ATAC-Seq Identifies Chromatin Landscapes Linked to the Regulation of Oxidative Stress in the Human Fungal Pathogen Candida albicans
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains, Media, and Growth Conditions
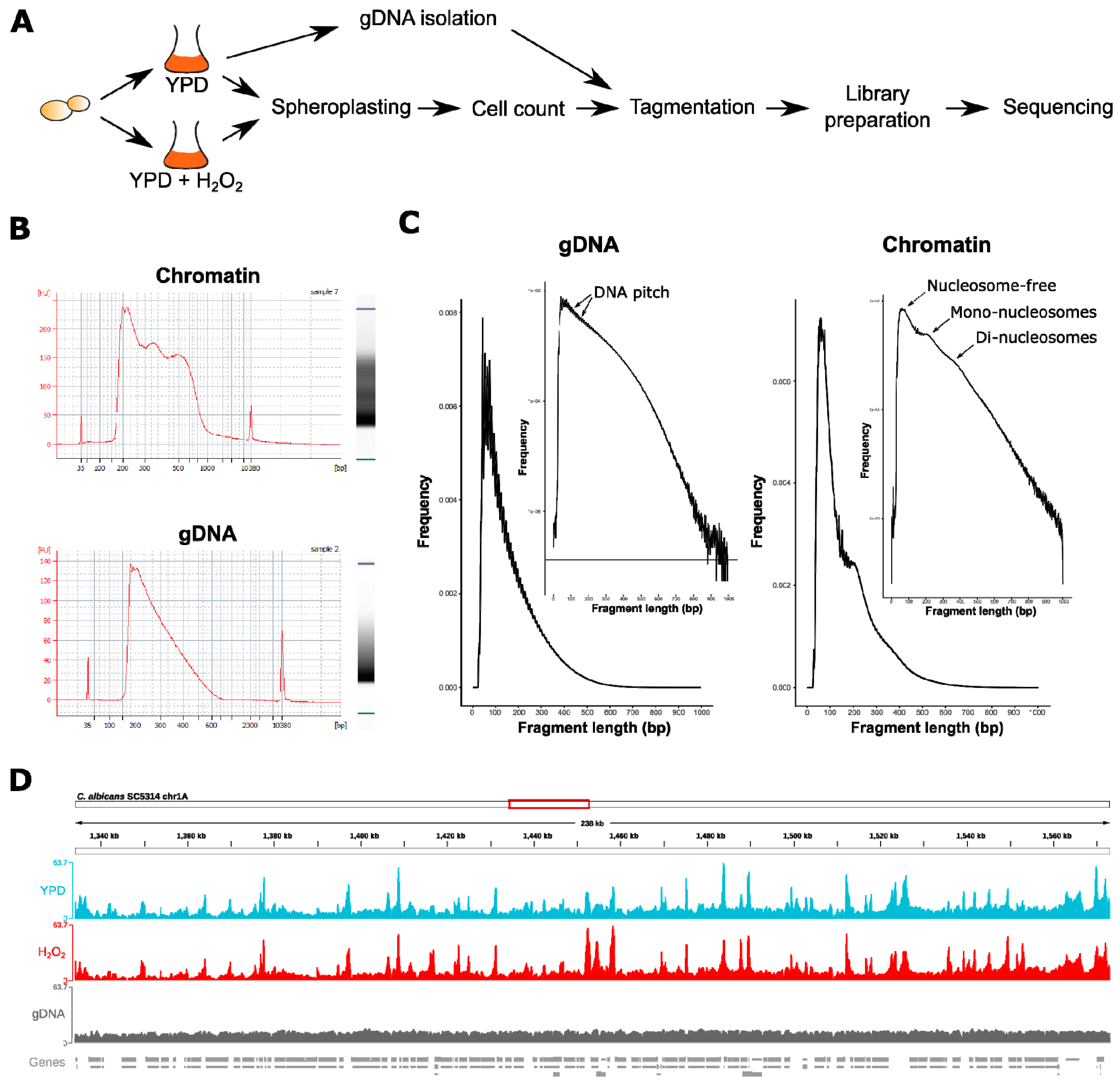
2.2. Nuclei and Genomic DNA (gDNA) Isolation, Tagmentation, ATAC-Seq Libraries, and Sequencing
2.2.1. gDNA Isolation
2.2.2. Tagmentation
2.2.3. ATAC-seq Library Amplification and Size Selection
2.2.4. Next-Generation Sequencing
2.3. ATAC-Seq Data Analysis Workflow
2.3.1. Pre-Processing and Read Alignment
2.3.2. Prediction of Nucleosomal Positions and Genomic Coverage of ATAC-seq Signals
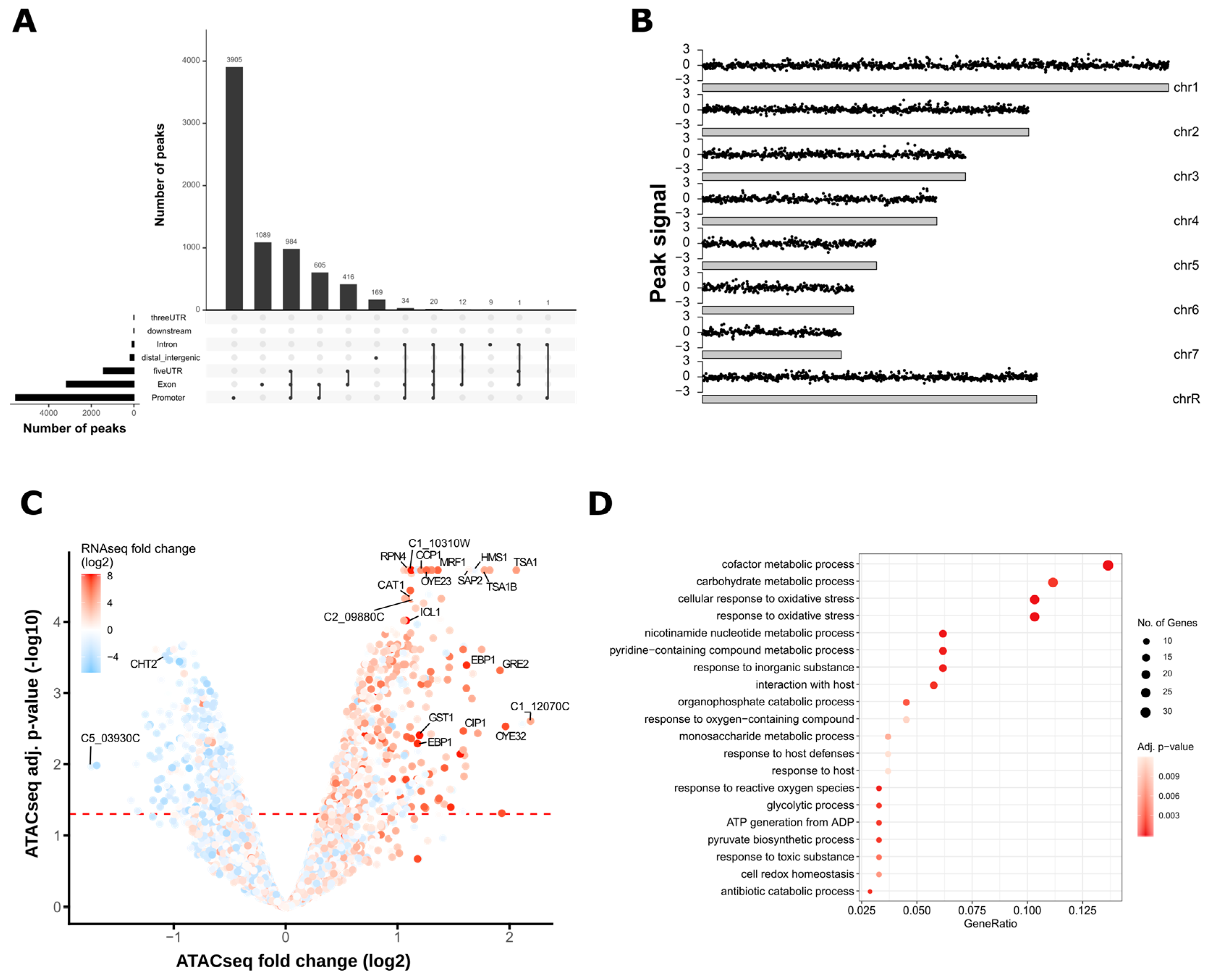
2.3.3. Peak Calling and Genomic Annotation of ATAC-seq Peaks
2.3.4. Analysis of Differential ATAC-seq Peaks
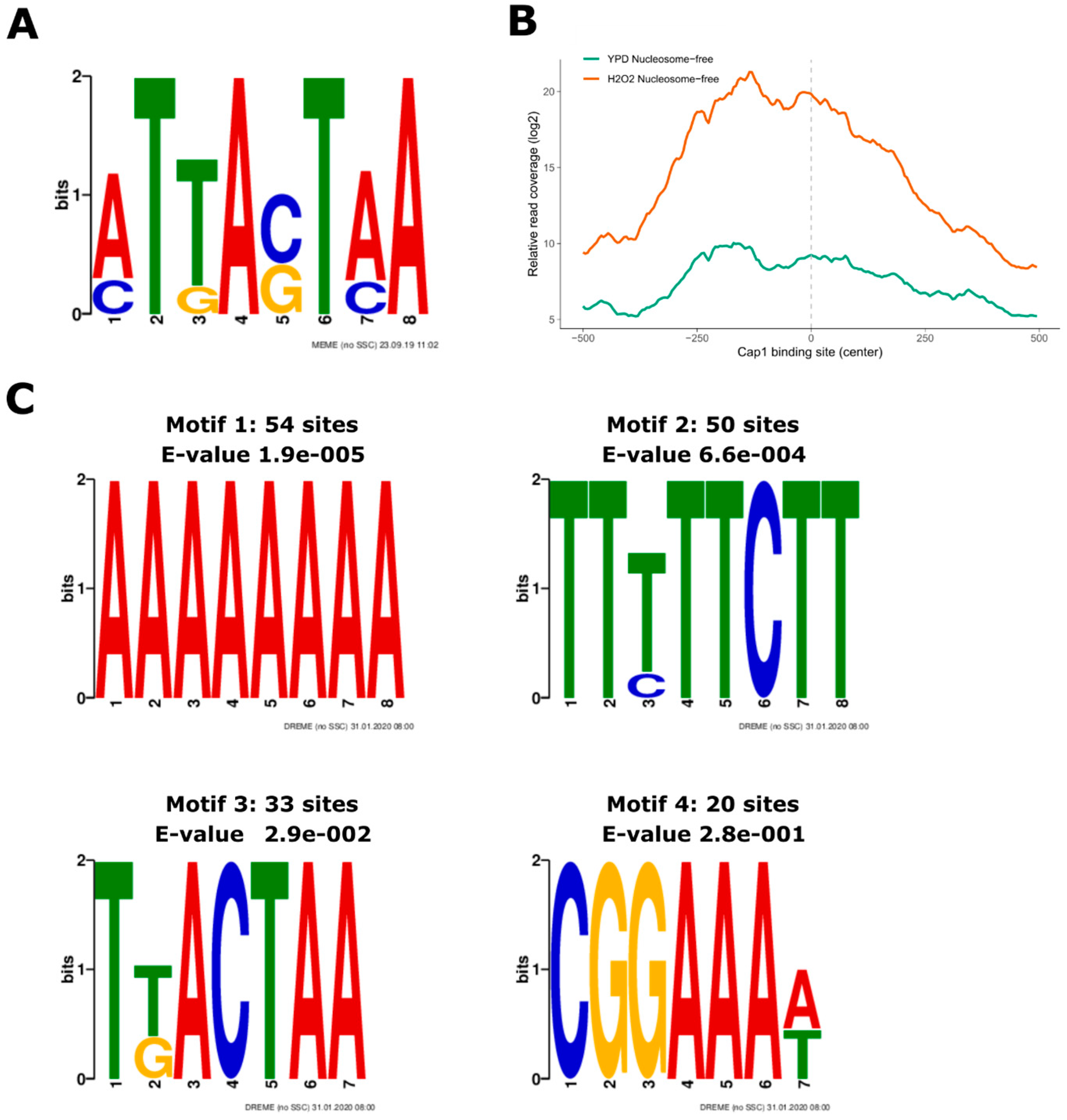
2.3.5. Motif Search
2.3.6. Data Plotting
2.4. External Datasets
2.5. Code Availability
2.6. Data Availability
3. Results and Discussion
3.1. ATAC-Seq in C. albicans Reflects Nucleosomal Organization Genome-Wide
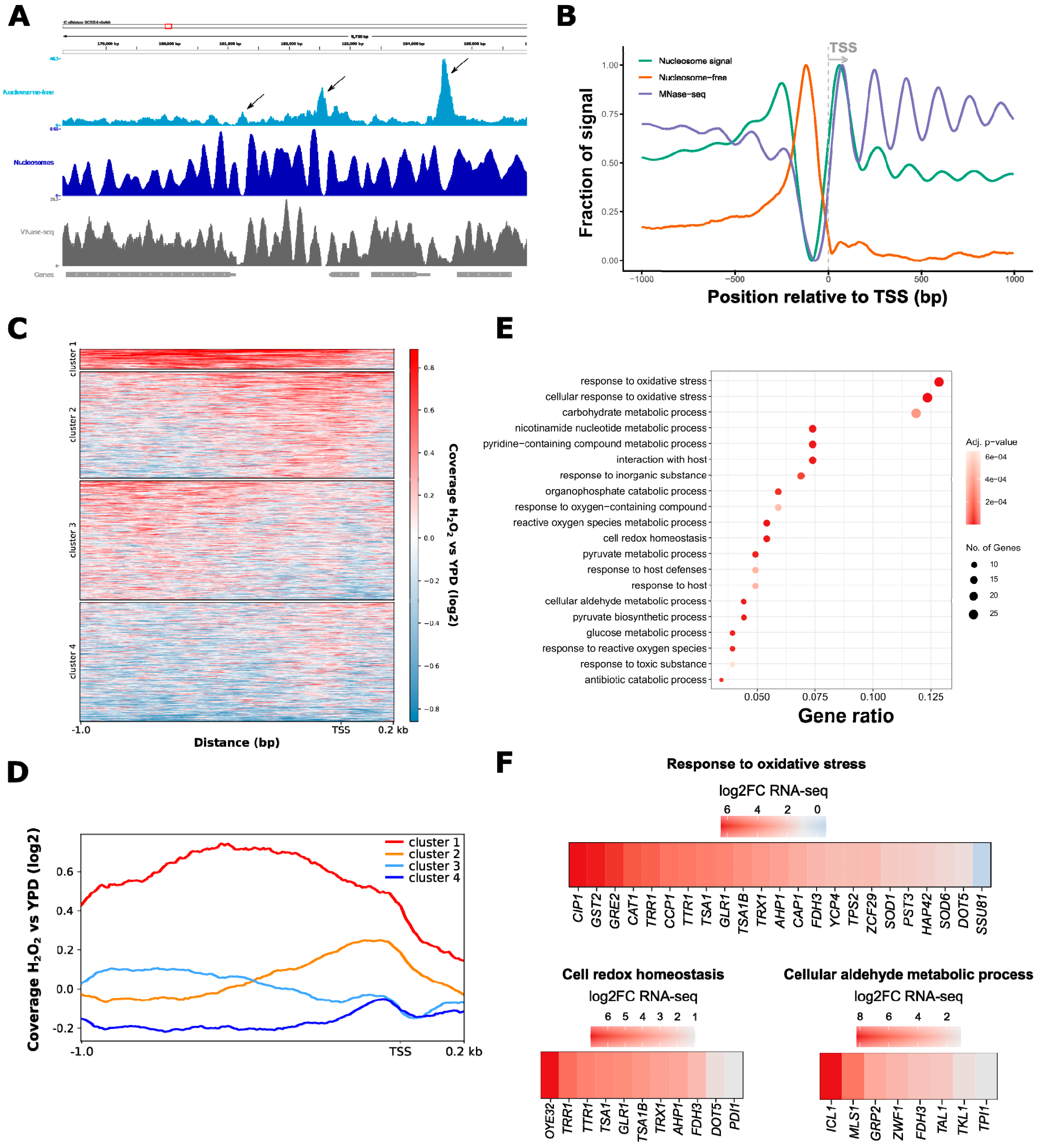
3.2. ATAC-Seq Detects Genome-Wide Changes in Chromatin Accessibility after Oxidative Stress
3.3. Oxidative Stress-Responsive ATAC-Seq Peaks Are Enriched for Cap1 Binding Sites
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rai, M.N.; Balusu, S.; Gorityala, N.; Dandu, L.; Kaur, R. Functional genomic analysis of Candida glabrata-macrophage interaction: Role of chromatin remodeling in virulence. PLoS Pathog. 2012, 8, e1002863. [Google Scholar] [CrossRef]
- Juárez-Reyes, A.; Castaño, I. Chromatin architecture and virulence-related gene expression in eukaryotic microbial pathogens. Curr. Genet. 2019, 65, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Dantas, A.; Lee, K.K.; Raziunaite, I.; Schaefer, K.; Wagener, J.; Yadav, B.; Gow, N.A. Cell biology of Candida albicans-host interactions. Curr. Opin. Microbiol. 2016, 34, 111–118. [Google Scholar] [CrossRef] [PubMed]
- López-Maury, L.; Marguerat, S.; Bähler, J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008, 9, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Cases, I.; de Lorenzo, V.; Ouzounis, C.A. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 2003, 11, 248–253. [Google Scholar] [CrossRef]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell. Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef]
- Voss, T.C.; Hager, G.L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014, 15, 69–81. [Google Scholar] [CrossRef]
- Brown, A.J.P.; Budge, S.; Kaloriti, D.; Tillmann, A.; Jacobsen, M.D.; Yin, Z.; Ene, I.V.; Bohovych, I.; Sandai, D.; Kastora, S.; et al. Stress adaptation in a pathogenic fungus. J. Exp. Biol. 2014, 217, 144–155. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Wilson, D.; Thewes, S.; Zakikhany, K.; Fradin, C.; Albrecht, A.; Almeida, R.; Brunke, S.; Grosse, K.; Martin, R.; Mayer, F.; et al. Identifying infection-associated genes of Candida albicans in the postgenomic era. FEMS Yeast Res. 2009, 9, 688–700. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.M.; Gianetti, B.A.; Witchley, J.N. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol. 2017, 15, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; Nantel, A.; Whiteway, M. Stress-induced gene expression in Candida albicans: Absence of a general stress response. Mol. Biol. Cell. 2002, 14, 1460–1467. [Google Scholar] [CrossRef] [PubMed]
- Shivarathri, R.; Tscherner, M.; Zwolanek, F.; Singh, N.K.; Chauhan, N.; Kuchler, K. The fungal histone acetyl transferase Gcn5 controls virulence of the human pathogen Candida albicans through multiple pathways. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Tscherner, M.; Zwolanek, F.; Jenull, S.; Sedlazeck, F.J.; Petryshyn, A.; Frohner, I.E.; Mavrianos, J.; Chauhan, N.; von Haeseler, A.; Kuchler, K. The Candida albicans histone acetyltransferase Hat1 regulates stress resistance and virulence via distinct chromatin assembly pathways. PLoS Pathog. 2015, 11, e1005218. [Google Scholar] [CrossRef]
- Kuchler, K.; Jenull, S.; Shivarathri, R.; Chauhan, N. Fungal KATs/KDACs: A new highway to better antifungal drugs? PLoS Pathog. 2016, 12, e1005938. [Google Scholar] [CrossRef]
- Da Rosa, J.L.; Boyartchuk, V.L.; Zhu, L.J.; Kaufman, P.D. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 1594–1599. [Google Scholar] [CrossRef]
- Wurtele, H.; Tsao, S.; Lépine, G.; Mullick, A.; Tremblay, J.; Drogaris, P.; Lee, E.H.; Thibault, P.; Verreault, A.; Raymond, M. Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat Med. 2010, 16, 774–780. [Google Scholar] [CrossRef]
- Hnisz, D.; Bardet, A.F.; Nobile, C.J.; Petryshyn, A.; Glaser, W.; Schöck, U.; Stark, A.; Kuchler, K. A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 2012, 8, e1003118. [Google Scholar] [CrossRef]
- Garnaud, C.; Champleboux, M.; Maubon, D.; Cornet, M.; Govin, J. Histone deacetylases and their inhibition in Candida species. Front. Microbiol. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.; Wright, G.D.; Cowen, L.E. Antifungal drugs: The current armamentarium and development of new agents. In The Fungal Kingdom; ASM Press: Washington, DC, USA, 2016; pp. 903–922. [Google Scholar]
- Lockhart, S.R. Candida auris and multidrug resistance: Defining the new normal. Fungal Genet. Biol. 2019, 131, 103243. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.F.; Gade, L.; Chow, N.A.; Loparev, V.N.; Juieng, P.; Berkow, E.L.; Farrer, R.A.; Litvintseva, A.P.; Cuomo, C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018, 9, 5346. [Google Scholar] [CrossRef]
- Kean, R.; Brown, J.; Gulmez, D.; Ware, A.; Ramage, G. Candida auris: A decade of understanding of an enigmatic pathogenic yeast. J. Fungi 2020, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.K.M.; Pugh, B.F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell. Biol. 2017, 18, 548–562. [Google Scholar] [CrossRef]
- Brogaard, K.; Xi, L.; Wang, J.-P.; Widom, J. A map of nucleosome positions in yeast at base-pair resolution. Nature 2012, 486, 496–501. [Google Scholar] [CrossRef]
- Thurman, R.E.; Rynes, E.; Humbert, R.; Vierstra, J.; Maurano, M.T.; Haugen, E.; Sheffield, N.C.; Stergachis, A.B.; Wang, H.; Vernot, B.; et al. The accessible chromatin landscape of the human genome. Nature 2012, 489, 75–82. [Google Scholar] [CrossRef]
- Boyle, A.P.; Davis, S.; Shulha, H.P.; Meltzer, P.; Margulies, E.H.; Weng, Z.; Furey, T.S.; Crawford, G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell 2008, 132, 311–322. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Schep, A.N.; Buenrostro, J.D.; Denny, S.K.; Schwartz, K.; Sherlock, G.; Greenleaf, W.J. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015, 25, 1757–1770. [Google Scholar] [CrossRef]
- Lu, Z.; Hofmeister, B.T.; Vollmers, C.; DuBois, R.M.; Schmitz, R.J. Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res. 2017, 45, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Huang, B.; Chen, H.; Yin, Q.; Liu, Y.; Xiang, Y.; Zhang, B.; Liu, B.; Wang, Q.; Xia, W.; et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016, 534, 652–657. [Google Scholar] [CrossRef]
- Maezawa, S.; Yukawa, M.; Alavattam, K.G.; Barski, A.; Namekawa, S.H. Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res. 2017, 46, 593–608. [Google Scholar] [CrossRef]
- Liu, C.; Wang, M.; Wei, X.; Wu, L.; Xu, J.; Dai, X.; Xia, J.; Cheng, M.; Yuan, Y.; Zhang, P.; et al. An ATAC-seq atlas of chromatin accessibility in mouse tissues. Sci. Data 2019, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- Corces, M.R.; Trevino, A.E.; Hamilton, E.G.; Greenside, P.G.; Sinnott-Armstrong, N.A.; Vesuna, S.; Satpathy, A.T.; Rubin, A.J.; Montine, K.S.; Wu, B.; et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 2017, 14, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Lareau, C.A.; Duarte, F.M.; Chew, J.G.; Kartha, V.K.; Burkett, Z.D.; Kohlway, A.S.; Pokholok, D.; Aryee, M.J.; Steemers, F.J.; Lebofsky, R.; et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 2019, 37, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Toenhake, C.G.; Fraschka, S.A.K.; Vijayabaskar, M.S.; Westhead, D.R.; van Heeringen, S.J.; Bártfai, R. Chromatin accessibility-based characterization of the gene regulatory network underlying plasmodium falciparum blood-stage development. Cell Host Microbe 2018, 23, 557–569. [Google Scholar] [CrossRef]
- Rogerson, C.; Britton, E.; Withey, S.; Hanley, N.; Ang, Y.S.; Sharrocks, A.D. Identification of a primitive intestinal transcription factor network shared between esophageal adenocarcinoma and its precancerous precursor state. Genome Res. 2019, 29, 723–736. [Google Scholar] [CrossRef]
- Li, Z.; Schulz, M.H.; Look, T.; Begemann, M.; Zenke, M.; Costa, I.G. Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 2019, 20, 45. [Google Scholar] [CrossRef]
- Liu, L.; Leng, L.; Liu, C.; Lu, C.; Yuan, Y.; Wu, L.; Gong, F.; Zhang, S.; Wei, X.; Wang, M.; et al. An integrated chromatin accessibility and transcriptome landscape of human pre-implantation embryos. Nat. Commun. 2019, 10, 364. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; MacCallum, D.M.; Odds, F.C.; Brown, A.J.P. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect. Immun. 2007, 75, 2143–2151. [Google Scholar] [CrossRef] [PubMed]
- Kaloriti, D.; Jacobsen, M.; Yin, Z.; Patterson, M.; Tillmann, A.; Smith, D.A.; Cook, E.; You, T.; Grimm, M.J.; Bohovych, I.; et al. Mechanisms underlying the exquisite sensitivity of Candida albicans to combinatorial cationic and oxidative stress that enhances the potent fungicidal activity of phagocytes. MBio 2014, 5, e01334-14. [Google Scholar] [CrossRef] [PubMed]
- Znaidi, S.; Barker, K.S.; Weber, S.; Alarco, A.-M.; Liu, T.T.; Boucher, G.; Rogers, P.D.; Raymond, M. Identification of the Candida albicans Cap1p regulon. Eukaryot. Cell 2009, 8, 806–820. [Google Scholar] [CrossRef] [PubMed]
- Gillum, A.M.; Tsay, E.Y.H.; Kirsch, D.R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. MGG Mol. Gen. Genet. 1984, 198, 179–182. [Google Scholar] [CrossRef]
- Goryshin, I.Y.; Miller, J.A.; Kil, Y.V.; Lanzov, V.A.; Reznikoff, W.S. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. USA 1998, 95, 10716–10721. [Google Scholar] [CrossRef]
- Hoffman, C.S.; Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformaion of Escherichia coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015, 109, 21–29. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Sedlazeck, F.J.; Rescheneder, P.; von Haeseler, A. NextGenMap: Fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 2013, 29, 2790–2791. [Google Scholar] [CrossRef]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef] [PubMed]
- Tsankov, A.M.; Thompson, D.A.; Socha, A.; Regev, A.; Rando, O.J. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010, 8, e1000414. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMISC: J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; He, Q.-Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Gel, B.; Serra, E. karyoploteR: An R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 2017, 33, 3088–3090. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. S2), W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics 2011, 27, 1653–1659. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Adey, A.; Morrison, H.G.; Asan Xun, X.; Kitzman, J.O.; Turner, E.H.; Stackhouse, B.; MacKenzie, A.P.; Caruccio, N.C.; Zhang, X.; Shendure, J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010, 11, R119. [Google Scholar] [CrossRef]
- Bai, L.; Morozov, A.V. Gene regulation by nucleosome positioning. Trends Genet. 2010, 26, 476–483. [Google Scholar] [CrossRef]
- Cottier, F.; Tan, A.S.M.; Chen, J.; Lum, J.; Zolezzi, F.; Poidinger, M.; Pavelka, N. The transcriptional stress response of Candida albicans to weak organic acids. G3 Genes Genomes Genet. 2015, 5, 497–505. [Google Scholar]
- Lorenz, M.C.; Bender, J.A.; Fink, G.R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell. 2004, 3, 1076–1087. [Google Scholar] [CrossRef]
- Enjalbert, B.; Smith, D.A.; Cornell, M.J.; Alam, I.; Nicholls, S.; Brown, A.J.P.; Quinn, J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell. 2006, 17, 1018–1032. [Google Scholar] [CrossRef]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Albert, B.; Kos-Braun, I.C.; Henras, A.K.; Dez, C.; Rueda, M.P.; Zhang, X.; Gadal, O.; Kos, M.; Shore, D. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. Elife 2019, 8, e45002. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, M.M.; Ernst, J.; Wilder, S.P.; Kundaje, A.; Harris, R.S.; Libbrecht, M.; Giardine, B.; Ellenbogen, P.M.; Bilmes, J.A.; Birney, E.; et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013, 41, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Gómez-Rodríguez, M.; Mieczkowski, J.; Tolstorukov, M.Y.; Kundu, S.; Sadreyev, R.I.; Jansen, L.E.; Kingston, R.E. Enhancer regions show high histone H3.3 turnover that changes during differentiation. Elife 2016, 5, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, D.G.; Soifer, I.; Wranik, B.J.; Botstein, D.; Scott McIsaac, R. Simultaneous profiling of DNA accessibility and gene expression dynamics with ATAC-seq and RNA-seq. Methods Mol. Biol. 2018, 1819, 317–333. [Google Scholar]
- Wysong, D.R.; Christin, L.; Sugar, A.M.; Robbins, P.W.; Diamond, R.D. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 1998, 66, 1953–1961. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, Y.Y.; Jia, X.M.; Cao, Y.B.; Gao, P.H.; Fu, X.P.; Ying, K.; Chen, W.S.; Jiang, Y.Y. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic. Biol. Med. 2006, 40, 1201–1209. [Google Scholar] [CrossRef]
- Piekarska, K.; Hardy, G.; Mol, E.; van den Burg, J.; Strijbis, K.; van Roermund, C.; van den Berg, M.; Distel, B. The activity of the glyoxylate cycle in peroxisomes of Candida albicans depends on a functional beta-oxidation pathway: Evidence for reduced metabolite transport across the peroxisomal membrane. Microbiology 2008, 154, 3061–3072. [Google Scholar] [CrossRef]
- Yan, F.; Powell, D.R.; Curtis, D.J.; Wong, N.C. From reads to insight: A hitchhiker’s guide to ATAC-seq data analysis. Genome Biol. 2020, 21, 22. [Google Scholar] [CrossRef]
- Urban, C.; Xiong, X.; Sohn, K.; Schröppel, K.; Brunner, H.; Rupp, S. The moonlighting protein Tsa1p is implicated in oxidative stress response and in cell wall biogenesis in Candida albicans. Mol. Microbiol. 2005, 57, 1318–1341. [Google Scholar] [CrossRef]
- Lee, K.; Kim, S.C.; Jung, I.; Kim, K.; Seo, J.; Lee, H.-S.; Bogu, G.K.; Kim, D.; Lee, S.; Lee, B.; et al. Genetic landscape of open chromatin in yeast. PLoS Genet. 2013, 9, e1003229. [Google Scholar] [CrossRef]
- Hernday, A.D.; Lohse, M.B.; Fordyce, P.M.; Nobile, C.J.; Derisi, J.L.; Johnson, A.D. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol. Microbiol. 2013, 90, 22–35. [Google Scholar]
- Nobile, C.J.; Fox, E.P.; Nett, J.E.; Sorrells, T.R.; Mitrovich, Q.M.; Hernday, A.D.; Tuch, B.B.; Andes, D.R.; Johnson, A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 2012, 148, 126–138. [Google Scholar] [CrossRef]
- Segal, E.; Widom, J. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009, 19, 65–71. [Google Scholar] [CrossRef]
- Rhee, H.S.; Pugh, B.F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 2012, 483, 295–301. [Google Scholar] [CrossRef]
- Wang, J.R.; Quach, B.; Furey, T.S. Correcting nucleotide-specific biases in high-throughput sequencing data. BMC Bioinformatics 2017, 18, 357. [Google Scholar] [CrossRef]
- Martins, A.L.; Walavalkar, N.M.; Anderson, W.D.; Zang, C.; Guertin, M.J. Universal correction of enzymatic sequence bias reveals molecular signatures of protein/DNA interactions. Nucleic Acids Res. 2017, 46, e9. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jenull, S.; Tscherner, M.; Mair, T.; Kuchler, K. ATAC-Seq Identifies Chromatin Landscapes Linked to the Regulation of Oxidative Stress in the Human Fungal Pathogen Candida albicans. J. Fungi 2020, 6, 182. https://doi.org/10.3390/jof6030182
Jenull S, Tscherner M, Mair T, Kuchler K. ATAC-Seq Identifies Chromatin Landscapes Linked to the Regulation of Oxidative Stress in the Human Fungal Pathogen Candida albicans. Journal of Fungi. 2020; 6(3):182. https://doi.org/10.3390/jof6030182
Chicago/Turabian StyleJenull, Sabrina, Michael Tscherner, Theresia Mair, and Karl Kuchler. 2020. "ATAC-Seq Identifies Chromatin Landscapes Linked to the Regulation of Oxidative Stress in the Human Fungal Pathogen Candida albicans" Journal of Fungi 6, no. 3: 182. https://doi.org/10.3390/jof6030182
APA StyleJenull, S., Tscherner, M., Mair, T., & Kuchler, K. (2020). ATAC-Seq Identifies Chromatin Landscapes Linked to the Regulation of Oxidative Stress in the Human Fungal Pathogen Candida albicans. Journal of Fungi, 6(3), 182. https://doi.org/10.3390/jof6030182




