Differential Gene Expression of Mucor lusitanicus under Aerobic and Anaerobic Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Culture Conditions
2.2. RNA Extraction and Sequencing
2.3. Analysis of the RNA-Seq Data
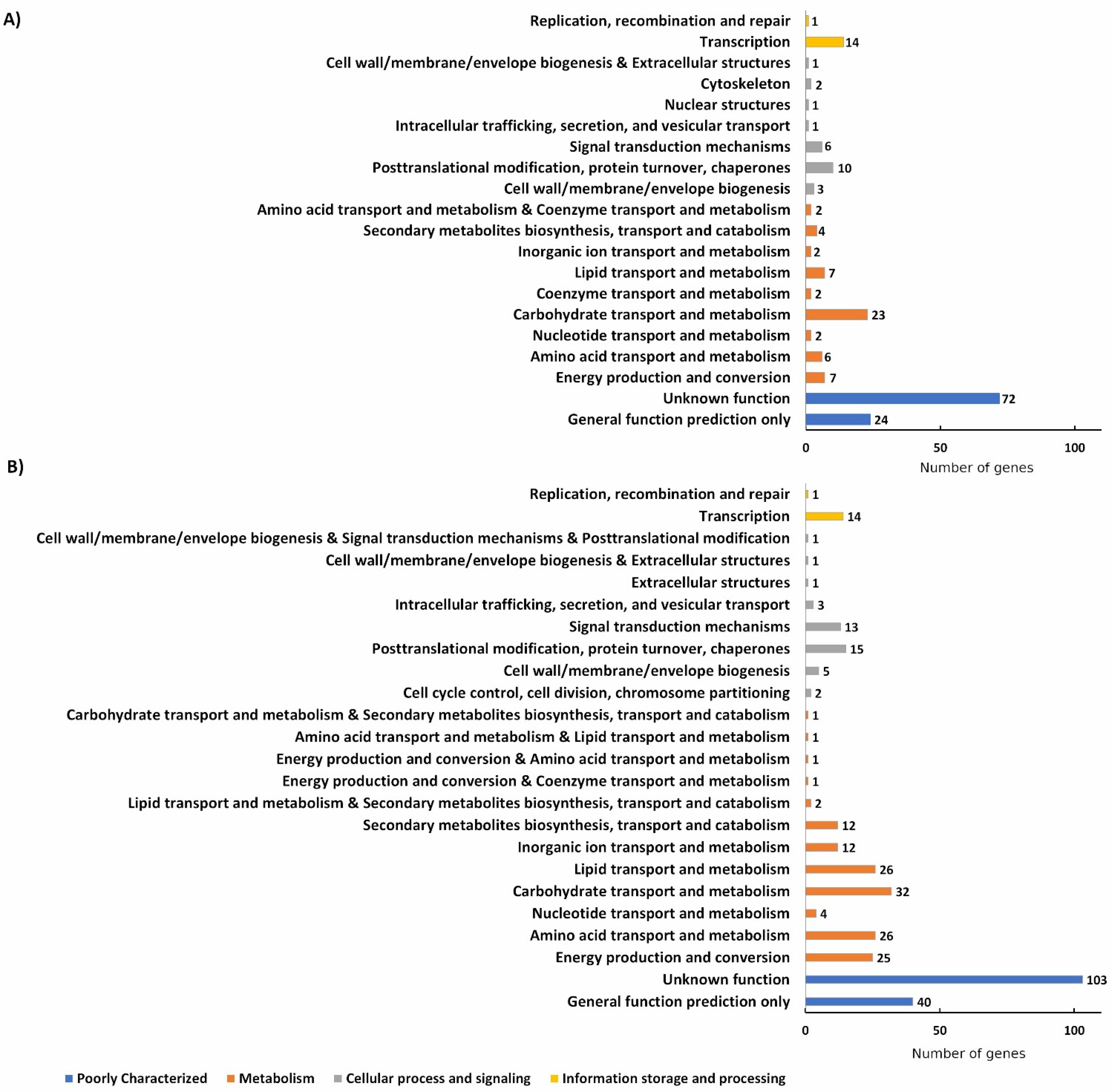
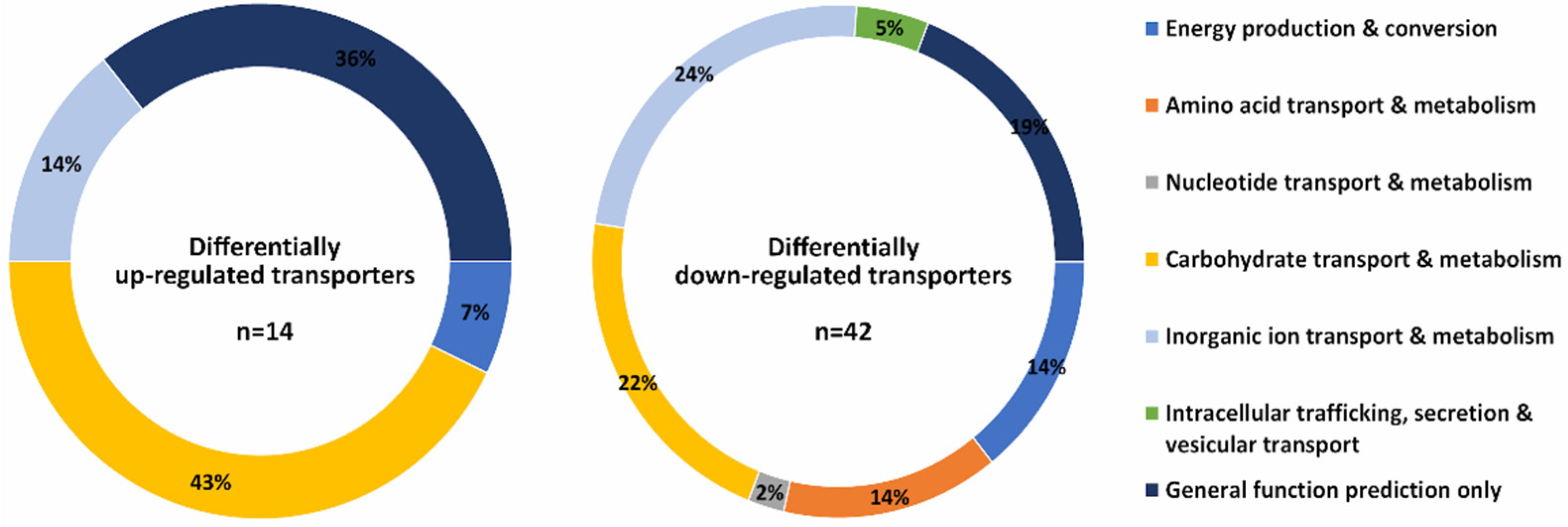
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wagner, L.; Stielow, J.B.; de Hoog, G.S.; Bensch, K.; Schwartze, V.U.; Voigt, K.; Alastruey-Izquierdo, A.; Kurzai, O.; Walther, G. A new species concept for the clinically relevant Mucor circinelloides complex. Pers. Mol. Phylogeny Evol. Fungi 2020, 44, 67–97. [Google Scholar] [CrossRef] [PubMed]
- Voigt, K.; Wolf, T.; Ochsenreiter, K.; Nagy, G.; Kaerger, K.; Shelest, E.; Papp, T. 15 Genetic and metabolic aspects of primary and secondary metabolism of the Zygomycetes. Biochem. Mol. Biol. 2016, 3, 361–385. [Google Scholar] [CrossRef]
- Avalos, J.; Nordzieke, S.; Parra, O.; Pardo-Medina, J.; Carmen Limón, M. Carotenoid Production by Filamentous Fungi and Yeasts BT—Biotechnology of Yeasts and Filamentous Fungi; Sibirny, A.A., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 225–279. ISBN 978-3-319-58829-2. [Google Scholar]
- Tang, X.; Zan, X.; Zhao, L.; Chen, H.; Chen, Y.Q.; Chen, W.; Song, Y.; Ratledge, C. Proteomics analysis of high lipid-producing strain Mucor circinelloides WJ11: An explanation for the mechanism of lipid accumulation at the proteomic level. Microb. Cell Fact. 2016, 15, 35. [Google Scholar] [CrossRef] [PubMed]
- Londoño-Hernández, L.; Ramírez-Toro, C.; Ruiz, H.A.; Ascacio-Valdés, J.A.; Aguilar-Gonzalez, M.A.; Rodríguez-Herrera, R.; Aguilar, C.N. Rhizopus oryzae—Ancient microbial resource with importance in modern food industry. Int. J. Food Microbiol. 2017, 257, 110–127. [Google Scholar] [CrossRef]
- Papp, T.; Nyilasi, I.; Csernetics, Á.; Nagy, G.; Takó, M.; Vágvölgyi, C. Improvement of Industrially Relevant Biological Activities in Mucoromycotina Fungi BT—Gene Expression Systems in Fungi: Advancements and Applications; Schmoll, M., Dattenböck, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 97–118. ISBN 978-3-319-27951-0. [Google Scholar]
- Riley, T.T.; Muzny, C.A.; Swiatlo, E.; Legendre, D.P. Breaking the mold: A review of mucormycosis and current pharmacological treatment options. Ann. Pharmacother. 2016, 50, 747–757. [Google Scholar] [CrossRef]
- Jeong, W.; Keighley, C.; Wolfe, R.; Lee, W.L.; Slavin, M.A.; Kong, D.C.M.; Chen, S.C.-A. The epidemiology and clinical manifestations of mucormycosis: A systematic review and meta-analysis of case reports. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2019, 25, 26–34. [Google Scholar] [CrossRef]
- Skiada, A.; Lanternier, F.; Groll, A.H.; Pagano, L.; Zimmerli, S.; Herbrecht, R.; Lortholary, O.; Petrikkos, G.L. Diagnosis and treatment of mucormycosis in patients with hematological malignancies: Guidelines from the 3rd European Conference on Infections in Leukemia (ECIL 3). Haematologica 2013, 98, 492–504. [Google Scholar] [CrossRef]
- Iturriaga, E.A.; Alvarez, M.I.; Eslava, A.P.; Papp, T. Expression Vectors and Gene Fusions for the Directed Modification of the Carotenoid Biosynthesis Pathway in Mucor circinelloides BT—Microbial Carotenoids: Methods and Protocols; Barreiro, C., Barredo, J.-L., Eds.; Springer New York: New York, NY, USA, 2018; pp. 239–256. ISBN 978-1-4939-8742-9. [Google Scholar]
- Naz, T.; Nosheen, S.; Li, S.; Nazir, Y.; Mustafa, K.; Liu, Q.; Garre, V.; Song, Y. Comparative analysis of β-carotene production by Mucor circinelloides strains CBS 277.49 and WJ11 under light and dark conditions. Metabolites 2020, 10, 38. [Google Scholar] [CrossRef]
- Carvalho, A.K.F.; Bento, H.B.S.; Izário Filho, H.J.; de Castro, H.F. Approaches to convert Mucor circinelloides lipid into biodiesel by enzymatic synthesis assisted by microwave irradiations. Renew. Energy 2018, 125, 747–754. [Google Scholar] [CrossRef]
- Carvalho, A.K.F.; da Conceição, L.R.V.; Silva, J.P.V.; Perez, V.H.; de Castro, H.F. Biodiesel production from Mucor circinelloides using ethanol and heteropolyacid in one and two-step transesterification. Fuel 2017, 202, 503–511. [Google Scholar] [CrossRef]
- Valle-Maldonado, M.I.; Jácome-Galarza, I.E.; Gutiérrez-Corona, F.; Ramírez-Díaz, M.I.; Campos-García, J.; Meza-Carmen, V. Selection of reference genes for quantitative real time RT-PCR during dimorphism in the zygomycete Mucor circinelloides. Mol. Biol. Rep. 2015, 42, 705–711. [Google Scholar] [CrossRef]
- Lee, S.C.; Li, A.; Calo, S.; Heitman, J. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog. 2013, 9, e1003625. [Google Scholar] [CrossRef]
- Lee, S.C.; Li, A.; Calo, S.; Inoue, M.; Tonthat, N.K.; Bain, J.M.; Louw, J.; Shinohara, M.L.; Erwig, L.P.; Schumacher, M.A.; et al. Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host-pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides. Mol. Microbiol. 2015, 97, 844–865. [Google Scholar] [CrossRef]
- Torres-Martínez, S.; Ruiz-Vazquez, R.M. RNAi pathways in Mucor: A tale of proteins, small RNAs and functional diversity. Fungal Genet. Biol. 2016, 90, 44–52. [Google Scholar] [CrossRef]
- Li, C.H.; Cervantes, M.; Springer, D.J.; Boekhout, T.; Ruiz-Vazquez, R.M.; Torres-Martinez, S.R.; Heitman, J.; Lee, S.C. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 2011, 7, e1002086. [Google Scholar] [CrossRef]
- Chang, Z.; Heitman, J. Drug-resistant epimutants exhibit organ-specific stability and induction during murine infections caused by the human fungal pathogen Mucor circinelloides. mBio 2019, 10, e02579-19. [Google Scholar] [CrossRef]
- Vellanki, S.; Billmyre, R.B.; Lorenzen, A.; Campbell, M.; Turner, B.; Huh, E.Y.; Heitman, J.; Lee, S.C. A novel resistance pathway for calcineurin inhibitors in the human-pathogenic mucorales mucor circinelloides. mBio 2020, 11, e02949-19. [Google Scholar] [CrossRef]
- Nagy, G.; Vaz, A.G.; Szebenyi, C.; Takó, M.; Tóth, E.J.; Csernetics, Á.; Bencsik, O.; Szekeres, A.; Homa, M.; Ayaydin, F.; et al. CRISPR-Cas9-mediated disruption of the HMG-CoA reductase genes of Mucor circinelloides and subcellular localization of the encoded enzymes. Fungal Genet. Biol. 2019, 129, 30–39. [Google Scholar] [CrossRef]
- Nagy, G.; Szebenyi, C.; Csernetics, Á.; Vaz, A.G.; Tóth, E.J.; Vágvölgyi, C.; Papp, T. Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci. Rep. 2017, 7, 16800. [Google Scholar] [CrossRef]
- Garcia, A.; Adedoyin, G.; Heitman, J.; Lee, S.C. Construction of a recyclable genetic marker and serial gene deletions in the human pathogenic Mucorales Mucor circinelloides. G3 2017, 7, 2047–2054. [Google Scholar] [CrossRef]
- Iturriaga, A.E.; Papp, T.; Breum, J.; Arnau, J. Strain and Culture Conditions Improvement for β-Carotene Production with Mucor BT—Microbial Processes and Products; Barredo, J.-L., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 239–256. ISBN 978-1-59259-847-2. [Google Scholar]
- McIntyre, M.; Breum, J.; Arnau, J.; Nielsen, J. Growth physiology and dimorphism of Mucor circinelloides (syn. racemosus) during submerged batch cultivation. Appl. Microbiol. Biotechnol. 2002, 58, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, M. Mucor dimorphism. Microbiol. Rev. 1991, 55, 234–258. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz, J.O.; García-Soto, J.; Uriostegui, C.; Carranza, L.; Novoa, G.; Reyna, G.; Martínez-Cadena, G. Differential expression of Rho1GTPase and Rho3GTPase during isotropic and polarized growth of Mucor circinelloides. Can. J. Microbiol. 2007, 53, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.M.; Appel, K.F.; Petersen, J.B.; Poulsen, U.; Arnau, J. Identification and analysis of genes involved in the control of dimorphism in Mucor circinelloides (syn. racemosus). FEMS Yeast Res. 2002, 2, 203–213. [Google Scholar] [CrossRef]
- Lübbehüsen, T.L.; Nielsen, J.; McIntyre, M. Morphology and physiology of the dimorphic fungus Mucor circinelloides (syn. M. racemosus) during anaerobic growth. Mycol. Res. 2003, 107, 223–230. [Google Scholar] [CrossRef]
- Brunke, S.; Mogavero, S.; Kasper, L.; Hube, B. Virulence factors in fungal pathogens of man. Curr. Opin. Microbiol. 2016, 32, 89–95. [Google Scholar] [CrossRef]
- Benito, E.P.; Díaz-Mínguez, J.M.; Iturriaga, E.A.; Campuzano, V.; Eslava, A.P. Cloning and sequence analysis of the Mucor circinelloides pyrG gene encoding orotidine-5’-monophosphate decarboxylase: Use of pyrG for homologous transformation. Gene 1992, 116, 59–67. [Google Scholar] [CrossRef]
- Corrochano, L.M.; Kuo, A.; Marcet-Houben, M.; Polaino, S.; Salamov, A.; Villalobos-Escobedo, J.M.; Grimwood, J.; Álvarez, M.I.; Avalos, J.; Bauer, D.; et al. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr. Biol. 2016, 26, 1577–1584. [Google Scholar] [CrossRef]
- Wood, V.; Gwilliam, R.; Rajandream, M.-A.; Lyne, M.; Lyne, R.; Stewart, A.; Sgouros, J.; Peat, N.; Hayles, J.; Baker, S.; et al. The genome sequence of Schizosaccharomyces pombe. Nature 2002, 415, 871–880. [Google Scholar] [CrossRef]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef]
- Jones, T.; Federspiel, N.A.; Chibana, H.; Dungan, J.; Kalman, S.; Magee, B.B.; Newport, G.; Thorstenson, Y.R.; Agabian, N.; Magee, P.T.; et al. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 2004, 101, 7329–7334. [Google Scholar] [CrossRef]
- Janbon, G.; Ormerod, K.L.; Paulet, D.; Byrnes III, E.J.; Yadav, V.; Chatterjee, G.; Mullapudi, N.; Hon, C.-C.; Billmyre, R.B.; Brunel, F.; et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014, 10, e1004261. [Google Scholar] [CrossRef]
- Lee, S.C.; Billmyre, R.B.; Li, A.; Carson, S.; Sykes, S.M.; Huh, E.Y.; Mieczkowski, P.; Ko, D.C.; Cuomo, C.A.; Heitman, J. Analysis of a food-borne fungal pathogen outbreak: Virulence and genome of a Mucor circinelloides isolate from yogurt. mBio 2014, 5, e01390-14. [Google Scholar] [CrossRef]
- Galagan, J.E.; Calvo, S.E.; Borkovich, K.A.; Selker, E.U.; Read, N.D.; Jaffe, D.; FitzHugh, W.; Ma, L.-J.; Smirnov, S.; Purcell, S.; et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature 2003, 422, 859–868. [Google Scholar] [CrossRef]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.-J.; Wortman, J.R.; Batzoglou, S.; Lee, S.-I.; Baştürkmen, M.; Spevak, C.C.; Clutterbuck, J.; et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar] [CrossRef]
- Vandeputte, P.; Ghamrawi, S.; Rechenmann, M.; Iltis, A.; Giraud, S.; Fleury, M.; Thornton, C.; Delhaès, L.; Meyer, W.; Papon, N.; et al. Draft genome sequence of the pathogenic fungus Scedosporium apiospermum. Genome Announc. 2014, 2, e00988-14. [Google Scholar] [CrossRef]
- Haitjema, C.H.; Gilmore, S.P.; Henske, J.K.; Solomon, K.V.; de Groot, R.; Kuo, A.; Mondo, S.J.; Salamov, A.A.; LaButti, K.; Zhao, Z.; et al. A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol. 2017, 2, 17087. [Google Scholar] [CrossRef]
- Hauser, M.; Steinegger, M.; Söding, J. MMseqs software suite for fast and deep clustering and searching of large protein sequence sets. Bioinformatics 2016, 32, 1323–1330. [Google Scholar] [CrossRef]
- van Dongen, S.M. Graph Clustering by Flow Simulation. Ph.D. Thesis, University of Utrecht, Utrecht, The Netherlands, 2000. [Google Scholar]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]
- Lax, C.; Pérez-Arques, C.; Navarro-Mendoza, M.I.; Cánovas-Márquez, J.T.; Tahiri, G.; Pérez-Ruiz, J.A.; Osorio-Concepción, M.; Murcia-Flores, L.; Navarro, E.; Garre, V.; et al. Genes, pathways, and mechanisms involved in the virulence of Mucorales. Genes 2020, 11, 317. [Google Scholar] [CrossRef] [PubMed]
- Kubo, H.; Mihara, H. cAMP promotes hyphal branching in Mucor globosus. Mycoscience 2007, 48, 187–189. [Google Scholar] [CrossRef]
- Lecointe, K.; Cornu, M.; Leroy, J.; Coulon, P.; Sendid, B. Polysaccharides cell wall architecture of Mucorales. Front. Microbiol. 2019, 10, 469. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Brown, H.E.; Esher, S.K.; Alspaugh, J.A. Chitin: A “hidden figure” in the fungal cell wall. Curr. Top. Microbiol. Immunol. 2020, 425, 83–111. [Google Scholar] [CrossRef]
- Doering, T.L. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 2009, 63, 223–247. [Google Scholar] [CrossRef]
- Bartnicki-Garcia, S.; Nickerson, W.J. Isolation, composition, and structure of cell walls of filamentous and yeast-like forms of Mucor rouxii. Biochim. Biophys. Acta 1962, 58, 102–119. [Google Scholar] [CrossRef]
- Dow, J.M.; Olona, P.M.; Villa, V.D. Glucuronosyl transferase from the dimorphic fungus Mucor rouxii. Exp. Mycol. 1982, 6, 329–334. [Google Scholar] [CrossRef]
- Dow, J.M.; Darnall, D.W.; Villa, V.D. Two distinct classes of polyuronide from the cell walls of a dimorphic fungus, Mucor rouxii. J. Bacteriol. 1983, 155, 1088–1093. [Google Scholar] [CrossRef]
- Bartnicki-Garcia, S.; Lindberg, B. Partial characterization of mucoran: The glucuronomannan component. Carbohydr. Res. 1972, 23, 75–85. [Google Scholar] [CrossRef]
- López-Fernández, L.; Sanchis, M.; Navarro-Rodríguez, P.; Nicolás, F.E.; Silva-Franco, F.; Guarro, J.; Garre, V.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Capilla, J. Understanding Mucor circinelloides pathogenesis by comparative genomics and phenotypical studies. Virulence 2018, 9, 707–720. [Google Scholar] [CrossRef]
- López-Matas, M.A.; Eslava, A.P.; Díaz-Mínguez, J.M. Mcchs1, a member of a chitin synthase gene family in Mucor circinelloides, is differentially expressed during dimorphism. Curr. Microbiol. 2000, 40, 169–175. [Google Scholar] [CrossRef]
- de Groot, P.W.J.; Brandt, B.W.; Horiuchi, H.; Ram, A.F.J.; de Koster, C.G.; Klis, F.M. Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet. Biol. 2009, 46 (Suppl. S1), S72–S81. [Google Scholar] [CrossRef]
- Rangel-Porras, R.A.; Meza-Carmen, V.; Martinez-Cadena, G.; Torres-Guzmán, J.C.; González-Hernández, G.A.; Arnau, J.; Gutiérrez-Corona, J.F. Molecular analysis of an NAD-dependent alcohol dehydrogenase from the zygomycete Mucor circinelloides. Mol. Genet. Genom. 2005, 274, 354–363. [Google Scholar] [CrossRef]
- Fleck, C.B.; Brock, M. Characterization of an acyl-CoA: Carboxylate CoA-transferase from Aspergillus nidulans involved in propionyl-CoA detoxification. Mol. Microbiol. 2008, 68, 642–656. [Google Scholar] [CrossRef]
- Arikawa, Y.; Enomoto, K.; Muratsubaki, H.; Okazaki, M. Soluble fumarate reductase isoenzymes from Saccharomyces cerevisiae are required for anaerobic growth. FEMS Microbiol. Lett. 1998, 165, 111–116. [Google Scholar] [CrossRef][Green Version]
- Navarro-Mendoza, M.I.; Pérez-Arques, C.; Murcia, L.; Martínez-García, P.; Lax, C.; Sanchis, M.; Capilla, J.; Nicolás, F.E.; Garre, V. Components of a new gene family of ferroxidases involved in virulence are functionally specialized in fungal dimorphism. Sci. Rep. 2018, 8, 7660. [Google Scholar] [CrossRef]
- Ocampo, J.; McCormack, B.; Navarro, E.; Moreno, S.; Garre, V.; Rossi, S. Protein kinase A regulatory subunit isoforms regulate growth and differentiation in Mucor circinelloides: Essential role of PKAR4. Eukaryot. Cell 2012, 11, 989–1002. [Google Scholar] [CrossRef]
- Moriwaki-Takano, M.; Iwakura, R.; Hoshino, K. Dimorphic mechanism on cAMP mediated signal pathway in Mucor circinelloides. Appl. Biochem. Biotechnol. 2021, 193, 1252–1265. [Google Scholar] [CrossRef]
- Fernández Núñez, L.; Ocampo, J.; Gottlieb, A.M.; Rossi, S.; Moreno, S. Multiple isoforms for the catalytic subunit of PKA in the basal fungal lineage Mucor circinelloides. Fungal Biol. 2016, 120, 1493–1508. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Homa, M.; Ibragimova, S.; Szebenyi, C.; Nagy, G.; Zsindely, N.; Bodai, L.; Vágvölgyi, C.; Nagy, G.; Papp, T. Differential Gene Expression of Mucor lusitanicus under Aerobic and Anaerobic Conditions. J. Fungi 2022, 8, 404. https://doi.org/10.3390/jof8040404
Homa M, Ibragimova S, Szebenyi C, Nagy G, Zsindely N, Bodai L, Vágvölgyi C, Nagy G, Papp T. Differential Gene Expression of Mucor lusitanicus under Aerobic and Anaerobic Conditions. Journal of Fungi. 2022; 8(4):404. https://doi.org/10.3390/jof8040404
Chicago/Turabian StyleHoma, Mónika, Sandugash Ibragimova, Csilla Szebenyi, Gábor Nagy, Nóra Zsindely, László Bodai, Csaba Vágvölgyi, Gábor Nagy, and Tamás Papp. 2022. "Differential Gene Expression of Mucor lusitanicus under Aerobic and Anaerobic Conditions" Journal of Fungi 8, no. 4: 404. https://doi.org/10.3390/jof8040404
APA StyleHoma, M., Ibragimova, S., Szebenyi, C., Nagy, G., Zsindely, N., Bodai, L., Vágvölgyi, C., Nagy, G., & Papp, T. (2022). Differential Gene Expression of Mucor lusitanicus under Aerobic and Anaerobic Conditions. Journal of Fungi, 8(4), 404. https://doi.org/10.3390/jof8040404






