Impacts of Sodium Arsenite on Wood Microbiota of Esca-Diseased Grapevines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Sampling
2.2. DNA Extraction and Analysis of Fungal and Bacterial Communities
2.3. Post-Run Analysis
2.4. Statistical Analysis
2.5. In Vitro Fungal Growth Inhibition Assays
3. Results
3.1. Sampling Grapevine Trunk Woody Tissues
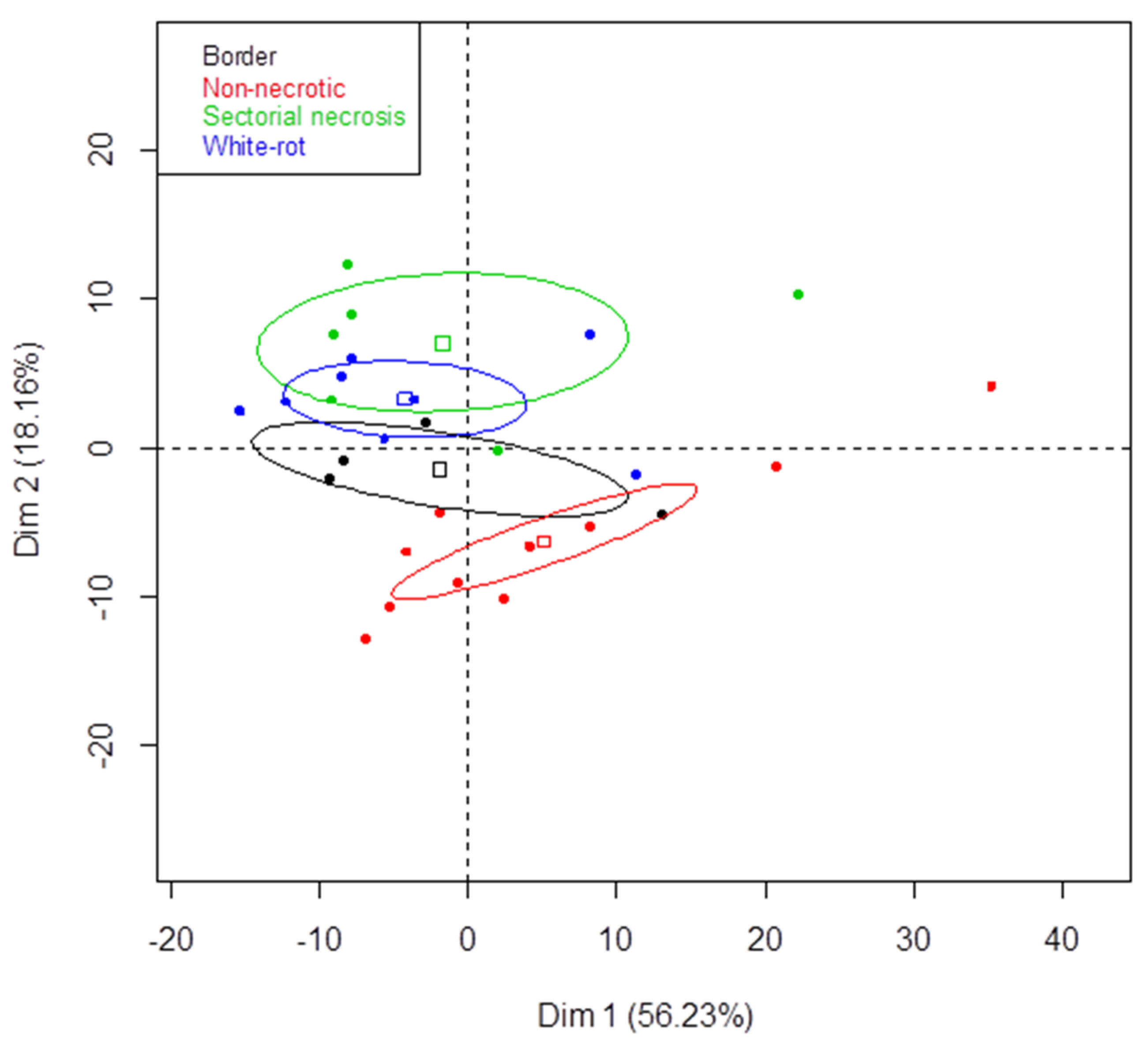
3.2. Metabarcoding Analysis of Microbial Taxa from Esca-Diseased Grapevines
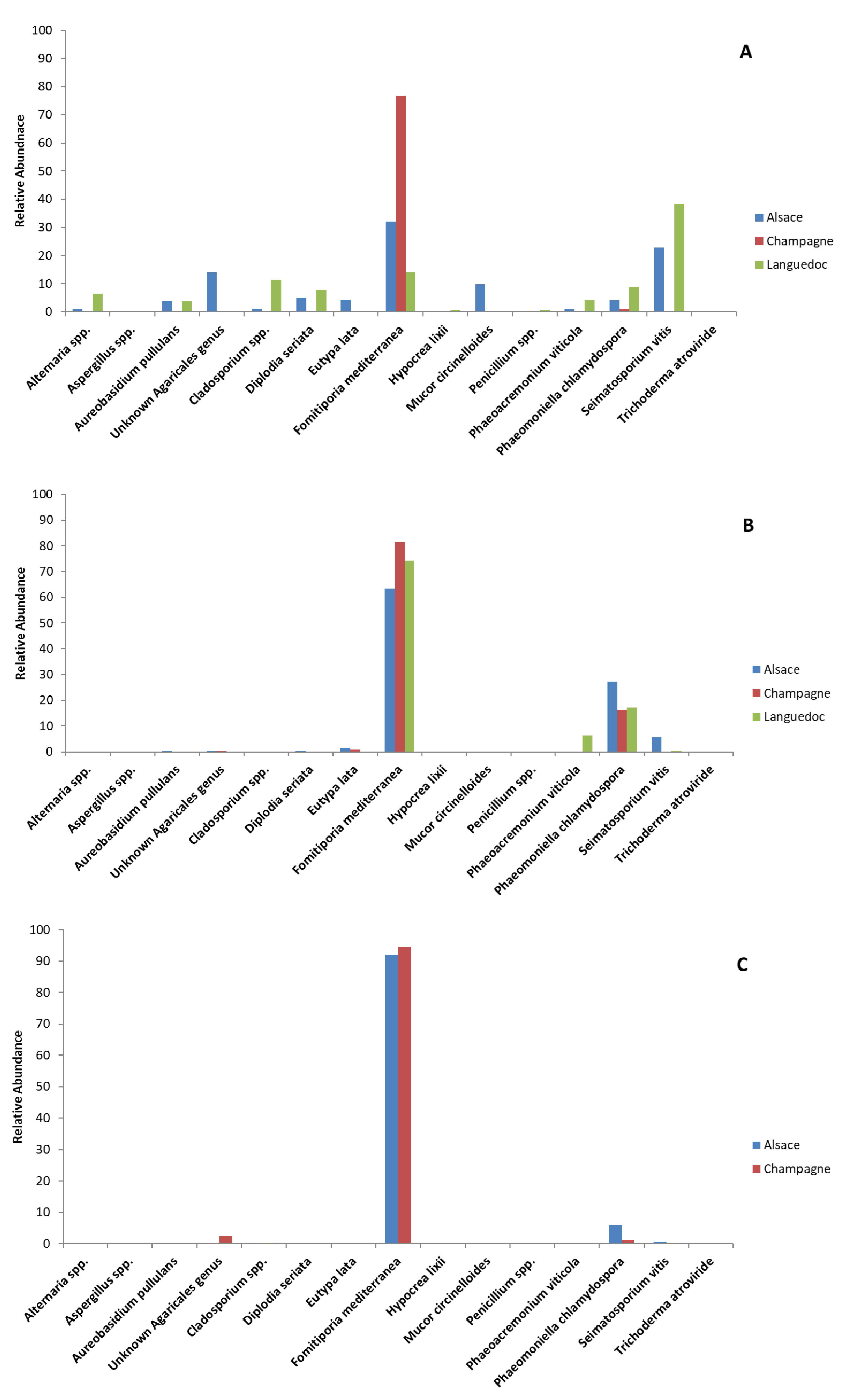
3.3. Metabarcoding Analysis of Fungal Taxa from Untreated Esca-Diseased Grapevines
3.4. Metabarcoding Analysis of Bacterial Taxa from Untreated Esca-Diseased Grapevines
3.5. Effects of Sodium Arsenite on Fungal Microbiota of Grapevine Woody Tissues
3.6. Sensitivity to Sodium Arsenite of Fungi Isolated from Grapevine Woody Tissues
4. Discussion
4.1. Fungal Microbiota Colonizing Woody Tissues from Esca-Diseased Grapevines
4.2. Bacterial Microbiota Colonizing Woody Tissues from Esca-Diseased Grapevines
4.3. Sodium Arsenite’s Impacts on Fungal Microbiota Colonizing Esca-Diseased Grapevine Woody Tissues
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Compant, S.; Brader, G.; Muzammil, S.; Sessitsch, A.; Lebrihi, A.; Mathieu, F. Use of beneficial bacteria and their secondary metabolites to control grapevine pathogen diseases. BioControl 2012, 58, 435–455. [Google Scholar] [CrossRef] [Green Version]
- Delmotte, F.; Mestre, P.; Schneider, C.; Kassemeyer, H.-H.; Kozma, P.; Richart-Cervera, S.; Rouxel, M.; Delière, L. Rapid and multiregional adaptation to host partial resistance in a plant pathogenic oomycete: Evidence from European populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Infect. Genet. Evol. 2014, 27, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Gramaje, D.; Úrbez-Torres, J.R.; Sosnowski, M.R. Managing grapevine trunk diseases with respect to etiology and epide-miology: Current strategies and future prospects. Plant Dis. 2018, 102, 12–39. [Google Scholar] [CrossRef] [Green Version]
- Guerin-Dubrana, L.; Fontaine, F.; Mugnai, L. Grapevine trunk disease in European and Mediterranean vineyards: Occur-rence, distribution and associated disease-affecting cultural factors. Phytopathol. Mediter. 2019, 58, 49–71. [Google Scholar]
- De La Fuente, M.; Fontaine, F.; Gramaje, D.; Armengol, J.; Smart, R.E.; Nagy, Z.A.; Borgo, M.; Rego, C.; Corio-Costet, M.-F. Grapevine Trunk Diseases—A Review, 1st ed.; OIV: Paris, France, 2016. [Google Scholar]
- Ùrbez-Torres, J.R. The status of Botryosphaeriaceae species infecting grapevines. Phytopathol. Med. 2011, 11, 5–45. [Google Scholar]
- Bertsch, C.; Ramírez-Suero, M.; Magninrobert, M.; Larignon, P.; Chong, J.; Mansour, E.A.; Spagnolo, A.; Clément, C.; Fontaine, F. Grapevine trunk diseases: Complex and still poorly understood. Plant Pathol. 2012, 62, 243–265. [Google Scholar] [CrossRef] [Green Version]
- Mondello, V.; Songy, A.; Battiston, E.; Pinto, C.; Coppin, C.; Trotel-Aziz, P.; Clément, C.; Mugnai, L.; Fontaine, F. Grapevine Trunk Diseases: A Review of Fifteen Years of Trials for Their Control with Chemicals and Biocontrol Agents. Plant Dis. 2018, 102, 1189–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusjan, D.; Persic, M.; Likar, M.; Binari, K.; Mikulic-Petkovsek, M. Phenolic responses to esca-associated fungi in differently decayed grapevine woods from different trunks parts of Caberne Sauvignon. J. Agric. Food Chem. 2017, 65, 6615–6624. [Google Scholar] [CrossRef]
- Fisher, M.A. New wood-decaying basidiomycete species associated with esca of grapevine: Fomitiporia mediterranea (Hy-menochaetales). Mycol. Prog. 2002, 1, 315–324. [Google Scholar] [CrossRef]
- Bruez, E.; Vallance, J.; Gerbore, J.; Lecomte, P.; Da Costa, J.-P.; Guérin-Dubrana, L.; Rey, P. Analyses of the Temporal Dynamics of Fungal Communities Colonizing the Healthy Wood Tissues of Esca Leaf-Symptomatic and Asymptomatic Vines. PLoS ONE 2014, 9, e95928. [Google Scholar] [CrossRef]
- Dupuy-Demportes, J.B. Le Gentilhomme Cultivateur ou Cours Complet D’agriculture; Chez, T., Simon, P.G., et Bauche, D., Eds.; Chapuis: Paris, France, 1763; 300p. [Google Scholar]
- Prévost, I.B. Mémoire sur la Cause Immédiate de la Carie ou Charbon des blés, et de Plusieurs Autres Maladies des Plantes, et sur les Préservatifs de la Carie; Fontanel: Paris, France, 1807; p. 67. [Google Scholar]
- Hewitt, W.B. Sodium arsenite, a promising control of Dead-arm disease of grapes. Phytopathol. Mediterr. 1947, 37, 362–370. [Google Scholar]
- Hewitt, W.B. Grape Dead-arm control. Plant Dis. 1951, 35, 132–143. [Google Scholar]
- Vergnes, A. Sur les traitements de l’anthracnose maculée. Prog. Agric. Vitic. 1957, 147, 304–309. [Google Scholar]
- Lafon, J.; Couillaud, P.; Hude, R. Anthracnose (Gleosporium ampelophagum). In Maladies et Parasites de la Vigne, 3rd ed.; Editeurs: Paris, France, 1966; Volume 1, pp. 229–234. [Google Scholar]
- Cucuzza, J.D.; Sall, M.A. Phomopsis cane and leaf spot disease of grape vine: Effect of chemical treatments on inoculum level, disease severity, and yield. Plant Dis. 1982, 66, 794–797. [Google Scholar] [CrossRef]
- Ravaz, L. Encore l’apoplexie de la vigne. Progrès Agric. Vitic. 1919, 52, 601–603. [Google Scholar]
- Bonnet, L.O. A promising remedy for black measles of the vine. In College of Agriculture Agricultural Experiment Station; Circular: Berkeley, CA, USA, 1926; 10p. [Google Scholar]
- Viala, P. Esca. In Annales des Epiphyties; Vinet: Paris, France, 1926; 180p. [Google Scholar]
- Rui, D.; Battel, C. Mise au point d’un nouveau moyen de lutte contre l’esca de la vigne. Not. Sulle Mal. Delle Piante 1963, 63, 9–18. [Google Scholar]
- Svampa, G.; Tosatti, E.M. Prove di lotta contro il “mal dell’esca” della vite. Infor. Fitipatol. 1977, 12, 21–24. [Google Scholar]
- Del Rivero, J.M.; García-Marí, F. Ensayo de productos contra la yesca de la vid y la piral de la vid en tratamientos de invierno. Bol. Serv. Plagas 1984, 10, 17–30. [Google Scholar]
- Larignon, P.; Darne, G.; Menard, E.; Desache, F.; Dubos, B. Comment agissait l’arsénite de sodium sur l’esca de la vigne? Progrès Agric. Vitic. 2008, 125, 642–651. [Google Scholar]
- Songy, A.; Fernandez, O.; Clément, C.; Larignon, P.; Fontaine, F. Grapevine trunk diseases under thermal and water stresses. Planta 2019, 249, 1655–1679. [Google Scholar] [CrossRef]
- Meier, U. Growth stages of mono-and dicotyledonous plants. In BBCH Monograph, 2nd ed.; Federal Biological Research Centre for Agriculture and Forestry: Bonn, Germany, 2001. [Google Scholar]
- Bruez, E.; Vallance, J.; Gautier, A.; Laval, V.; Compant, S.; Maurer, W.; Sessitsch, A.; Lebrun, M.; Rey, P. Major changes in grapevine wood microbiota are associated with the onset of esca, a devastating trunk disease. Environ. Microbiol. 2020, 22, 5189–5206. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phy-logenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, H.H., White, T.J., Eds.; Academic Press Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Redford, A.J.; Bowers, R.M.; Knight, R.; Linhart, Y.; Fierer, N. The ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010, 12, 2885–2893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtsson, J.; Eriksson, K.M.; Hartmann, M.; Wang, Z.; Shenoy, B.D.; Grelet, G.A.; Abarenkov, K.; Petri, A.; Rosenblad, M.A.; Nilsson, R.H. Metaxa: A software tool for au-tomated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea bacteria eukaryotes mitochondria and chloroplasts in metagenomes and environmental sequencing datasets. Antonie Leeuwenhoek 2011, 100, 471–475. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, M.; Howes, C.G.; Abarenkov, K.; Mohn, W.W.; Nilsson, R.H. V-Xtractor: An open-source, high-throughput software tool to identify and extract hypervariable regions of small subunit (16S/18S) ribosomal RNA gene sequences. J. Microbiol. Methods 2010, 83, 250–253. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Veldre, V.; Hartmann, M.; Unterseher, M.; Amend, A.; Bergsten, J.; Kristiansson, E.; Ryberg, M.; Jumpponen, A.; Abarenkov, K. An open source software package for automated extraction of ITS1 and ITS2 from fungal ITS sequences for use in high-throughput community assays and molecular ecology. Fungal Ecol. 2010, 3, 284–287. [Google Scholar] [CrossRef]
- Colwell, R.K.; Elsensohn, J.E. EstimateS turns 20: Statistical estimation of species richness and shared species from samples, with non-parametric extrapolation. Ecography 2014, 37, 609–613. [Google Scholar] [CrossRef]
- Guijarro, B.; Larena, I.; Vilanova, L.; Torres, R.; Balsells-Llauradó, M.; Teixidó, N.; Melgarejo, P.; De Cal, A. Dispersion, persistence, and stability of the biocontrol agent Penicillium frequentans strain 909 after stone fruit tree applications. Environ. Sci. Pollut. Res. 2019, 26, 29138–29156. [Google Scholar] [CrossRef]
- Bigot, G.; Sivilotti, P.; Stecchina, M.; Lujan, C.; Freccero, A.; Mosetti, D. Long-term effects of Trichoderma asperellum and Trichoderma gamsii on the prevention of esca in different vineyards of Northeastern Italy. Crop. Prot. 2020, 137, 105264. [Google Scholar] [CrossRef]
- Peet, R.K. The Measurement of Species Diversity. Annu. Rev. Ecol. Syst. 1974, 5, 285–307. [Google Scholar] [CrossRef]
- Del Frari, G.; Gobbi, A.; Aggerbeck, M.R.; Oliveira, H.; Hansen, L.H.; Ferreira, R.B. Characterization of the Wood Mycobiome of Vitis vinifera in a Vineyard Affected by Esca. Spatial Distribution of Fungal Communities and Their Putative Relation with Leaf Symptoms. Front. Plant Sci. 2019, 10, 910. [Google Scholar] [CrossRef] [Green Version]
- Senanayake, I.C.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, J.D.; Jones, E.B.G.; McKenzie, E.H.C.; Dai, D.Q.; Daranagama, D.A.; Dayarathne, M.C.; Goonasekara, I.D.; et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 2015, 73, 73–144. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Wanasinghe, D.N.; Papizadeh, M.; Goonasekara, I.D.; Camporesi, E.; Bhat, D.J.; McKenzie, E.H.C.; Phillips, A.J.L.; Diederich, P.; et al. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers. 2016, 77, 1–316. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Travadon, R.; Baumgartner, K. Novel Seimatosporium species from grapevine in northern California and their interactions with fungal pathogens involved in the trunk-disease complex. Plant Dis. 2018, 102, 1081–1092. [Google Scholar] [CrossRef] [Green Version]
- Váczy, K.Z. First Report of Seimatosporium vitis Associated with Grapevine Trunk Disease Symptoms in Hungary. Plant Dis. 2017, 101, 253. [Google Scholar] [CrossRef]
- Camele, I.; Mang, S.M. First Report of Seimatosporium vitis Associated with Grapevine Trunk Diseases on Vitis vinifera in Italy. Plant Dis. 2019, 103, 771. [Google Scholar] [CrossRef]
- Raimondo, M.L.; Carlucci, A.; Ciccarone, C.; Sadallah, A.; Lops, F. Identification and pathogenicity of lignicolous fungi associated with grapevine trunk diseases in southern Italy. Phytopathol. Mediter. 2019, 58, 639–662. [Google Scholar]
- Mehrabi, M.; Hemmati, R.; Abdollahzadeh, J. Description of the Sexual Morph of Seimatosporium vitis. Cryptogam. Mycol. 2017, 38, 3–11. [Google Scholar] [CrossRef]
- Bruez, E.; Haidar, R.; Alou, M.T.; Vallance, J.; Bertsch, C.; Mazet, F.; Fermaud, M.; Deschamps, A.; Guerin-Dubrana, L.; Compant, S.; et al. Bacteria in a wood fungal disease: Characterization of bacterial communities in wood tissues of esca-foliar symptomatic and asymptomatic grapevines. Front. Microbiol. 2015, 6, 1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. MBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larignon, P.; Fontaine, F. Comprendre le Mode D’action de L’arsénite de Sodium Afin de Proposer de Nouveaux Moyens de Lute; Presentation: Toulouse, France, 2018. [Google Scholar]

| Alsace | Champagne | Languedoc | ||||
|---|---|---|---|---|---|---|
| Treated | Untreated | Treated | Untreated | Treated | Untreated | |
| Non-necrotic (NN) | 100% * | 100% | 100% | 100% | 100% | 100% |
| Border (NN-WR) | 100% * | 100% | 100% | 80% | 75% | 80% |
| White-rot necrosis (WR) | 100% * | 100% | 80% | 80% | 50% ** | 60% ** |
| Sectorial necrosis (SN) | 0% * | 100% | 0% | 0% | 100% | 100% |
| Esca-foliar symptoms | 0% *** | 100% | 0% | 100% | 0% | 100% |
| Shannon (2014) | Alsace White-Rot | Champagne White-Rot | Languedoc Sectorial Necrosis | |
|---|---|---|---|---|
| Treated | Fungi | 1.53 | 1.17 | 1.80 |
| Bacteria | 1.39 | 1.34 | 3.27 | |
| Untreated | Fungi | 0.57 | 0.99 | 1.60 |
| Bacteria | 4.40 | 2.37 | 4.77 | |
| Shannon (2015) | Alsace White-rot | Champagne White-rot | Languedoc Sectorial necrosis | |
| Treated | Fungi | 1.79 | 1.51 | 2.03 |
| Bacteria | 1.59 | 1.97 | 4.48 | |
| Untreated | Fungi | 0.66 | 0.44 | 2.21 |
| Bacteria | 2.33 | 2.15 | 5.51 |
| Treated | Untreated | Treated | Untreated | Treated | Untreated | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-Necrotic | Border | White-Rot | Non-Necrotic | Border | White-Rot | Non-Necrotic | Border | White-Rot | Non-Necrotic | Border | White-Rot | Non-Necrotic | Border | Sectorial Necrosis | Non-Necrotic | Border | Sectorial Necrosis | |
| Alternaria spp. | 3.8 | 0.2 | 0.1 | 0.9 | 0.1 | 0.0 | 2.1 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 3.7 | 0.2 | 0.1 | 6.4 | 0.1 | 0.0 |
| Aspergillus spp. | 0.2 | 0.0 | 66.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Aureobasidium pullulans | 6.2 | 0.1 | 0.1 | 3.9 | 0.5 | 0.1 | 5.8 | 0.1 | 87.3 | 0.1 | 0.1 | 0.1 | 7.1 | 0.1 | 0.1 | 4.0 | 0.1 | 0.0 |
| Unknown Agaricales genus | 7.1 | 1.1 | 0.4 | 14.0 | 0.3 | 0.3 | 61.7 | 0.7 | 0.2 | 0.3 | 0.3 | 2.5 | 0.3 | 0.1 | 0.2 | 0.3 | 0.3 | 0.2 |
| Cladosporium spp. | 4.0 | 0.3 | 0.1 | 1.2 | 0.1 | 0.1 | 2.9 | 0.1 | 0.3 | 0.1 | 0.1 | 0.4 | 11.0 | 0.3 | 0.2 | 11.4 | 0.1 | 0.0 |
| Diplodia seriata | 2.3 | 0.1 | 0.5 | 5.2 | 0.3 | 0.2 | 3.1 | 0.1 | 0.8 | 0.2 | 0.1 | 0.2 | 1.1 | 0.1 | 14.0 | 7.8 | 0.2 | 18.0 |
| Eutypa lata | 1.0 | 0.1 | 0.4 | 4.4 | 1.5 | 0.2 | 0.1 | 16.5 | 0.8 | 0.1 | 0.8 | 0.1 | 0.1 | 0.1 | 0.1 | 0.2 | 0.1 | 0.1 |
| Fomitiporia mediterranea | 14.7 | 1.2 | 7.2 | 32.2 | 63.5 | 92.1 | 3.1 | 2.3 | 6.2 | 76.9 | 81.5 | 94.5 | 8.7 | 1.2 | 39.6 | 14.1 | 74.2 | 16.8 |
| Hypocrea lixii | 32.9 | 0.1 | 0.2 | 0.1 | 0.0 | 0.0 | 4.7 | 0.0 | 0.1 | 0.1 | 0.0 | 0.1 | 0.1 | 0.0 | 0.0 | 0.6 | 0.0 | 0.0 |
| Inonotus hispidus | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 16.6 | 0.0 | 2.6 | 0.2 | 0.0 |
| Lepiota brunneoincarnata | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 21.6 | 0.0 | 0.0 | 0.3 | 0.0 | 0.0 |
| Mucor circinelloides | 0.4 | 0.0 | 0.0 | 9.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Mycena maurella | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 20.4 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Penicillium spp. | 0.1 | 0.0 | 21.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.7 | 0.0 | 0.0 | 0.5 | 0.0 | 0.0 |
| Phaeoacremonium viticola | 12.5 | 0.4 | 0.7 | 1.0 | 1.0 | 0.3 | 1.1 | 1.0 | 1.0 | 0.3 | 0.2 | 0.3 | 20.3 | 27.3 | 7.0 | 4.1 | 6.5 | 3.1 |
| Phaeomoniella chlamydospora | 10.8 | 96.1 | 1.5 | 4.2 | 42.4 | 5.8 | 1.4 | 42.4 | 1.6 | 1.0 | 16.2 | 1.1 | 18.4 | 33.6 | 9.6 | 8.8 | 17.2 | 5.7 |
| Phaeoacremonium fraxinopennsylvanicum | 0.4 | 0.0 | 0.0 | 0.0 | 36.5 | 0.1 | 0.4 | 36.5 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 |
| Sebacina spp. | 0.3 | 0.0 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.1 | 4.9 | 19.9 | 0.0 | 0.0 | 0.5 | 0.0 |
| Seimatosporium vitis | 1.6 | 0.2 | 0.8 | 23.0 | 0.2 | 0.6 | 0.2 | 0.2 | 1.3 | 0.3 | 0.2 | 0.3 | 1.8 | 0.3 | 28.9 | 38.5 | 0.3 | 55.8 |
| Trichoderma atroviride | 1.3 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 13.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.2 | 0.1 | 0.1 |
| Alsace | Champagne | Languedoc | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treated | Untreated | Treated | Untreated | Treated | Untreated | |||||||||||||
| Non-Necrotic | Border | White-Rot | Non-Necrotic | Border | White-Rot | Non-Necrotic | Border | White-Rot | Non-Necrotic | Border | White-Rot | Non-Necrotic | Border | Sectorial Necrosis | Non-Necrotic | Border | Sectorial Necrosis | |
| Amnibacterium sp. | 0.0 | 0.0 | 0.0 | 0.0 | 14.8 | 6.5 | 0.0 | 0.3 | 0.0 | 0.0 | 0.2 | 0.0 | 6.5 | ND | 0.5 | 22.3 | 0.0 | 26.0 |
| Badyrhizobium sp. | 2.1 | 1.0 | 6.4 | 0.0 | 0.7 | 2.9 | 1.8 | 0.5 | 4.9 | 0.4 | 11.0 | 6.0 | 2.6 | ND | 0.0 | 2.6 | 0.1 | 0.1 |
| Brevundimonas sp. | 26.6 | 0.7 | 33.5 | 0.3 | 9.7 | 18.3 | 84.0 | 5.5 | 53.8 | 98.7 | 4.1 | 29.1 | 34.8 | ND | 1.1 | 7.3 | 0.3 | 1.0 |
| Buttiauxella sp. | 0.3 | 1.2 | 0.0 | 1.2 | 0.4 | 0.1 | 0.0 | 0.5 | 0.0 | 0.0 | 0.3 | 0.0 | 0.8 | ND | 0.3 | 0.4 | 0.0 | 0.1 |
| Cellulomonas sp. | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.3 | 0.0 | 0.6 | 0.0 | 0.0 | 0.5 | 26.4 | 0.9 | ND | 0.1 | 0.5 | 0.1 | 0.1 |
| Chryseobacterium sp. | 0.1 | 13.5 | 0.0 | 0.0 | 0.3 | 6.4 | 4.2 | 0.4 | 0.0 | 0.2 | 0.2 | 3.1 | 1.8 | ND | 0.0 | 0.2 | 5.9 | 0.1 |
| Cloacibacterium sp. | 0.0 | 5.9 | 0.0 | 0.0 | 0.3 | 6.4 | 3.7 | 29.4 | 0.0 | 0.1 | 0.2 | 4.0 | 3.0 | ND | 0.0 | 0.2 | 5.8 | 0.1 |
| Curtobacterium sp. | 0.3 | 0.1 | 0.1 | 0.1 | 10.1 | 1.4 | 0.1 | 0.8 | 0.1 | 0.1 | 0.7 | 0.1 | 16.9 | ND | 28.7 | 37.6 | 0.1 | 35.7 |
| Enterobacter sp. | 0.1 | 0.2 | 0.0 | 39.2 | 0.2 | 0.1 | 0.0 | 0.5 | 0.0 | 0.0 | 1.6 | 0.0 | 0.4 | ND | 0.1 | 0.4 | 0.1 | 0.1 |
| Erwinia sp. | 2.4 | 3.9 | 0.0 | 0.4 | 1.8 | 0.1 | 0.1 | 2.2 | 0.0 | 0.0 | 2.2 | 0.0 | 0.9 | ND | 39.0 | 1.2 | 0.2 | 11.1 |
| Methylobacterium sp. | 0.5 | 0.1 | 3.8 | 0.0 | 0.6 | 0.7 | 0.1 | 0.6 | 7.8 | 0.0 | 0.2 | 4.4 | 0.7 | ND | 0.1 | 0.8 | 0.0 | 0.2 |
| Microbacterium sp. | 7.3 | 0.2 | 0.2 | 0.0 | 0.2 | 12.6 | 0.1 | 0.2 | 0.0 | 0.0 | 0.7 | 0.3 | 5.4 | ND | 0.2 | 3.2 | 0.1 | 0.1 |
| Nocardia sp. | 0.9 | 0.7 | 0.0 | 0.0 | 1.5 | 40.6 | 4.5 | 21.3 | 0.2 | 0.0 | 0.6 | 3.6 | 1.4 | ND | 0.0 | 1.6 | 0.0 | 0.1 |
| Pantoea sp. | 45.1 | 68.8 | 0.1 | 13.7 | 23.7 | 0.2 | 0.1 | 2.5 | 0.1 | 0.1 | 1.2 | 0.1 | 3.9 | ND | 21.9 | 5.6 | 0.2 | 6.9 |
| Pseudomonas sp. | 0.1 | 1.3 | 0.1 | 0.2 | 5.8 | 0.2 | 0.7 | 30.8 | 0.0 | 0.0 | 29.7 | 0.0 | 7.6 | ND | 5.0 | 9.1 | 0.1 | 2.6 |
| Raoultella sp. | 1.3 | 1.3 | 0.1 | 44.4 | 1.5 | 0.2 | 0.1 | 1.0 | 0.1 | 0.1 | 32.4 | 0.1 | 1.0 | ND | 1.3 | 1.7 | 0.9 | 0.5 |
| Rhizobium sp. | 1.6 | 0.8 | 4.9 | 0.1 | 27.0 | 1.2 | 0.2 | 0.7 | 2.2 | 0.1 | 12.7 | 6.9 | 9.0 | ND | 1.4 | 1.4 | 0.2 | 14.4 |
| Sodalis sp. | 0.0 | 0.1 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.3 | 0.0 | 0.0 | 0.2 | 0.0 | 0.3 | ND | 0.0 | 0.5 | 45.8 | 0.1 |
| Sphingomonas sp. | 11.2 | 0.2 | 50.7 | 0.1 | 0.9 | 1.7 | 0.1 | 1.7 | 30.7 | 0.1 | 1.2 | 15.8 | 1.7 | ND | 0.2 | 2.2 | 0.1 | 0.4 |
| Yersinia sp. | 0.0 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.2 | 0.0 | 0.5 | ND | 0.0 | 0.8 | 40.1 | 0.1 |
| Strains | IC 50 | Relative IC50 Fm | IC 90 | LD 100 |
|---|---|---|---|---|
| Fomitiporia mediterranea | 0.4 * | 1 ** | 7 * | 50 *** |
| Phaeomoniella chlamydospora | 6 | 15 | 53 | 100 |
| Eutypa lata | 8 | 20 | 46 | 500 |
| Trichoderma sp. | 29 | 72 | 210 | ND |
| Penicillium sp. strain 1 | 139 | 347 | 18 | ND |
| Penicillium sp. strain 2 | 261 | 652 | 442 | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruez, E.; Larignon, P.; Bertsch, C.; Robert-Siegwald, G.; Lebrun, M.-H.; Rey, P.; Fontaine, F. Impacts of Sodium Arsenite on Wood Microbiota of Esca-Diseased Grapevines. J. Fungi 2021, 7, 498. https://doi.org/10.3390/jof7070498
Bruez E, Larignon P, Bertsch C, Robert-Siegwald G, Lebrun M-H, Rey P, Fontaine F. Impacts of Sodium Arsenite on Wood Microbiota of Esca-Diseased Grapevines. Journal of Fungi. 2021; 7(7):498. https://doi.org/10.3390/jof7070498
Chicago/Turabian StyleBruez, Emilie, Philippe Larignon, Christophe Bertsch, Guillaume Robert-Siegwald, Marc-Henri Lebrun, Patrice Rey, and Florence Fontaine. 2021. "Impacts of Sodium Arsenite on Wood Microbiota of Esca-Diseased Grapevines" Journal of Fungi 7, no. 7: 498. https://doi.org/10.3390/jof7070498
APA StyleBruez, E., Larignon, P., Bertsch, C., Robert-Siegwald, G., Lebrun, M.-H., Rey, P., & Fontaine, F. (2021). Impacts of Sodium Arsenite on Wood Microbiota of Esca-Diseased Grapevines. Journal of Fungi, 7(7), 498. https://doi.org/10.3390/jof7070498








