Functional Analysis of a Novel ABL (Abnormal Browning Related to Light) Gene in Mycelial Brown Film Formation of Lentinula edodes
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Materials and Cultivation Conditions
2.2. Comparative Genetic Analysis of Chamaram
2.3. Genomic Analysis of Mycelial Browning
2.4. Plasmid Construction
2.5. PEG-Mediated Transformation of Protoplasts
2.6. Microstructure of Mycelial Layer
3. Results
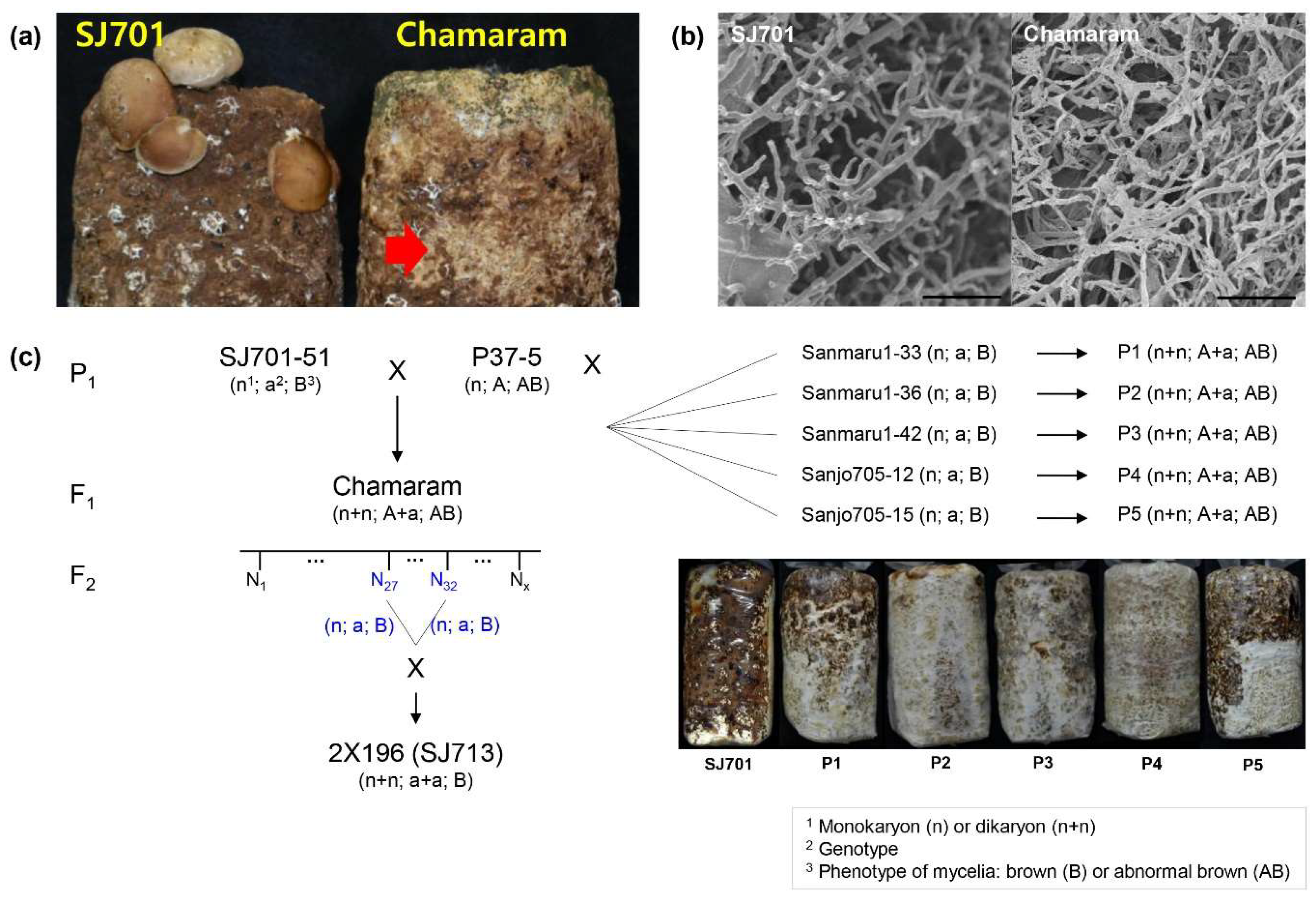
3.1. Genetic Analysis of Mycelium Browning in L. edodes
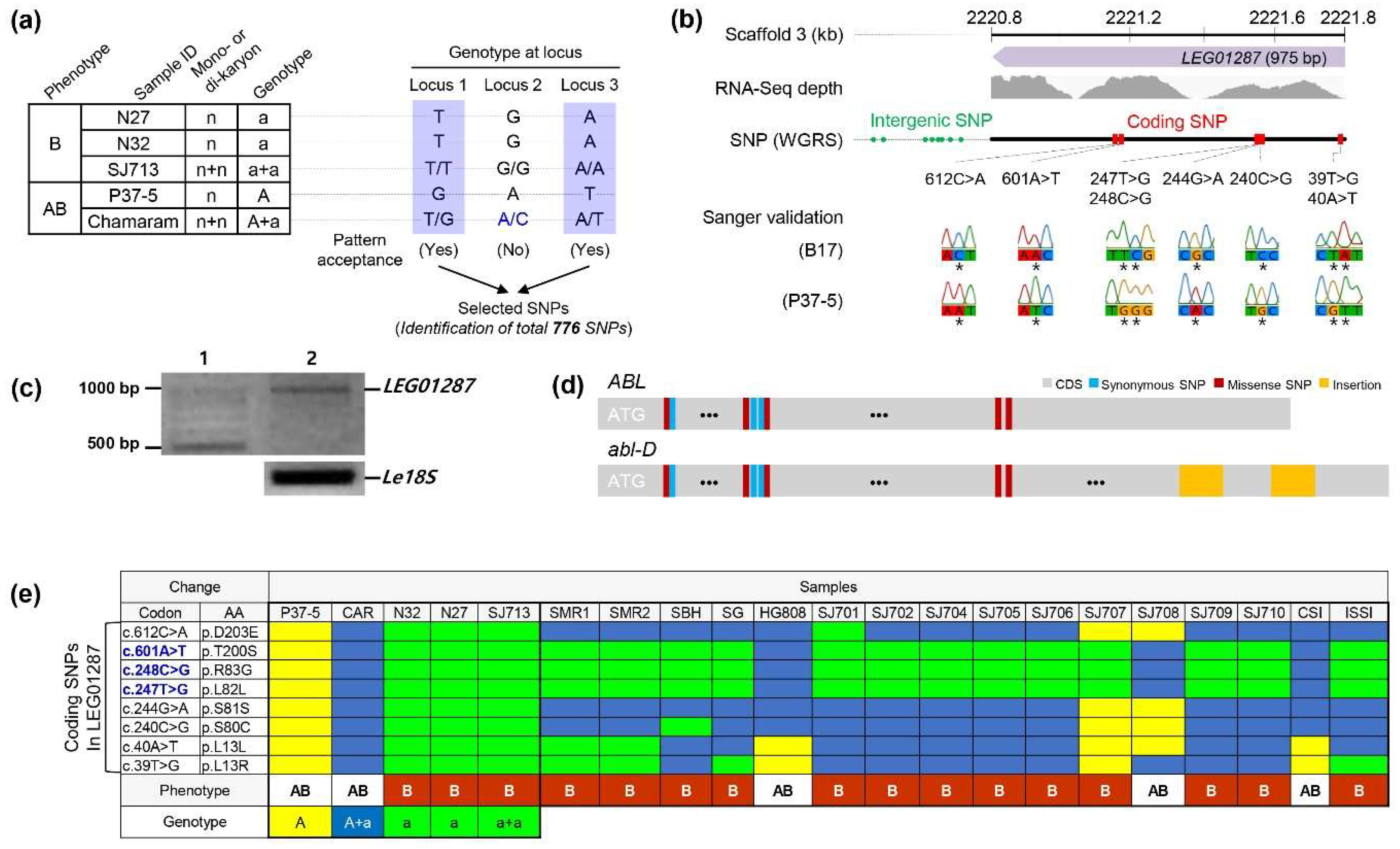
3.2. Identification of SNPs Associated with Mycelium Browning in L. edodes
3.3. Transformation of L. edodes with the ABL Gene
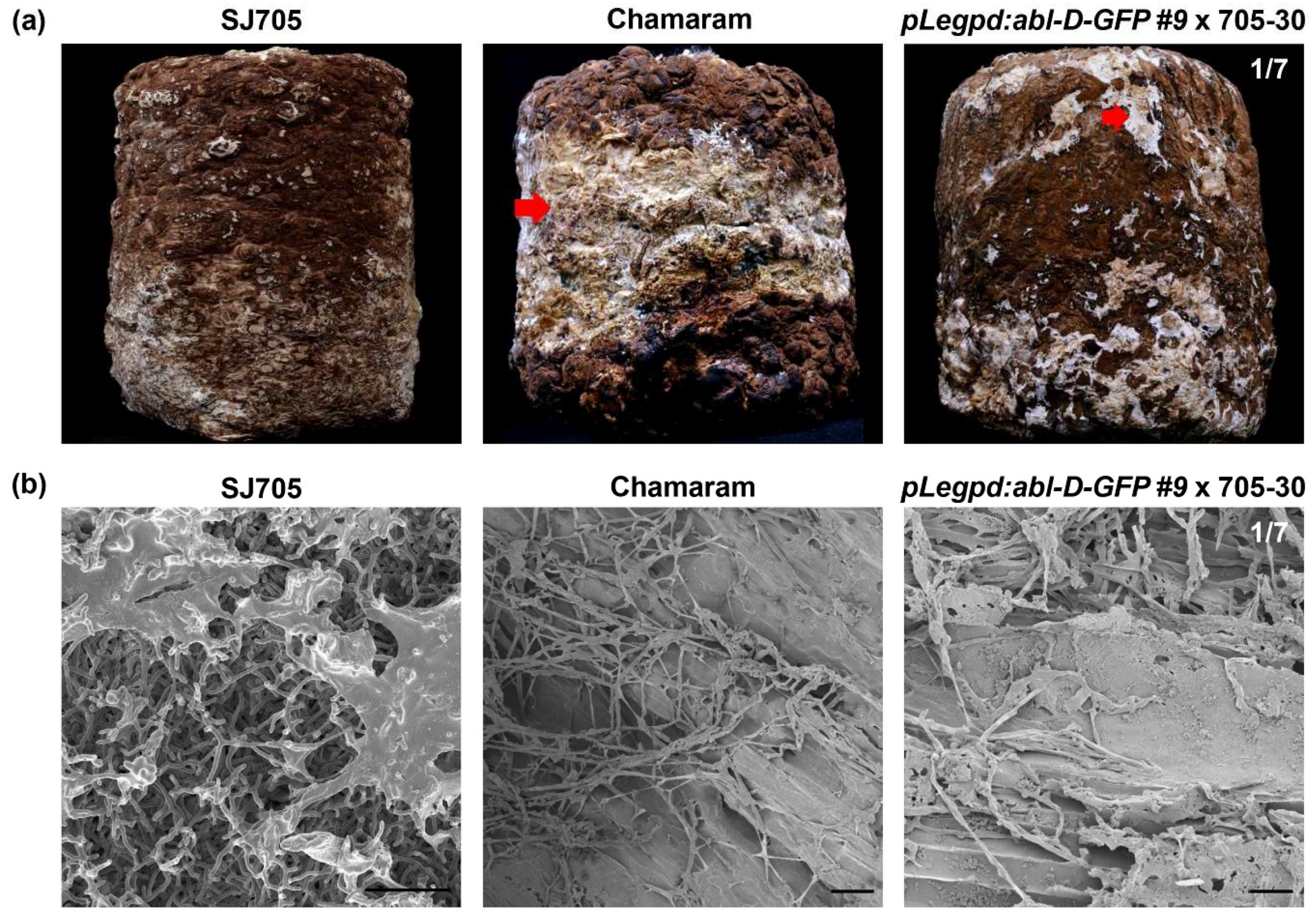
3.4. Functional Analysis of ABL in Brown Film Formation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Royse, D.J.; Baars, J.; Tan, Q. Current overview of mushroom production in the world. In Edible and Medicinal Mushrooms: Technology and Applications; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2017; pp. 5–13. [Google Scholar]
- Finimundy, T.C.; Dillon, A.J.P.; Henriques, J.A.P.; Ely, M.R. A review on general nutritional compounds and pharmacological properties of the Lentinula edodes mushroom. Food Nutr. Sci. 2014, 5, 1095–1105. [Google Scholar]
- Bisen, P.; Baghel, R.K.; Sanodiya, B.S.; Thakur, G.S.; Prasad, G. Lentinus edodes: A macrofungus with pharmacological activities. Curr. Med. Chem. 2010, 17, 2419–2430. [Google Scholar] [CrossRef] [PubMed]
- George, P.L.; Sripathi, V.R.; Nyaku, S.T.; Sharma, G.C.; Kantety, R.V. DNA-based identification of Lentinula edodes strains with species-specific primers. Afr. J. Biotechnol. 2016, 15, 191–198. [Google Scholar]
- Pire, D.; Wright, J.; Albertó, E. Cultivation of shiitake using sawdust from widely available local woods in Argentina. Micol. Apl. Int. 2001, 13, 87–91. [Google Scholar]
- Tang, L.-H.; Jian, H.-H.; Song, C.-Y.; Bao, D.-P.; Shang, X.-D.; Wu, D.-Q.; Tan, Q.; Zhang, X.-H. Transcriptome analysis of candidate genes and signaling pathways associated with light-induced brown film formation in Lentinula edodes. Appl. Microbiol. Biotechnol. 2013, 97, 4977–4989. [Google Scholar] [CrossRef]
- Yoo, S.-I.; Lee, H.-Y.; Markkandan, K.; Moon, S.; Ahn, Y.J.; Ji, S.; Ko, J.; Kim, S.-J.; Ryu, H.; Hong, C.P. Comparative transcriptome analysis identified candidate genes involved in mycelium browning in Lentinula edodes. BMC Genom. 2019, 20, 121. [Google Scholar] [CrossRef]
- Santoyo, F.; González, A.E.; Terrón, M.C.; Ramírez, L.; Pisabarro, A.G. Quantitative linkage mapping of lignin-degrading enzymatic activities in Pleurotus ostreatus. Enzym. Microb. Technol. 2008, 43, 137–143. [Google Scholar] [CrossRef]
- Li, C.; Gong, W.; Zhang, L.; Yang, Z.; Nong, W.; Bian, Y.; Kwan, H.-S.; Cheung, M.-K.; Xiao, Y. Association mapping reveals genetic loci associated with important agronomic traits in Lentinula edodes, shiitake mushroom. Front. Microbiol. 2017, 8, 237. [Google Scholar] [CrossRef][Green Version]
- Chen, L.; Gong, Y.; Cai, Y.; Liu, W.; Zhou, Y.; Xiao, Y.; Xu, Z.; Liu, Y.; Lei, X.; Wang, G. Genome sequence of the edible cultivated mushroom Lentinula edodes (Shiitake) reveals insights into lignocellulose degradation. PLoS ONE 2016, 11, e0160336. [Google Scholar] [CrossRef] [PubMed]
- Shim, D.; Park, S.-G.; Kim, K.; Bae, W.; Lee, G.W.; Ha, B.-S.; Ro, H.-S.; Kim, M.; Ryoo, R.; Rhee, S.-K. Whole genome de novo sequencing and genome annotation of the world popular cultivated edible mushroom, Lentinula edodes. J. Biotechnol. 2016, 223, 24–25. [Google Scholar] [CrossRef] [PubMed]
- Samuels, M.; Witmer, J. Statistics for the Life Sciences, 2nd ed.; Prentice Hall: Upper Saddle River, NJ, USA, 1999; pp. 383–388. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491. [Google Scholar] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar]
- Park, S.-G.; il Yoo, S.; Ryu, D.S.; Lee, H.; Ahn, Y.J.; Ryu, H.; Ko, J.; Hong, C.P. Long-read transcriptome data for improved gene prediction in Lentinula edodes. Data Brief 2017, 15, 454–458. [Google Scholar]
- Kuo, C.-Y.; Huang, C.-T. A reliable transformation method and heterologous expression of β-glucuronidase in Lentinula edodes. J. Microbiol. Methods 2008, 72, 111–115. [Google Scholar]
- Noh, J.-H.; Ko, H.-G.; Park, H.-S.; Koo, C.-D. Selection of parental strain on the sawdust cultivation and mycelial growth and cultural characteristics of Lentinula edodes hybrid strains. J. Mushroom 2015, 13, 41–49. [Google Scholar]
- Yan, D.; Liu, Y.; Rong, C.; Song, S.; Zhao, S.; Qin, L.; Wang, S.; Gao, Q. Characterization of brown film formed by Lentinula edodes. Fungal Biol. 2020, 124, 135–143. [Google Scholar]
- Yu, Z.; Fischer, R. Light sensing and responses in fungi. Nat. Rev. Microbiol. 2019, 17, 25–36. [Google Scholar]
- Ziemienowicz, A. Agrobacterium-mediated plant transformation: Factors, applications and recent advances. Biocatal. Agric. Biotechnol. 2014, 3, 95–102. [Google Scholar]
- Hooykaas, P.J.; van Heusden, G.P.H.; Niu, X.; Roushan, M.R.; Soltani, J.; Zhang, X.; van der Zaal, B.J. Agrobacterium-mediated transformation of yeast and fungi. In Agrobacterium Biology; Springer: Cham, Switzerland, 2018; pp. 349–374. [Google Scholar]
- Burns, C.; Leach, K.; Elliott, T.; Challen, M.; Foster, G.; Bailey, A. Evaluation of Agrobacterium-mediated transformation of Agricus bisporus using a range of promoters linked to hygromycin resistance. Mol. Biotechnol. 2006, 32, 129–138. [Google Scholar]
- Chen, X.; Stone, M.; Schlagnhaufer, C.; Romaine, C.P. A fruiting body tissue method for efficient Agrobacterium-mediated transformation of Agaricus bisporus. Appl. Environ. Microbiol. 2000, 66, 4510–4513. [Google Scholar] [CrossRef]
- Liu, J.; Song, C.; Li, Q.; Xu, Z.; Zhang, D.; Zhang, M.; Tan, Q.; Shang, X. A colonized millet grain method for Agrobacterium-mediated transformation of the button mushroom Agaricus bisporus. J. Microbiol. Methods 2018, 152, 148–153. [Google Scholar] [CrossRef]
- Cho, J.-H.; Lee, S.-E.; Chang, W.-B.; Cha, J.-S. Agrobacterium-mediated transformation of the winter mushroom, Flammulina velutipes. Mycobiology 2006, 34, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Yamada, M.; Sekiya, S.; Okuhara, T.; Taguchi, G.; Inatomi, S.; Shimosaka, M. Agrobacterium tumefaciens-mediated transformation of the vegetative dikaryotic mycelium of the cultivated mushroom Flammulina velutipes. Biosci. Biotechnol. Biochem. 2010, 74, 2327–2329. [Google Scholar] [CrossRef]
- Combier, J.-P.; Melayah, D.; Raffier, C.; Gay, G.; Marmeisse, R. Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in the symbiotic ectomycorrhizal fungus Hebeloma cylindrosporum. FEMS Microbiol. Lett. 2003, 220, 141–148. [Google Scholar] [CrossRef]
- Zhang, J.I.; Shi, L.; Chen, H.; Sun, Y.Q.; Zhao, M.W.; Ren, A.; Chen, M.j.; Wang, H.; Feng, Z.Y. An efficient Agrobacterium-mediated transformation method for the edible mushroom Hypsizygus marmoreus. Microbiol. Res. 2014, 169, 741–748. [Google Scholar] [CrossRef]
- Yan, L.; Xu, R.; Zhou, Y.; Gong, Y.; Dai, S.; Liu, H.; Bian, Y. Effects of Medium Composition and Genetic Background on Agrobacterium-Mediated Transformation Efficiency of Lentinula edodes. Genes 2019, 10, 467. [Google Scholar] [CrossRef]
- Lv, S.; Chen, X.; Mou, C.; Dai, S.; Bian, Y.; Kang, H. Agrobacterium-mediated transformation of the ascomycete mushroom Morchella importuna using polyubiquitin and glyceraldehyde-3-phosphate dehydrogenase promoter-based binary vectors. World J. Microbiol. Biotechnol. 2018, 34, 148. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, R.; Mao, W.J.; Gong, M.; Gao, Y.N.; Tang, L.H.; Yang, R.F.; Li, Y.; Zhou, C.L. A simple and efficient transformation system for the edible mushroom Pleurotus eryngii. Mycoscience 2016, 57, 356–360. [Google Scholar] [CrossRef]
- Ding, Y.; Liang, S.; Lei, J.; Chen, L.; Kothe, E.; Ma, A. Agrobacterium tumefaciens mediated fused egfp-hph gene expression under the control of gpd promoter in Pleurotus ostreatus. Microbiol. Res. 2011, 166, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, L.; Zhang, K.; Wu, Q.; Lin, J. Highly efficient Agrobacterium-mediated transformation of Volvariella volvacea. Bioresour. Technol. 2008, 99, 8524–8527. [Google Scholar] [CrossRef]
- Liu, G.; Cao, L.; Rao, Z.; Qiu, X.; Han, R. Identification of the genes involved in growth characters of medicinal fungus Ophiocordyceps sinensis based on Agrobacterium tumefaciens–mediated transformation. Appl. Microbiol. Biotechnol. 2020, 104, 2663–2674. [Google Scholar] [CrossRef]
- Li, D.; Tang, Y.; Lin, J.; Cai, W. Methods for genetic transformation of filamentous fungi. Microb. Cell Factories 2017, 16, 168. [Google Scholar] [CrossRef]
- Mello-Farias, P.C.D.; Chaves, A.L.S. Advances in Agrobacterium-mediated plant transformation with enphasys on soybean. Sci. Agric. 2008, 65, 95–106. [Google Scholar] [CrossRef]
- Liu, Z.; Friesen, T.L. Polyethylene glycol (PEG)-mediated transformation in filamentous fungal pathogens. In Plant Fungal Pathogens; Humana Press: Totowa, NJ, USA, 2012; pp. 365–375. [Google Scholar]

| Generation | Abnormal | Normal | Total | Expected Ratio | χ2 | p-Value |
|---|---|---|---|---|---|---|
| F2 | 80 | 28 | 108 | 3:1 | 0.049 | 0.824 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, C.P.; Moon, S.; Yoo, S.-i.; Noh, J.-H.; Ko, H.-G.; Kim, H.A.; Ro, H.-S.; Cho, H.; Chung, J.-W.; Lee, H.-Y.; et al. Functional Analysis of a Novel ABL (Abnormal Browning Related to Light) Gene in Mycelial Brown Film Formation of Lentinula edodes. J. Fungi 2020, 6, 272. https://doi.org/10.3390/jof6040272
Hong CP, Moon S, Yoo S-i, Noh J-H, Ko H-G, Kim HA, Ro H-S, Cho H, Chung J-W, Lee H-Y, et al. Functional Analysis of a Novel ABL (Abnormal Browning Related to Light) Gene in Mycelial Brown Film Formation of Lentinula edodes. Journal of Fungi. 2020; 6(4):272. https://doi.org/10.3390/jof6040272
Chicago/Turabian StyleHong, Chang Pyo, Suyun Moon, Seung-il Yoo, Jong-Hyun Noh, Han-Gyu Ko, Hyun A. Kim, Hyeon-Su Ro, Hyunwoo Cho, Jong-Wook Chung, Hwa-Yong Lee, and et al. 2020. "Functional Analysis of a Novel ABL (Abnormal Browning Related to Light) Gene in Mycelial Brown Film Formation of Lentinula edodes" Journal of Fungi 6, no. 4: 272. https://doi.org/10.3390/jof6040272
APA StyleHong, C. P., Moon, S., Yoo, S.-i., Noh, J.-H., Ko, H.-G., Kim, H. A., Ro, H.-S., Cho, H., Chung, J.-W., Lee, H.-Y., & Ryu, H. (2020). Functional Analysis of a Novel ABL (Abnormal Browning Related to Light) Gene in Mycelial Brown Film Formation of Lentinula edodes. Journal of Fungi, 6(4), 272. https://doi.org/10.3390/jof6040272





