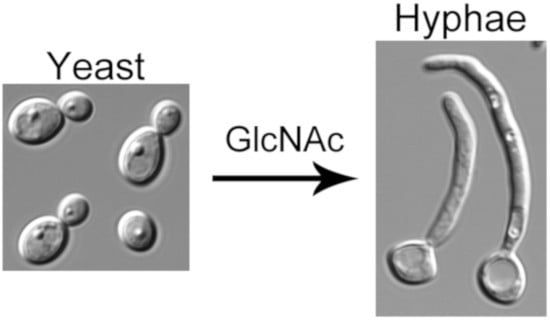
N-Acetylglucosamine Regulates Morphogenesis and Virulence Pathways in Fungi
Abstract
1. Introduction
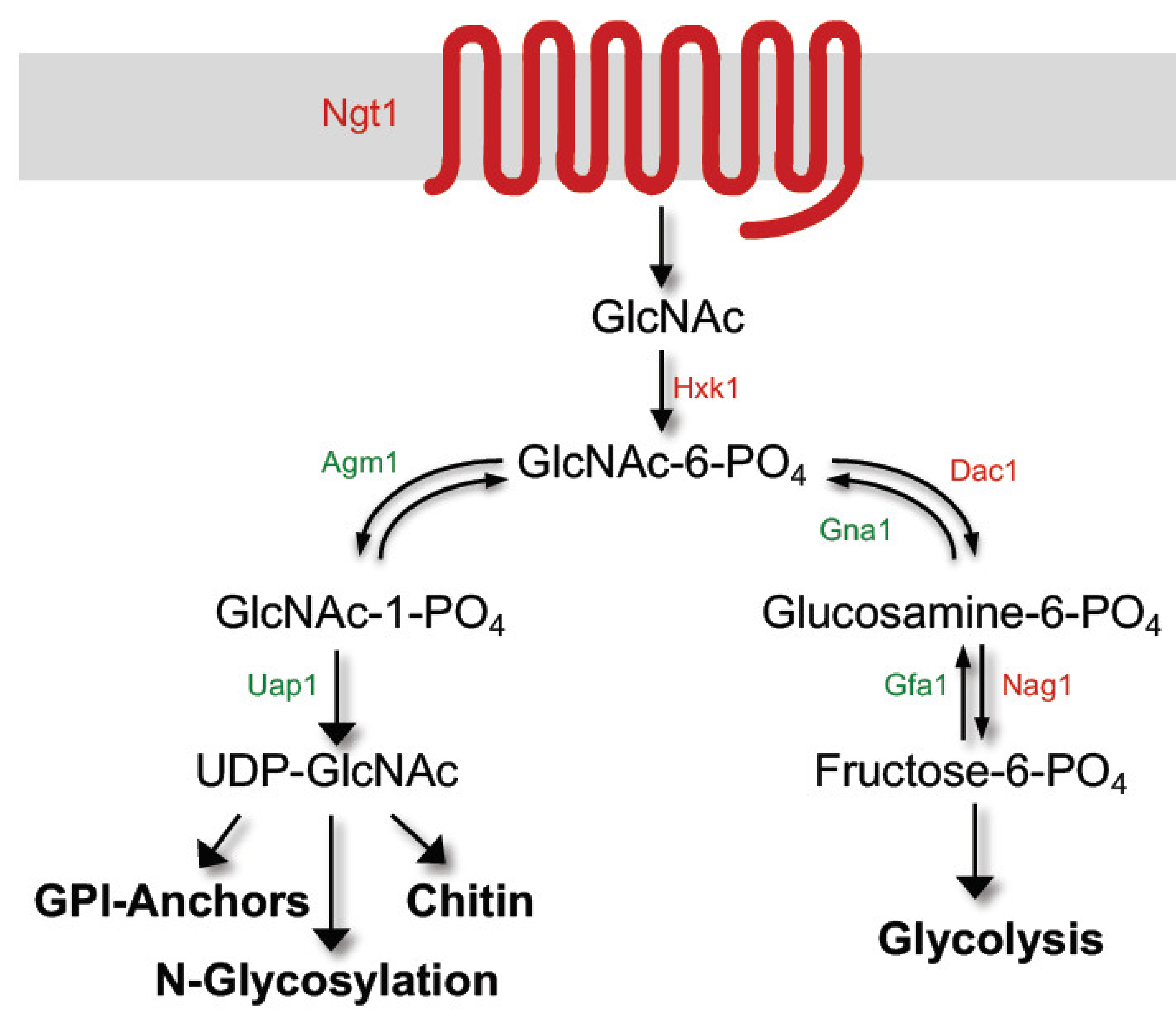
2. GlcNAc Induces Expression of Genes Needed for Its Catabolism
Open Questions
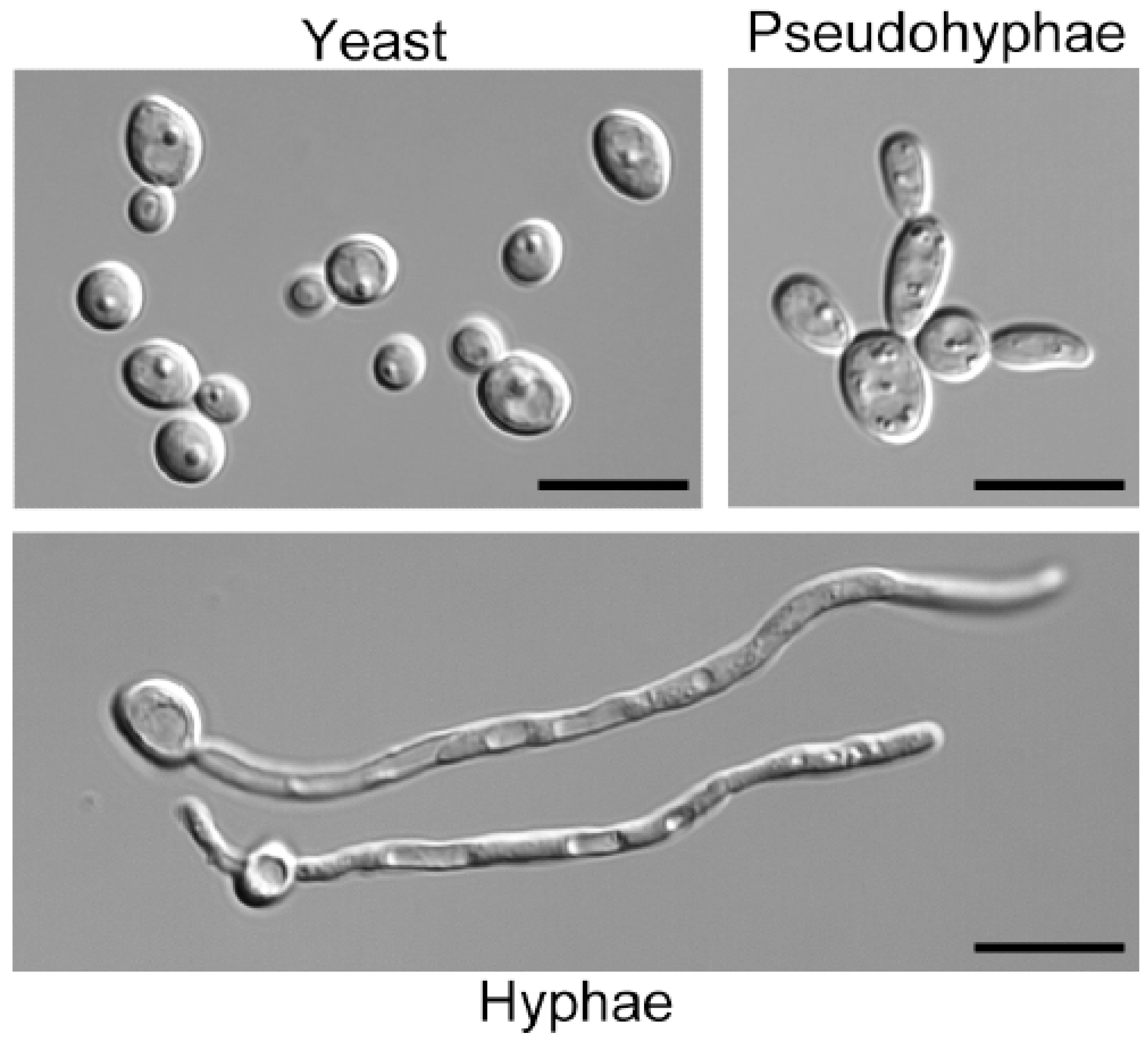
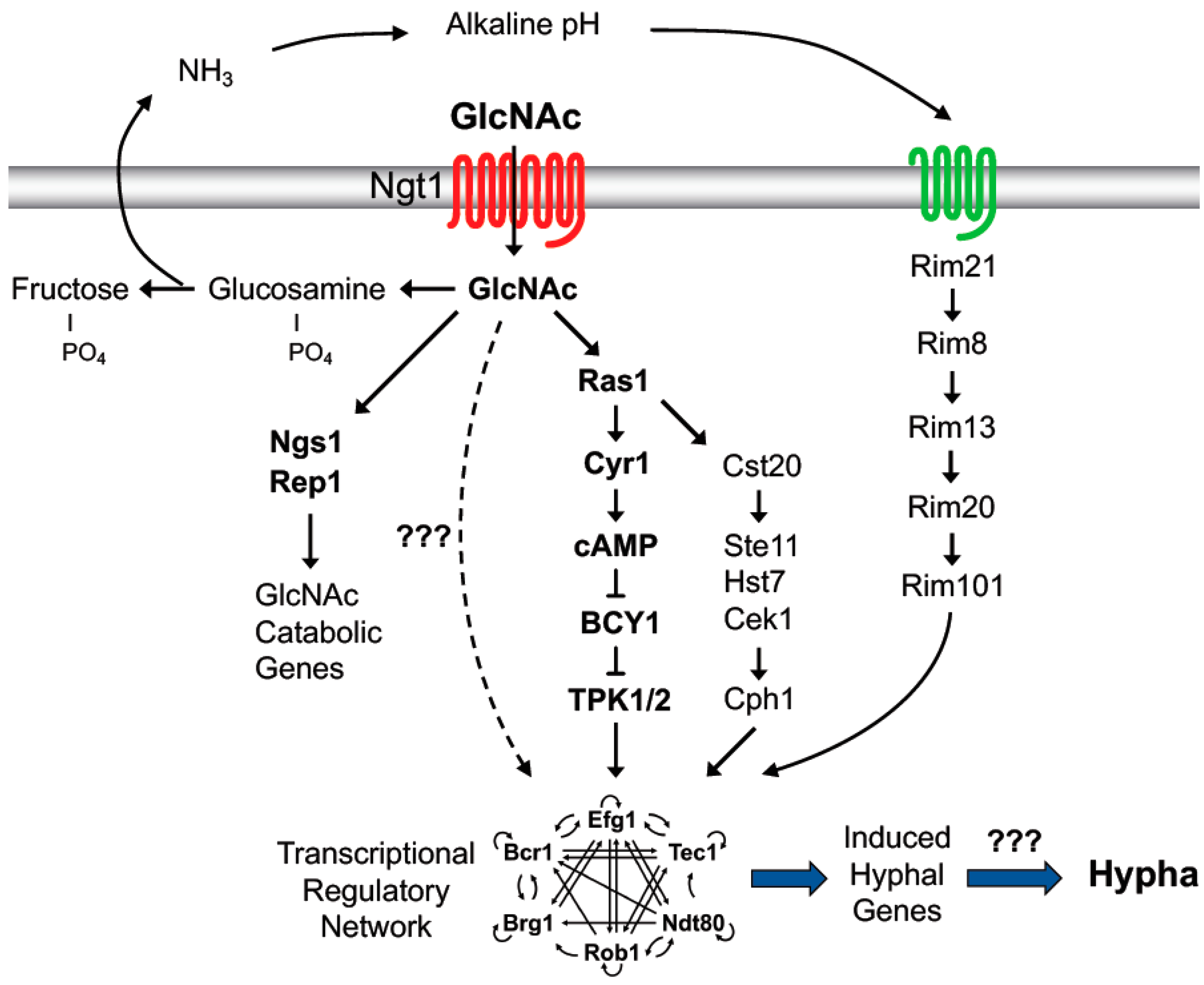
3. GlcNAc Stimulates a Switch to Hyphal Morphogenesis
3.1. GlcNAc Has to Be Taken Up by Cells to Induce Hyphal Growth
3.2. GlcNAc Metabolism Enhances Hyphal Gene Transcription by Alkalinizing the Extracellular Environment
3.3. GlcNAc Induces a cAMP-Independent Signal
3.4. Hyphal-Induced Genes Do Not Play an Obvious Role in Hyphal Morphogenesis
3.5. Protein Phosphorylation Promotes Hyphal Morphogenesis
3.6. Open Questions
4. GlcNAc Induces the White-Opaque Epigenetic Switch
4.1. GlcNAc Regulates White-Opaque Switching
4.2. Open Questions
5. Roles of GlcNAc in C. albicans Commensalism and Virulence
5.1. GlcNAc Influences Virulence Functions
5.2. Potential Roles of GlcNAc in Commensalism
5.3. Open Questions
6. Roles of GlcNAc in Other Fungal Species
6.1. GlcNAc Catabolism in Other Fungal Species
6.2. GlcNAc Can Stimulate or Inhibit Hyphal Morphogenesis in Other Species
6.3. GlcNAc Roles in Interspecies Communication
6.4. Open Questions
7. Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simonetti, N.; Strippoli, V.; Cassone, A. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature 1974, 250, 344–346. [Google Scholar] [CrossRef] [PubMed]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.M.; Gianetti, B.A.; Witchley, J.N. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol. 2017, 15, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Whiteway, M.; Bachewich, C. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 2007, 61, 529–553. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A.; Vinces, M.D. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 2005, 7, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Van Dijck, P.; Datta, A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 2007, 71, 348–376. [Google Scholar] [CrossRef]
- Davis, D.A. How human pathogenic fungi sense and adapt to pH: The link to virulence. Curr. Opin. Microbiol. 2009, 12, 365–370. [Google Scholar] [CrossRef]
- Kornitzer, D. Regulation of Candida albicans Hyphal Morphogenesis by Endogenous Signals. J. Fungi 2019, 5, 21. [Google Scholar] [CrossRef]
- Slutsky, B.; Staebell, M.; Anderson, J.; Risen, L.; Pfaller, M.; Soll, D.R. “White-opaque transition”: A second high-frequency switching system in Candida albicans. J. Bacteriol. 1987, 169, 189–197. [Google Scholar] [CrossRef]
- Huang, G.; Yi, S.; Sahni, N.; Daniels, K.J.; Srikantha, T.; Soll, D.R. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 2010, 6, e1000806. [Google Scholar] [CrossRef]
- Sun, Y.; Cao, C.; Jia, W.; Tao, L.; Guan, G.; Huang, G. pH Regulates White-Opaque Switching and Sexual Mating in Candida albicans. Eukaryot. Cell 2015, 14, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Srikantha, T.; Sahni, N.; Yi, S.; Soll, D.R. CO2 regulates white-to-opaque switching in Candida albicans. Curr. Biol. 2009, 19, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Ene, I.V.; Lohse, M.B.; Vladu, A.V.; Morschhauser, J.; Johnson, A.D.; Bennett, R.J. Phenotypic Profiling Reveals that Candida albicans Opaque Cells Represent a Metabolically Specialized Cell State Compared to Default White Cells. MBio 2016, 7, e01269-16. [Google Scholar] [CrossRef]
- Kvaal, C.; Lachke, S.A.; Srikantha, T.; Daniels, K.; McCoy, J.; Soll, D.R. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect. Immun. 1999, 67, 6652–6662. [Google Scholar]
- Soll, D.R. The role of phenotypic switching in the basic biology and pathogenesis of Candida albicans. J. Oral. Microbiol. 2014, 6, 22993. [Google Scholar] [CrossRef]
- Miller, M.G.; Johnson, A.D. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 2002, 110, 293–302. [Google Scholar] [CrossRef]
- Zordan, R.E.; Miller, M.G.; Galgoczy, D.J.; Tuch, B.B.; Johnson, A.D. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007, 5, e256. [Google Scholar] [CrossRef]
- Kumar, M.J.; Jamaluddin, M.S.; Natarajan, K.; Kaur, D.; Datta, A. The inducible N-acetylglucosamine catabolic pathway gene cluster in Candida albicans: Discrete N-acetylglucosamine-inducible factors interact at the promoter of NAG1. Proc. Natl. Acad. Sci. USA 2000, 97, 14218–14223. [Google Scholar] [CrossRef]
- Gunasekera, A.; Alvarez, F.J.; Douglas, L.M.; Wang, H.X.; Rosebrock, A.P.; Konopka, J.B. Identification of GIG1, a GlcNAc-induced gene in Candida albicans needed for normal sensitivity to the chitin synthase inhibitor nikkomycin Z. Eukaryot. Cell 2010, 9, 1476–1483. [Google Scholar] [CrossRef]
- Yamada-Okabe, T.; Sakamori, Y.; Mio, T.; Yamada-Okabe, H. Identification and characterization of the genes for N-acetylglucosamine kinase and N-acetylglucosamine-phosphate deacetylase in the pathogenic fungus Candida albicans. Eur. J. Biochem. 2001, 268, 2498–2505. [Google Scholar] [CrossRef]
- Alvarez, F.J.; Konopka, J.B. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol. Biol. Cell 2007, 18, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Naseem, S.; Gunasekera, A.; Araya, E.; Konopka, J.B. N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. J. Biol. Chem. 2011, 286, 28671–28680. [Google Scholar] [CrossRef] [PubMed]
- Naseem, S.; Araya, E.; Konopka, J.B. Hyphal growth in Candida albicans does not require induction of hyphal-specific gene expression. Mol. Biol. Cell 2015, 26, 1174–1187. [Google Scholar] [CrossRef] [PubMed]
- Hart, G.W.; Slawson, C.; Ramirez-Correa, G.; Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011, 80, 825–858. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, S.A.; Naseem, S.; Konopka, J.B.; Sil, A. N-acetylglucosamine (GlcNAc) Triggers a Rapid, Temperature-Responsive Morphogenetic Program in Thermally Dimorphic Fungi. PLoS Genet. 2013, 9, e1003799. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Cheon, S.A.; Park, S.; Song, Y.; Kim, J.Y. Serum-induced hypha formation in the dimorphic yeast Yarrowia lipolytica. FEMS Microbiol. Lett. 2000, 190, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Kappel, L.; Gaderer, R.; Flipphi, M.; Seidl-Seiboth, V. The N-acetylglucosamine catabolic gene cluster in Trichoderma reesei is controlled by the Ndt80-like transcription factor RON1. Mol. Microbiol. 2016, 99, 640–657. [Google Scholar] [CrossRef]
- Nadal, M.; Sawers, R.; Naseem, S.; Bassin, B.; Kulicke, C.; Sharman, A.; An, G.; An, K.; Ahern, K.R.; Romag, A.; et al. An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat. Plants 2017, 3, 17073. [Google Scholar] [CrossRef]
- Naseem, S.; Min, K.; Spitzer, D.; Gardin, J.; Konopka, J.B. Regulation of Hyphal Growth and N-Acetylglucosamine Catabolism by Two Transcription Factors in Candida albicans. Genetics 2017, 206, 299–314. [Google Scholar] [CrossRef]
- Sullivan, P.A.; McHugh, N.J.; Romana, L.K.; Shepherd, M.G. The secretion of N-acetylglucosaminidase during germ-tube formation in Candida albicans. Microbiology 1984, 130, 2213–2218. [Google Scholar] [CrossRef]
- Ruhela, D.; Kamthan, M.; Saha, P.; Majumdar, S.S.; Datta, K.; Abdin, M.Z.; Datta, A. In vivo role of Candida albicans beta-hexosaminidase (HEX1) in carbon scavenging. MicrobiologyOpen 2015, 4, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Milewski, S.; Gabriel, I.; Olchowy, J. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast 2006, 23, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Naseem, S.; Parrino, S.; Beuenten, D.; Konopka, J.B. Novel roles for GlcNAc in cell signaling. Commun. Integr. Biol. 2012, 5, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Van Ende, M.; Wijnants, S.; Van Dijck, P. Sugar Sensing and Signaling in Candida albicans and Candida glabrata. Front. Microbiol. 2019, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Konopka, J.B. N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica 2012, 2012, 489208. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Lu, Y.; Liu, H. N-acetylglucosamine sensing by a GCN5-related N-acetyltransferase induces transcription via chromatin histone acetylation in fungi. Nat. Commun. 2016, 7, 12916. [Google Scholar] [CrossRef]
- Su, C.; Yu, J.; Sun, Q.; Liu, Q.; Lu, Y. Hyphal induction under the condition without inoculation in Candida albicans is triggered by Brg1-mediated removal of NRG1 inhibition. Mol. Microbiol. 2018, 108, 410–423. [Google Scholar] [CrossRef]
- Su, C.; Yu, J.; Lu, Y. Hyphal development in Candida albicans from different cell states. Curr. Genet. 2018, 64, 1239–1243. [Google Scholar] [CrossRef]
- White, R.J. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem. J. 1968, 106, 847–858. [Google Scholar] [CrossRef]
- Bernheim, N.J.; Dobrogosz, W.J. Amino sugar sensitivity in Escherichia coli mutants unable to grow on N-acetylglucosamine. J. Bacteriol. 1970, 101, 384–391. [Google Scholar]
- Du, H.; Guan, G.; Li, X.; Gulati, M.; Tao, L.; Cao, C.; Johnson, A.D.; Nobile, C.J.; Huang, G. N-Acetylglucosamine-Induced Cell Death in Candida albicans and Its Implications for Adaptive Mechanisms of Nutrient Sensing in Yeasts. MBio 2015, 6, e01376-15. [Google Scholar] [CrossRef] [PubMed]
- Kamthan, M.; Kamthan, A.; Ruhela, D.; Maiti, P.; Bhavesh, N.S.; Datta, A. Upregulation of galactose metabolic pathway by N-acetylglucosamine induced endogenous synthesis of galactose in Candida albicans. Fungal Genet. Biol. 2013, 54, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Vylkova, S.; Carman, A.J.; Danhof, H.A.; Collette, J.R.; Zhou, H.; Lorenz, M.C. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio 2011, 2, e00055-11. [Google Scholar] [CrossRef] [PubMed]
- Rocha, C.R.; Schroppel, K.; Harcus, D.; Marcil, A.; Dignard, D.; Taylor, B.N.; Thomas, D.Y.; Whiteway, M.; Leberer, E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 2001, 12, 3631–3643. [Google Scholar] [CrossRef] [PubMed]
- Kamthan, M.; Mukhopadhyay, G.; Chakraborty, N.; Chakraborty, S.; Datta, A. Quantitative proteomics and metabolomics approaches to demonstrate N-acetyl-D-glucosamine inducible amino acid deprivation response as morphological switch in Candida albicans. Fungal Genet. Biol. 2012, 49, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y. Fungal adenylyl cyclase acts as a signal sensor and integrator and plays a central role in interaction with bacteria. PLoS Pathog. 2013, 9, e1003612. [Google Scholar] [CrossRef] [PubMed]
- Sabie, F.T.; Gadd, G.M. Effect of nucleosides and nucleotides and the relationship between cellular adenosine 3’:5’-cyclic monophosphate (cyclic AMP) and germ tube formation in Candida albicans. Mycopathologia 1992, 119, 147–156. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Staab, J.; Sundstrom, P. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol. Microbiol. 2003, 50, 391–409. [Google Scholar] [CrossRef]
- Davis-Hanna, A.; Piispanen, A.E.; Stateva, L.I.; Hogan, D.A. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol. Microbiol. 2008, 67, 47–62. [Google Scholar] [CrossRef]
- Bai, C.; Xu, X.L.; Wang, H.S.; Wang, Y.M.; Chan, F.Y.; Wang, Y. Characterization of a hyperactive Cyr1 mutant reveals new regulatory mechanisms for cellular cAMP levels in Candida albicans. Mol. Microbiol. 2011, 82, 879–893. [Google Scholar] [CrossRef]
- Maidan, M.M.; De Rop, L.; Serneels, J.; Exler, S.; Rupp, S.; Tournu, H.; Thevelein, J.M.; Van Dijck, P. The G Protein-coupled Receptor Gpr1 and the Ga Protein Gpa2 Act through the cAMP-PKA Pathway to Induce Morphogenesis in Candida albicans. Mol. Biol. Cell 2005, 16, 1971–1986. [Google Scholar] [CrossRef] [PubMed]
- Parrino, S.M.; Si, H.; Naseem, S.; Groudan, K.; Gardin, J.; Konopka, J.B. cAMP-independent signal pathways stimulate hyphal morphogenesis in Candida albicans. Mol. Microbiol. 2017, 103, 764–779. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Su, C.; Wang, A.; Liu, H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 2011, 9, e1001105. [Google Scholar] [CrossRef]
- Harcus, D.; Nantel, A.; Marcil, A.; Rigby, T.; Whiteway, M. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell 2004, 15, 4490–4499. [Google Scholar] [CrossRef]
- Lu, Y.; Su, C.; Solis, N.V.; Filler, S.G.; Liu, H. Synergistic Regulation of Hyphal Elongation by Hypoxia, CO2, and Nutrient Conditions Controls the Virulence of Candida albicans. Cell Host Microbe 2013, 14, 499–509. [Google Scholar] [CrossRef]
- Carlisle, P.L.; Banerjee, M.; Lazzell, A.; Monteagudo, C.; Lopez-Ribot, J.L.; Kadosh, D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. USA 2009, 106, 599–604. [Google Scholar] [CrossRef]
- Kadosh, D. Regulatory mechanisms controlling morphology and pathogenesis in Candida albicans. Curr. Opin. Microbiol. 2019, 52, 27–34. [Google Scholar] [CrossRef]
- Martin, R.; Albrecht-Eckardt, D.; Brunke, S.; Hube, B.; Hunniger, K.; Kurzai, O. A core filamentation response network in Candida albicans is restricted to eight genes. PLoS ONE 2013, 8, e58613. [Google Scholar] [CrossRef]
- Giusani, A.D.; Vinces, M.; Kumamoto, C.A. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 2002, 160, 1749–1753. [Google Scholar]
- Min, K.; Biermann, A.; Hogan, D.A.; Konopka, J.B. Genetic Analysis of NDT80 Family Transcription Factors in Candida albicans Using New CRISPR-Cas9 Approaches. mSphere 2018, 3, e00545-18. [Google Scholar] [CrossRef]
- Woolford, C.A.; Lagree, K.; Xu, W.; Aleynikov, T.; Adhikari, H.; Sanchez, H.; Cullen, P.J.; Lanni, F.; Andes, D.R.; Mitchell, A.P. Bypass of Candida albicans Filamentation/Biofilm Regulators through Diminished Expression of Protein Kinase Cak1. PLoS Genet. 2016, 12, e1006487. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y. Hgc1-Cdc28-how much does a single protein kinase do in the regulation of hyphal development in Candida albicans? J. Microbiol. 2016, 54, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.D.; Lee, R.T.; Wang, Y.M.; Lin, Q.S.; Wang, Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007, 26, 3760–3769. [Google Scholar] [CrossRef] [PubMed]
- Bishop, A.; Lane, R.; Beniston, R.; Chapa-y-Lazo, B.; Smythe, C.; Sudbery, P. Hyphal growth in Candida albicans requires the phosphorylation of Sec2 by the Cdc28-Ccn1/Hgc1 kinase. EMBO J. 2010, 29, 2930–2942. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Lima, D.; Sudbery, P.E. In Candida albicans, phosphorylation of Exo84 by Cdk1-Hgc1 is necessary for efficient hyphal extension. Mol. Biol. Cell 2014, 25, 1097–1110. [Google Scholar] [CrossRef]
- Sinha, I.; Wang, Y.M.; Philp, R.; Li, C.R.; Yap, W.H.; Wang, Y. Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev. Cell 2007, 13, 421–432. [Google Scholar] [CrossRef]
- Zeng, G.; Wang, Y.M.; Wang, Y. Cdc28-Cln3 phosphorylation of Sla1 regulates actin patch dynamics in different modes of fungal growth. Mol. Biol. Cell 2012, 23, 3485–3497. [Google Scholar] [CrossRef]
- Gutierrez-Escribano, P.; Gonzalez-Novo, A.; Suarez, M.B.; Li, C.R.; Wang, Y.; de Aldana, C.R.; Correa-Bordes, J. CDK-dependent phosphorylation of Mob2 is essential for hyphal development in Candida albicans. Mol. Biol. Cell 2011, 22, 2458–2469. [Google Scholar] [CrossRef]
- Gonzalez-Novo, A.; Correa-Bordes, J.; Labrador, L.; Sanchez, M.; Vazquez de Aldana, C.R.; Jimenez, J. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol. Biol. Cell 2008, 19, 1509–1518. [Google Scholar] [CrossRef]
- Wang, A.; Raniga, P.P.; Lane, S.; Lu, Y.; Liu, H. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol. Cell. Biol. 2009, 29, 4406–4416. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004, 23, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Willger, S.D.; Liu, Z.; Olarte, R.A.; Adamo, M.E.; Stajich, J.E.; Myers, L.C.; Kettenbach, A.N.; Hogan, D.A. Analysis of the Candida albicans Phosphoproteome. Eukaryot. Cell 2015, 14, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Moussian, B. The role of GlcNAc in formation and function of extracellular matrices. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 149, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Vesely, E.M.; Williams, R.B.; Konopka, J.B.; Lorenz, M.C. N-Acetylglucosamine Metabolism Promotes Survival of Candida albicans in the Phagosome. mSphere 2017, 2, e00357-17. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.M.; Soll, D.R. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J. Bacteriol. 1987, 169, 5579–5588. [Google Scholar] [CrossRef]
- Xie, J.; Tao, L.; Nobile, C.J.; Tong, Y.; Guan, G.; Sun, Y.; Cao, C.; Hernday, A.D.; Johnson, A.D.; Zhang, L.; et al. White-opaque switching in natural MTLa/alpha isolates of Candida albicans: Evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol. 2013, 11, e1001525. [Google Scholar] [CrossRef]
- Tao, L.; Du, H.; Guan, G.; Dai, Y.; Nobile, C.J.; Liang, W.; Cao, C.; Zhang, Q.; Zhong, J.; Huang, G. Discovery of a “white-gray-opaque” tristable phenotypic switching system in candida albicans: Roles of non-genetic diversity in host adaptation. PLoS Biol. 2014, 12, e1001830. [Google Scholar] [CrossRef]
- Lohse, M.B.; Johnson, A.D. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS ONE 2008, 3, e1473. [Google Scholar] [CrossRef]
- Tsong, A.E.; Miller, M.G.; Raisner, R.M.; Johnson, A.D. Evolution of a combinatorial transcriptional circuit: A case study in yeasts. Cell 2003, 115, 389–399. [Google Scholar] [CrossRef]
- Huang, G.; Wang, H.; Chou, S.; Nie, X.; Chen, J.; Liu, H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc. Natl. Acad. Sci. USA 2006, 103, 12813–12818. [Google Scholar] [CrossRef]
- Zordan, R.E.; Galgoczy, D.J.; Johnson, A.D. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. USA 2006, 103, 12807–12812. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.N.; Conway, K.; Conway, T.P.; Daniels, K.J.; Soll, D.R. Roles of the Transcription Factors Sfl2 and Efg1 in White-Opaque Switching in a/alpha Strains of Candida albicans. mSphere 2019, 4, e00703-18. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Guan, G.; Du, H.; Tao, L.; Huang, G. Role of the N-acetylglucosamine kinase (Hxk1) in the regulation of white-gray-opaque tristable phenotypic transitions in C. albicans. Fungal Genet. Biol. 2016, 92, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.H.; Ruhela, D.; Ghosh, S.; Abdin, M.Z.; Datta, A. N-acetylglucosamine kinase, HXK1 contributes to white-opaque morphological transition in Candida albicans. Biochem. Biophys. Res. Commun. 2014, 445, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Pande, K.; Chen, C.; Noble, S.M. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat. Genet. 2013, 45, 1088–1091. [Google Scholar] [CrossRef]
- Liang, S.H.; Anderson, M.Z.; Hirakawa, M.P.; Wang, J.M.; Frazer, C.; Alaalm, L.M.; Thomson, G.J.; Ene, I.V.; Bennett, R.J. Hemizygosity Enables a Mutational Transition Governing Fungal Virulence and Commensalism. Cell Host Microbe 2019, 25, 418–431.e6. [Google Scholar] [CrossRef]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Hofs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef]
- Kadosh, D. Shaping up for battle: Morphological control mechanisms in human fungal pathogens. PLoS Pathog. 2013, 9, e1003795. [Google Scholar] [CrossRef][Green Version]
- Gleason, J.E.; Galaleldeen, A.; Peterson, R.L.; Taylor, A.B.; Holloway, S.P.; Waninger-Saroni, J.; Cormack, B.P.; Cabelli, D.E.; Hart, P.J.; Culotta, V.C. Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc. Natl. Acad. Sci. USA 2014, 111, 5866–5871. [Google Scholar] [CrossRef]
- Westman, J.; Moran, G.; Mogavero, S.; Hube, B.; Grinstein, S. Candida albicans Hyphal Expansion Causes Phagosomal Membrane Damage and Luminal Alkalinization. MBio 2018, 9, e01226-18. [Google Scholar] [CrossRef]
- Williams, R.B.; Lorenz, M.C. Multiple alternative carbon pathways combine to promote C. albicans stress resistance, immune interactions, and virulence. MBio 2020, 11, e03070-19. [Google Scholar]
- Singh, P.; Ghosh, S.; Datta, A. Attenuation of virulence and changes in morphology in Candida albicans by disruption of the N-acetylglucosamine catabolic pathway. Infect. Immun. 2001, 69, 7898–7903. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rao, K.H.; Ghosh, S.; Natarajan, K.; Datta, A. N-acetylglucosamine kinase, HXK1 is involved in morphogenetic transition and metabolic gene expression in Candida albicans. PLoS ONE 2013, 8, e53638. [Google Scholar] [CrossRef]
- Pierce, J.V.; Kumamoto, C.A. Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations. MBio 2012, 3, e00117-12. [Google Scholar] [CrossRef]
- Witchley, J.N.; Penumetcha, P.; Abon, N.V.; Woolford, C.A.; Mitchell, A.P.; Noble, S.M. Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host Microbe 2019, 25, 432–443.e6. [Google Scholar] [CrossRef]
- Naderer, T.; Heng, J.; McConville, M.J. Evidence that intracellular stages of Leishmania major utilize amino sugars as a major carbon source. PLoS Pathog. 2010, 6, e1001245. [Google Scholar] [CrossRef]
- Camacho, E.; Chrissian, C.; Cordero, R.J.B.; Liporagi-Lopes, L.; Stark, R.E.; Casadevall, A. N-acetylglucosamine affects Cryptococcus neoformans cell-wall composition and melanin architecture. Microbiology 2017, 163, 1540–1556. [Google Scholar] [CrossRef]
- Dennis, J.W.; Nabi, I.R.; Demetriou, M. Metabolism, cell surface organization, and disease. Cell 2009, 139, 1229–1241. [Google Scholar] [CrossRef]
- Gaderer, R.; Seidl-Seiboth, V.; de Vries, R.P.; Seiboth, B.; Kappel, L. N-acetylglucosamine, the building block of chitin, inhibits growth of Neurospora crassa. Fungal Genet. Biol. 2017, 107, 1–11. [Google Scholar] [CrossRef]
- Felice, M.R.; Gulati, M.; Giuffre, L.; Giosa, D.; Di Bella, L.M.; Criseo, G.; Nobile, C.J.; Romeo, O.; Scordino, F. Molecular Characterization of the N-Acetylglucosamine Catabolic Genes in Candida africana, a Natural N-Acetylglucosamine Kinase (HXK1) Mutant. PLoS ONE 2016, 11, e0147902. [Google Scholar] [CrossRef][Green Version]
- Perez-Campo, F.M.; Dominguez, A. Factors affecting the morphogenetic switch in Yarrowia lipolytica. Curr. Microbiol. 2001, 43, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Du, H.; Guan, G.; Tong, Y.; Kourkoumpetis, T.K.; Zhang, L.; Bai, F.Y.; Huang, G. N-acetylglucosamine induces white-to-opaque switching and mating in Candida tropicalis, providing new insights into adaptation and fungal sexual evolution. Eukaryot. Cell 2012, 11, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tao, L.; Guan, G.; Yue, H.; Liang, W.; Cao, C.; Dai, Y.; Huang, G. Regulation of filamentation in the human fungal pathogen Candida tropicalis. Mol. Microbiol. 2016, 99, 528–545. [Google Scholar] [CrossRef] [PubMed]
- Gilfillan, G.D.; Sullivan, D.J.; Haynes, K.; Parkinson, T.; Coleman, D.C.; Gow, N.A. Candida dubliniensis: Phylogeny and putative virulence factors. Microbiology 1998, 144, 829–838. [Google Scholar] [CrossRef]
- Xu, X.; Lin, J.; Zhao, Y.; Kirkman, E.; So, Y.S.; Bahn, Y.S.; Lin, X. Glucosamine stimulates pheromone-independent dimorphic transition in Cryptococcus neoformans by promoting Crz1 nuclear translocation. PLoS Genet. 2017, 13, e1006982. [Google Scholar] [CrossRef]
- Choi, J.; Summers, W.; Paszkowski, U. Mechanisms Underlying Establishment of Arbuscular Mycorrhizal Symbioses. Annu. Rev. Phytopathol. 2018, 56, 135–160. [Google Scholar] [CrossRef]
- Lefebvre, B. Arbuscular mycorrhiza: A new role for N-acetylglucosamine. Nat. Plants 2017, 3, 17085. [Google Scholar] [CrossRef]
- Kobae, Y.; Kawachi, M.; Saito, K.; Kikuchi, Y.; Ezawa, T.; Maeshima, M.; Hata, S.; Fujiwara, T. Up-regulation of genes involved in N-acetylglucosamine uptake and metabolism suggests a recycling mode of chitin in intraradical mycelium of arbuscular mycorrhizal fungi. Mycorrhiza 2015, 25, 411–417. [Google Scholar] [CrossRef]
- Gust, A.A.; Willmann, R.; Desaki, Y.; Grabherr, H.M.; Nurnberger, T. Plant LysM proteins: Modules mediating symbiosis and immunity. Trends Plant Sci. 2012, 17, 495–502. [Google Scholar] [CrossRef]
- Fitzpatrick, D.A.; O’Gaora, P.; Byrne, K.P.; Butler, G. Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genom. 2010, 11, 290. [Google Scholar] [CrossRef]
- Shank, E.A.; Kolter, R. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 2009, 12, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Park, J.T.; Uehara, T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 2008, 72, 211–227. [Google Scholar] [CrossRef] [PubMed]
- Naseem, S.; Konopka, J.B. N-acetylglucosamine regulates virulence properties in microbial pathogens. PLoS Pathog. 2015, 11, e1004947. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.J.; Reyes, C.N.; Liang, W.; Becker, C.; Shimada, K.; Wheeler, M.L.; Cho, H.C.; Popescu, N.I.; Coggeshall, K.M.; Arditi, M.; et al. Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan. Cell 2016, 166, 624–636. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Min, K.; Naseem, S.; Konopka, J.B. N-Acetylglucosamine Regulates Morphogenesis and Virulence Pathways in Fungi. J. Fungi 2020, 6, 8. https://doi.org/10.3390/jof6010008
Min K, Naseem S, Konopka JB. N-Acetylglucosamine Regulates Morphogenesis and Virulence Pathways in Fungi. Journal of Fungi. 2020; 6(1):8. https://doi.org/10.3390/jof6010008
Chicago/Turabian StyleMin, Kyunghun, Shamoon Naseem, and James B. Konopka. 2020. "N-Acetylglucosamine Regulates Morphogenesis and Virulence Pathways in Fungi" Journal of Fungi 6, no. 1: 8. https://doi.org/10.3390/jof6010008
APA StyleMin, K., Naseem, S., & Konopka, J. B. (2020). N-Acetylglucosamine Regulates Morphogenesis and Virulence Pathways in Fungi. Journal of Fungi, 6(1), 8. https://doi.org/10.3390/jof6010008