the Cytosol and a CTR-Like Protein Likely Involved in Copper Tolerance. Front. Plant Sci. 2019, 10, 604. [Google Scholar] [CrossRef] [Green Version]
- Tamayo, E.; Gómez-Gallego, T.; Azcón-Aguilar, C.; Ferrol, N. Genome-wide analysis of copper, iron and zinc transporters in the arbuscular mycorrhizal fungus Rhizophagus irregularis. Front. Plant Sci. 2014, 5, 547. [Google Scholar] [CrossRef] [Green Version]
- Ouziad, F.; Hildebrandt, U.; Schmelzer, E.; Bothe, H. Differential gene expressions in arbuscular mycorrhizal-colonized tomato grown under heavy metal stress. J. Plant Physiol. 2005, 162, 634–649. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.d.A.; Parvin, M.; Das, U.; Ela, E.J.; Lee, S.H.; Lee, K.W.; Kabir, A.H. Arbuscular mycorrhizal symbiosis mitigates iron (Fe)-deficiency retardation in alfalfa (Medicago sativa L.) through the enhancement of Fe accumulation and sulfur-assisted antioxidant defense. Int. J. Mol. Sci. 2020, 21, 2219. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.C.H.; Morin, E.; Beaudet, D.; Noel, J.; Yildirir, G.; Ndikumana, S.; Charron, P.; St-Onge, C.; Giorgi, J.; Krüger, M.; et al. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 2018, 220, 1161–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, T.; Holmer, R.; Hontelez, J.; Lintel-Hekkert, B.; Marufu, L.; Zeeuw, T.; Wu, F.; Schijlen, E.; Bisseling, T.; Limpens, E. Host- and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J. 2018, 94, 411–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, T.; Rodriguez-Moreno, L.; Mansurkhodzaev, A.; Wang, P.; Berg, W.; Gasciolli, V.; Cottaz, S.; Fort, S.; Thomma, B.P.H.J.; Bono, J.-J.; et al. A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 2020, 225, 448–460. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chabot, S.; Bécard, G.; Piché, Y. Life cycle of Glomus intraradix in root organ culture. Mycologia 1992, 84, 315–321. [Google Scholar] [CrossRef]
- St-Arnaud, M.; Hamel, C.; Vimard, B.; Caron, M.; Fortin, J.A. Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol. Res. 1996, 100, 328–332. [Google Scholar] [CrossRef]
- Pepe, A.; Sbrana, C.; Ferrol, N.; Giovannetti, M. An in vivo whole-plant experimental system for the analysis of gene expression in extraradical mycorrhizal mycelium. Mycorrhiza 2017, 27, 659–668. [Google Scholar] [CrossRef]
- Cohen, A.; Nelson, H.; Nelson, N. The family of SMF metal ion transporters in yeast cells. J. Biol. Chem. 2000, 275, 33388–33394. [Google Scholar] [CrossRef] [Green Version]
- Eide, D.J.; Broderius, M.A.; Fett, J.; Guerinot, M.L. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 5624–5628. [Google Scholar] [CrossRef] [Green Version]
- García-Rodríguez, S.; Azcón-Aguilar, C.; Ferrol, N. Transcriptional regulation of host enzymes involved in the cleavage of sucrose during arbuscular mycorrhizal symbiosis. Physiol. Plant. 2007, 129, 737–746. [Google Scholar] [CrossRef]
- Schiestl, R.H.; Gietz, R.D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989, 16, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Trouvelot, A.; Kough, J.L.; Gianinazzi-Pearson, V. Estimation of vesicular arbuscular mycorrhizal infection levels. Research for methods having a functional significance. In Physiological and Genetical Aspects of Mycorrhizae = Aspects Physiologiques et Genetiques des Mycorhizes: Proceedings of the 1st European Symposium on Mycorrhizae, Dijon, France, 1–5 July 1985; Institut National de le Recherche Agronomique: Paris, France, 1986. [Google Scholar]
- Marsh, B.B. Measurement of length in random arrangements of lines. J. Appl. Ecol. 1971, 8, 265–267. [Google Scholar] [CrossRef]
- Supek, F.; Supekova, L.; Nelson, H.; Nelson, N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc. Natl. Acad. Sci. USA 1996, 93, 5105–5110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dix, D.R.; Bridgham, J.T.; Broderius, M.A.; Byersdorfer, C.A.; Eide, D.J. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 26092–26099. [Google Scholar] [CrossRef]
- Kauffman, K.J.; Pridgen, E.M.; Doyle, F.J.; Dhurjati, P.S.; Robinson, A.S. Decreased protein expression and intermittent recoveries in BiP levels result from cellular stress during heterologous protein expression in Saccharomyces cerevisiae. Biotechnol. Prog. 2002, 18, 942–950. [Google Scholar] [CrossRef]
- Perls, M. Nachweis von Eisenoxyd in gewissen Pigmenten. Arch. Pathol. Anat. Physiol. Klin. Med. 1867, 39, 42–48. [Google Scholar] [CrossRef]
- Helber, N.; Wippel, K.; Sauer, N.; Schaarschmidt, S.; Hause, B.; Requena, N. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 2011, 23, 3812–3823. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, Y.; Hijikata, N.; Ohtomo, R.; Handa, Y.; Kawaguchi, M.; Saito, K.; Masuta, C.; Ezawa, T. Aquaporin-mediated long-distance polyphosphate translocation directed towards the host in arbuscular mycorrhizal symbiosis: Application of virus-induced gene silencing. New Phytol. 2016, 211, 1202–1208. [Google Scholar] [CrossRef] [Green Version]
- Bozzi, A.T.; Gaudet, R. Molecular mechanism of Nramp-family transition metal transport. J. Mol. Biol. 2021, 433, 166991. [Google Scholar] [CrossRef]
- Ehrnstorfer, I.A.; Geertsma, E.R.; Pardon, E.; Steyaert, J.; Dutzler, R. Crystal structure of a SLC11 (NRAMP) transporter reveals the basis for transition-metal ion transport. Nat. Struct. Mol. Biol. 2014, 21, 990–996. [Google Scholar] [CrossRef]
- Bozzi, A.T.; Bane, L.B.; Weihofen, W.A.; McCabe, A.L.; Singharoy, A.; Chipot, C.J.; Schulten, K.; Gaudet, R. Conserved methionine dictates substrate preference in Nramp-family divalent metal transporters. Proc. Natl. Acad. Sci. USA 2016, 113, 10310–10315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaloupka, R.; Courville, P.; Veyrier, F.; Knudsen, B.; Tompkins, T.A.; Cellier, M.F.M. Identification of functional amino acids in the Nramp family by a combination of evolutionary analysis and biophysical studies of metal and proton cotransport in vivo. Biochemistry 2005, 44, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Frommer, W.B.; Ninnemann, O. Heterologous Expression of genes in bacterial, fungal, animal, and plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 419–444. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, H.; Li, Y.; Chen, Z.; Chen, T.; Zeng, B.; Zhang, Z. Identification and characterization of Nramp transporter AoNramp1 in Aspergillus oryzae. Biotech 2021, 11, 452. [Google Scholar] [CrossRef]
- Zhang, H.; Li, G.; Cao, N.; Yang, H.; Zhu, F. Genome-wide identification and expression analysis of NRAMP transporter genes in Cucumis sativus and Citrullus lanatus. Can. J. Plant Sci. 2021, 101, 377–392. [Google Scholar] [CrossRef]
- Stearman, R.; Yuan, D.S.; Yamaguchi-Iwai, Y.; Klausner, R.D.; Dancis, A. A Permease-Oxidase Complex Involved in High-Affinity Iron Uptake in Yeast. Science 1996, 271, 1552–1557. [Google Scholar] [CrossRef]
- Liu, X.F.; Culotta, V.C. Post-translation Control of Nramp Metal Transport in Yeast: Role of Metal Ions and the BDSD2 Gene. J. Biol. Chem. 1999, 274, 4863–4868. [Google Scholar] [CrossRef] [Green Version]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [Green Version]
- Johns, L.E.; Goldman, G.H.; Ries, L.N.A.; Brown, N.A. Nutrient sensing and acquisition in fungi: Mechanisms promoting pathogenesis in plant and human hosts. Fungal Biol. Rev. 2021, 36, 1–14. [Google Scholar] [CrossRef]
- Crowley, J.D.; Traynor, D.A.; Weatherburn, D.C. Enzymes and Protein Containing Manganese: An Overview. In Metal Ions in Biological Systems; CRC Press: Boca Ratón, FL, USA, 2000; pp. 209–278. [Google Scholar]
- Luk, E.E.-C.; Culotta, V.C. Manganese Superoxide Dismutase in Saccharomyces cerevisiae Acquires Its Metal Co-factor through a Pathway Involving the Nramp Metal Transporter, Smf2p. J. Biol. Chem. 2001, 276, 47556–47562. [Google Scholar] [CrossRef] [Green Version]
- Eivazi, F.; Weir, C.C. Phosphorus and mycorrhizal interaction on uptake of P and trace elements by maize. Fertil. Res. 1989, 21, 19–22. [Google Scholar] [CrossRef]
- Lehmann, A.; Rillig, M.C. Arbuscular mycorrhizal contribution to copper, manganese and iron nutrient concentrations in crops—A meta-analysis. Soil Biol. Biochem. 2015, 81, 147–158. [Google Scholar] [CrossRef]
- Kothari, S.K.; Marschner, H.; Römheld, V. Effect of a vesicular-arbuscular mycorrhizal fungus and rhizosphere micro-organisms on manganese reduction in the rhizosphere and manganese concentrations in maize (Zea mays L.). New Phytol. 1991, 117, 649–655. [Google Scholar] [CrossRef]
- Posta, K.; Marschner, H.; Römheld, V. Manganese reduction in the rhizosphere of mycorrhizal and nonmycorrhizal maize. Mycorrhiza 1994, 5, 119–124. [Google Scholar] [CrossRef]
- Arines, J.; Porto, M.E.; Vilariño, A. Effect of manganese on vesicular-arbuscular mycorrhizal development in red clover plants and on soil Mn-oxidizing bacteria. Mycorrhiza 1992, 1, 127–131. [Google Scholar] [CrossRef]
- Bago, B.; Cano, C.; Azcón-Aguilar, C.; Samson, J.; Coughlan, A.P.; Piché, Y. Differential morphogenesis of the extraradical mycelium of an arbuscular mycorrhizal fungus grown monoxenically on spatially heterogeneous culture media. Mycologia 2004, 96, 452–462. [Google Scholar] [CrossRef]
- Olsson, O.; Olsson, P.A.; Hammer, E.C. Phosphorus and carbon availability regulate structural composition and complexity of AM fungal mycelium. Mycorrhiza 2014, 24, 443–451. [Google Scholar] [CrossRef]
- Lippard, S.J.; Berg, J.M. Principles of Bioinorganic Chemistry; University Science Books: Mill Valley, CA, USA, 1994. [Google Scholar]
- Hänsch, R.; Mendel, R.R. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 2009, 12, 259–266. [Google Scholar] [CrossRef]
- González-Guerrero, M.; Melville, L.H.; Ferrol, N.; Lott, J.N.A.; Azcón-Aguilar, C.; Peterson, R.L. Ultrastructural localization of heavy metals in the extraradical mycelium and spores of the arbuscular mycorrhizal fungus Glomus intraradices. Can. J. Microbiol. 2008, 54, 103–110. [Google Scholar] [CrossRef]
- Cornejo, P.; Pérez-Tienda, J.; Meier, S.; Valderas, A.; Borie, F.; Azcón-Aguilar, C.; Ferrol, N. Copper compartmentalization in spores as a survival strategy of arbuscular mycorrhizal fungi in Cu-polluted environments. Soil Biol. Biochem. 2013, 57, 925–928. [Google Scholar] [CrossRef]
- Garcia, K.G.V.; Mendes Filho, P.F.; Pinheiro, J.I.; do Carmo, J.F.; de Araújo Pereira, A.P.; Martins, C.M.; de Abreu, M.G.P.; Filho, J.D.S.O. Attenuation of manganese-induced toxicity in Leucaena leucocephala colonized by arbuscular mycorrhizae. Water Air Soil Pollut. 2020, 231, 22. [Google Scholar] [CrossRef]
- Zanchi, C.S.; Batista, É.R.; Silva, A.O.; Barbosa, M.V.; Pinto, F.A.; dos Santos, J.V.; Carneiro, M.A.C. Recovering soils affected by iron mining tailing using herbaceous species with mycorrhizal inoculation. Water Air Soil Pollut. 2021, 232, 110. [Google Scholar] [CrossRef]
- Zago, V.C.P.; das Dores, N.C.; Watts, B.A. Strategy for phytomanagement in an area affected by iron ore dam rupture: A study case in Minas Gerais State, Brazil. Environ. Pollut. 2019, 249, 1029–1037. [Google Scholar] [CrossRef] [PubMed]

| Control | -Mn | |
|---|---|---|
| Spores/cm² | 98.67 ± 21.75a | 96.28 ± 24.86a |
| Hyphal length (cm) | 93.76 ± 14.07b | 197.97 ± 14.22a |
| RiEF1α | 0.20 ± 0.03a | 0.15 ± 0.02a |
| F% | 98.60 ± 0.50a | 93.09 ± 1.54a |
| M% | 61.93 ± 2.22a | 64.94 ± 2.47a |
| m% | 62.71 ± 2.21a | 64.62 ± 2.91a |
| A% | 46.61 ± 1.59a | 45.77 ± 3.98a |
| a% | 74.98 ± 0.63a | 74.66 ± 2.7a |
| Control | -Fe | |
|---|---|---|
| Spores/cm² | 100.21 ± 20.89a | 42.50 ± 10.78b |
| Hyphal length (cm) | 95.80 ± 6.01a | 115.01 ± 6.12a |
| RiEF1α | 0.18 ± 0.05a | 0.20 ± 0.01a |
| F% | 97.12 ± 2.00a | 90.88 ± 2.69a |
| M% | 61.93 ± 4.34a | 49.55 ± 8.42a |
| m% | 60.75 ± 3.50a | 54.27 ± 8.70a |
| A% | 39.36 ± 6.81a | 32.04 ± 7.05a |
| a% | 71.95 ± 7.31a | 59.72 ± 9.36a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Lorca, V.M.; Molina-Luzón, M.J.; Ferrol, N. Characterization of the NRAMP Gene Family in the Arbuscular Mycorrhizal Fungus Rhizophagus irregularis. J. Fungi 2022, 8, 592. https://doi.org/10.3390/jof8060592
López-Lorca VM, Molina-Luzón MJ, Ferrol N. Characterization of the NRAMP Gene Family in the Arbuscular Mycorrhizal Fungus Rhizophagus irregularis. Journal of Fungi. 2022; 8(6):592. https://doi.org/10.3390/jof8060592
Chicago/Turabian StyleLópez-Lorca, Víctor Manuel, María Jesús Molina-Luzón, and Nuria Ferrol. 2022. "Characterization of the NRAMP Gene Family in the Arbuscular Mycorrhizal Fungus Rhizophagus irregularis" Journal of Fungi 8, no. 6: 592. https://doi.org/10.3390/jof8060592
APA StyleLópez-Lorca, V. M., Molina-Luzón, M. J., & Ferrol, N. (2022). Characterization of the NRAMP Gene Family in the Arbuscular Mycorrhizal Fungus Rhizophagus irregularis. Journal of Fungi, 8(6), 592. https://doi.org/10.3390/jof8060592







